Genetic Prediction of Erectile Dysfunction Caused by Statins Among Malaysian Cardiac Patients
Abstract
1. Introduction
2. Results
2.1. Demographic Characteristics
2.2. Statin Therapy and Dosing Regimens
2.3. ED and Alleles
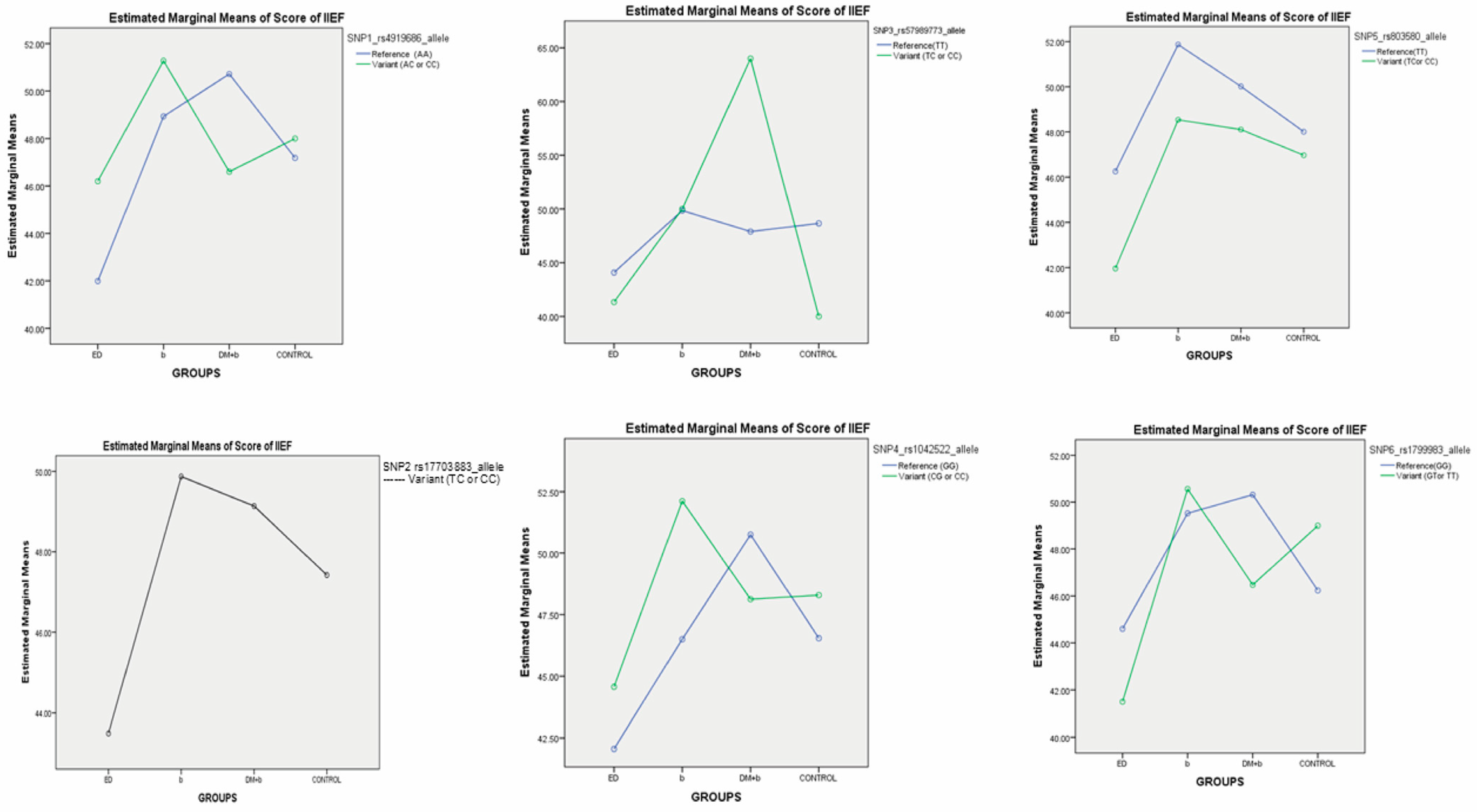
2.4. Association Analysis Between SNPs and IIEF Score of ED
2.5. Association Between Statin Types and IIEF Scores
2.6. Interaction Effects of Statin Types and Marker SNPs on the IIEF Score
3. Discussion
3.1. Prediction of Genetic Effects on ED
3.2. Statin Types and ED
4. Materials and Methods
4.1. Study Design
4.2. Data Collection
4.3. DNA Extraction
4.4. DNA Sequencing
4.5. Selection of Genotypes and Primer Design
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| ED | Erectile dysfunction |
| IIEF | International Index of Erectile Function |
| SD | Standard deviation |
| SNP | Single-nucleotide polymorphism |
| SPSS | Statistical Package for the Social Sciences |
| CI | Confidence interval |
References
- Chen, L.; Huang, D.-D.; Li, Y.; Ma, C.-C.; Shi, M.; Su, B.-X.; Shi, G.-J. Male sexual dysfunction: A review of literature on its pathological mechanisms, potential risk factors, and herbal drug intervention. Biomed. Pharmacother. 2019, 112, 108585. [Google Scholar] [CrossRef]
- Nordin, R.B.; Soni, T.; Kaur, A.; Loh, K.; Miranda, S. Prevalence and predictors of erectile dysfunction in adult male outpatient clinic attendees in Johor, Malaysia. Singap. Med. J. 2019, 60, 40–47. [Google Scholar] [CrossRef]
- El-Ganainy, S.O.; El-Mallah, A.; Abdallah, D.; Khattab, M.M.; El-Din, M.M.M.; El-Khatib, A.S. Elucidation of the mechanism of atorvastatin-induced myopathy in a rat model. Toxicology 2016, 359-360, 29–38. [Google Scholar] [CrossRef]
- Adhyaru, B.B.; Jacobson, T.A. Safety and efficacy of statin therapy. Nat. Rev. Cardiol. 2018, 15, 757–769. [Google Scholar] [CrossRef]
- Bustan, A.A.; Jawad, A.M. The Effect of Two Types of Statins (Rosuvastatin and Atorvastatin) on the Fertility of Male and Female Mice. Br. J. Med. Med. Res. 2017, 19, 1–11. [Google Scholar] [CrossRef]
- Kostis, J.B.; Dobrzynski, J.M. Statins and Erectile Dysfunction. Korean Soc. Sex. Med. Androl. 2019, 37, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Keihani, S.; Martin, C.; Craig, J.R.; Zhang, C.; Presson, A.P.; Myers, J.B.; Aston, K.I.; Emery, B.R.; Carrell, D.T.; Hotaling, J.M. Semen parameters are unaffected by statin use in men evaluated for infertility. Andrologia 2018, 50, e12995. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Long, C.; Zhu, J.; Tian, J. Protective Effects of Fluvastatin on Reproductive Function in Obese Male Rats Induced by High-Fat Diet through Enhanced Signaling of mTOR. Cell. Physiol. Biochem. 2017, 41, 598–608. [Google Scholar] [CrossRef]
- Zhao, S.-G.; Han, Q.-F.; Li, T.; Yao, H.-C. Association between statins use and erectile dysfunction. Int. J. Cardiol. 2017, 239, 16. [Google Scholar] [CrossRef]
- Aguirre, L.E.; Colleluori, G.; Robbins, D.; Dorin, R.; Shah, V.O.; Chen, R.; Jan, I.Z.; Qualls, C.; Villareal, D.T.; Armamento-Villareal, R. Bone and body composition response to testosterone therapy vary according to polymorphisms in the CYP19A1 gene. Endocrine 2019, 65, 692–706. [Google Scholar] [CrossRef]
- Roosenboom, J.; Indencleef, K.; Lee, M.K.; Hoskens, H.; White, J.D.; Liu, D.; Hecht, J.T.; Wehby, G.L.; Moreno, L.M.; Hodges-Simeon, C.; et al. SNPs Associated With Testosterone Levels Influence Human Facial Morphology. Front. Genet. 2018, 9, 497. [Google Scholar] [CrossRef] [PubMed]
- NCBI. CYP19A1 Cytochrome P450 Family 19 Subfamily A Member 1 [Homo Sapiens (Human)]. National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/gene/1588 (accessed on 15 June 2025).
- Ortega, I.; Cress, A.B.; Wong, D.H.; Villanueva, J.A.; Sokalska, A.; Moeller, B.C.; Stanley, S.D.; Duleba, A.J. Simvastatin Reduces Steroidogenesis by Inhibiting Cyp17a1 Gene Expression in Rat Ovarian Theca-Interstitial Cells. Biol. Reprod. 2012, 86, 1–9. [Google Scholar] [CrossRef]
- NCBI. CYP17A1 Cytochrome P450 Family 17 Subfamily A Member 1 [Homo Sapiens (Human)]. National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/gene/1586 (accessed on 15 June 2025).
- Jorgenson, E.; Matharu, N.; Palmer, M.R.; Yin, J.; Shan, J.; Hoffmann, T.J.; Thai, K.K.; Zhou, X.; Hotaling, J.M.; Jarvik, G.P.; et al. Genetic variation in the SIM1 locus is associated with erectile dysfunction. Proc. Natl. Acad. Sci. USA 2018, 115, 11018–11023. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, X.; Gao, X.; Mei, Y.; Wu, M. A new role of p53 in regulating lipid metabolism. J. Mol. Cell Biol. 2013, 5, 147–150. [Google Scholar] [CrossRef]
- Wankanit, S.; Zidoune, H.; Bignon-Topalovic, J.; Schlick, L.; Houzelstein, D.; Fusée, L.; Boukri, A.; Nouri, N.; McElreavey, K.; Bashamboo, A.; et al. Evidence for NR2F2/COUP-TFII involvement in human testis development. Sci. Rep. 2024, 17869. [Google Scholar] [CrossRef]
- Yao, H.X.; Ma, F.Z.; Tan, Y.Y.; Liu, L.Y. Endothelial nitric oxide synthase gene polymorphisms and risk of erectile dysfunction: An updated meta-analysis of genetic association studies. Int. J. Surg. 2018, 54, 141–148. [Google Scholar] [CrossRef]
- Burris-Hiday, S.D.; Scott, E.E. Steroidogenic cytochrome P450 17A1 structure and function. Mol. Cell. Endocrinol. 2021, 528, 111261. [Google Scholar] [CrossRef]
- Bai, S.; Li, M.-Z.; Wan, Y.-Y.; Hu, X.-C.; Liu, Y.-X.; Tong, X.-H.; Guo, T.-H.; Zong, L.; Liu, R.; Zhao, Y.-Q.; et al. Association between MTHFR c.677C>T variant and erectile dysfunction among males attending fertility clinic. Asian J. Androl. 2024, 26, 41–45. [Google Scholar] [CrossRef]
- Ferezin, L.P.; Kayzuka, C.; Pereira, V.C.R.; de Andrade, M.F.; Molina, C.A.F.; Tucci, S.; Tanus-Santos, J.E.; Lacchini, R. The rs2682826 Polymorphism of the NOS1 Gene Is Associated with the Degree of Disability of Erectile Dysfunction. Life 2023, 13, 1082. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Wang, R.; Luo, Y.; Tang, Q.; Wang, K. Genetic association of lipid-lowering drug target genes with erectile dysfunction and male reproductive health. Front. Endocrinol. 2024, 15, 1362499. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Chen, Y.; Sun, T.; Cui, L.; Wei, Y.; Li, T.; Meng, Q. Exploring potential genes and mechanisms linking erectile dysfunction and depression. Front. Endocrinol. 2023, 14, 1221043. [Google Scholar] [CrossRef]
- Oliveira-Paula, G.H.; Lacchini, R.; Tanus-Santos, J.E. Endothelial nitric oxide synthase: From biochemistry and gene structure to clinical implications of NOS3 polymorphisms. Gene 2016, 575, 584–599. [Google Scholar] [CrossRef]
- Hattori, A.; Fukami, M. Nuclear Receptor Gene Variants Underlying Disorders/Differences of Sex Development through Abnormal Testicular Development. Biomolecules 2023, 13, 691. [Google Scholar] [CrossRef]
- Kataoka, T.; Hotta, Y.; Kimura, K. A review of experimental techniques for erectile function researches and development of medical technology using animal erectile dysfunction models in sexual and reproductive medicine. Reprod. Med. Biol. 2023, 22, e12513. [Google Scholar] [CrossRef]
- Nakayama, A.; Morita, H.; Kawahara, T.; Itoh, H.; Komuro, I. Association between testosterone and lipid profiles under statin therapy and its clinical impact on the cardiovascular event risk. Heart Vessels. 2021, 36, 1794–1803. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-J.; Huang, B. Rosuvastatin decreases free testosterone levels but does not influence sexual function in men with type 2 diabetes. Diabetes Res. Clin. Pract. 2016, 120, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, T.; Taymour, M. Gene Polymorphisms Affecting Erectile Dysfunction. Sex. Med. Rev. 2020, 8, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Mykoniatis, I.; Pyrgidis, N.; Sokolakis, I.; Ouranidis, A.; Sountoulides, P.; Haidich, A.-B.; van Renterghem, K.; Hatzichristodoulou, G.; Hatzichristou, D. Assessment of Combination Therapies vs Monotherapy for Erectile Dysfunction: A Systematic Review and Meta-analysis. JAMA Netw. Open 2021, 4, e2036337. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Lu, Y.; Ma, C.; Li, H.; Su, X. Impact of atorvastatin on erectile dysfunction: A meta-analysis and systematic review. Andrologia 2022, 54, e14408. [Google Scholar] [CrossRef]
- Omolaoye, T.S.; Halabi, M.O.; Mubarak, M.; Cyril, A.C.; Duvuru, R.; Radhakrishnan, R.; Du Plessis, S.S. Statins and Male Fertility: Is There a Cause for Concern? Toxics 2022, 10, 627. [Google Scholar] [CrossRef]
- Pons-Rejraji, H.; Brugnon, F.; Sion, B.; Maqdasy, S.; Gouby, G.; Pereira, B.; Marceau, G.; Gremeau, A.-S.; Drevet, J.; Grizard, G.; et al. Evaluation of atorvastatin efficacy and toxicity on spermatozoa, accessory glands and gonadal hormones of healthy men: A pilot prospective clinical trial. Reprod. Biol. Endocrinol. 2014, 12, 65. [Google Scholar] [CrossRef]
- Pautz, A.; Li, H.; Kleinert, H. Regulation of NOS expression in vascular diseases. Front. Biosci. 2021, 26, 85–101. [Google Scholar] [CrossRef]
- Şahin, T.D.; Yazir, Y.; Utkan, T.; Gacar, G.; Rençber, S.F.; Gocmez, S.S. TNF-α antagonism with etanercept enhances penile NOS expression, cavernosal reactivity, and testosterone levels in aged rats. Can. J. Physiol. Pharmacol. 2018, 96, 200–207. [Google Scholar] [CrossRef]
- Freudenstein, D.; Litchfield, C.; Caramia, F.; Wright, G.; Solomon, B.J.; Ball, D.; Keam, S.P.; Neeson, P.; Haupt, Y.; Haupt, S. TP53 Status, Patient Sex, and the Immune Response as Determinants of Lung Cancer Patient Survival. Cancers 2020, 12, 1535. [Google Scholar] [CrossRef]
- Ganiyani, M.A.; Prabhakar, P.; Khosla, A.A.; Bellur, S.S.; Avudaiappan, A.P.; Bond, L.; Marada, S.; McCracken, A.; Hurmiz, C.; Bastos, B.R.; et al. Impact of TP53, RB1, and PTEN mutations on overall survival in metastatic prostate cancer: A multi-center study via the Guardian Research Network. J. Clin. Oncol. 2024, 42, 82. [Google Scholar] [CrossRef]
- Mędraś, M.; Kubicka, E.; Jóźkow, P.; Słowińska-Lisowska, M.; Trzmiel-Bira, A.; Filus, A. Treatment with statins and testosterone levels in men. Endokrynol. Polska 2014, 65, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Saeed, R.; Amin, F.; Durrani, N.; Saif, S.M.A.; Zafar, M.T. Prevalence of erectile dysfunction and associated factors among males visiting family medicine clinics in a Tertiary Care Hospital in Karachi, Pakistan. J. Fam. Med. Prim. Care 2021, 10, 1294–1300. [Google Scholar] [CrossRef] [PubMed]
- Quek, K.F.; Low, W.Y.; Razack, A.H.; Chua, C.B.; Loh, C.S. Reliability and validity of the Malay version of the International Index of Erectile Function (IIEF-15) in the Malaysian population. Int. J. Impot. Res. 2002, 14, 310–315. [Google Scholar] [CrossRef] [PubMed]
| SNPs | Erectile Dysfunction | |||
|---|---|---|---|---|
| Total (N = 246, %) | NO = 186 (%) | YES = 60 (%) | ||
| CYP17A1: rs4919686 genotype | AA | 221 (89.8) | 166 (89.2) | 55 (91.7) |
| AC | 13 (5.3) | 10 (5.4) | 3 (5) | |
| CC | 12 (4.9) | 10 (5.4) | 2 (3.3) | |
| CYP19A1: rs17703883 genotype | TC | 224 (91.1) | 170 (91.4) | 54 (90) |
| CC | 22 (8.9) | 16 (8.6) | 6 (10) | |
| SIM1: rs57989773 genotype | TT | 237 (96.3) | 181 (97.3) | 56 (93.3) |
| TC | 9 (3.7) | 5 (2.7) | 4 (6.7) | |
| TP53: rs1042522 genotype | GG | 57 (23.2) | 44 (23.7) | 13 (21.7) |
| CG | 127 (51.6) | 97 (52.2) | 30 (50) | |
| CC | 62 (25.2) | 45 (24.2) | 17 (28.3) | |
| NR2F2-AS1: rs803580 genotype | TT | 96 (39) | 76 (40.9) | 20 (33.3) |
| TC | 128 (52) | 91 (48.9) | 37 (61.7) | |
| CC | 22 (9) | 19 (10.2) | 3 (5) | |
| NOS3: rs1799983 genotype | GG | 174 (70.7) | 129 (69.4) | 45 (75) |
| GT | 65 (26.4) | 51 (27.4) | 14 (23.3) | |
| TT | 7 (2.8) | 6 (3.2) | 1 (1.7) | |
| Total | 246 | 186 | 60 | |
| Variants | Groups | Unstandardized Coefficients | Standardized Coefficients | t | Sig. | 95.0% CI for B | ||
|---|---|---|---|---|---|---|---|---|
| B | Std. Error | Beta | Lower Bound | Upper Bound | ||||
| SNP1 rs4919686 * | (Constant) | 48.284 | 3.208 | 15.051 | 0.000 | 41.965 | 54.604 | |
| Group 1 | −6.037 | 3.195 | −0.207 | −1.890 | 0.060 | −12.33 | 0.256 | |
| Group 2 | 2.410 | 2.964 | 0.094 | 0.813 | 0.417 | −3.429 | 8.249 | |
| Group 3 | 0.555 | 3.055 | 0.020 | 0.182 | 0.856 | −5.462 | 6.572 | |
| Group 4 | 0.174 | 3.280 | 0.006 | 0.053 | 0.958 | −6.286 | 6.635 | |
| SNP2 rs17703883 ** | (Constant) | 41.606 | 9.141 | 4.551 | 0.000 | 23.599 | 59.613 | |
| Group 1 | −0.301 | 4.334 | −0.019 | −0.070 | 0.945 | −8.839 | 8.236 | |
| Group 2 | 4.940 | 4.638 | 0.325 | 1.065 | 0.288 | −4.195 | 14.076 | |
| Group 3 | 3.730 | 4.653 | 0.230 | 0.802 | 0.424 | −5.436 | 12.896 | |
| Group 4 | 3.456 | 4.662 | 0.205 | 0.741 | 0.459 | −5.728 | 12.640 | |
| SNP3 rs57989773 *** | (Constant) | 46.830 | 4.631 | 10.113 | 0.000 | 37.708 | 55.952 | |
| Group 1 | −5.270 | 4.406 | −0.177 | −1.196 | 0.233 | −13.950 | 3.410 | |
| Group 2 | 4.387 | 4.652 | 0.151 | 0.943 | 0.347 | −4.778 | 13.551 | |
| Group 3 | 2.403 | 4.797 | 0.076 | 0.501 | 0.617 | −7.047 | 11.852 | |
| Group 4 | 1.292 | 4.662 | 0.041 | 0.277 | 0.782 | −7.891 | 10.475 | |
| SNP4 rs1042522 † | (Constant) | 47.168 | 3.603 | 13.091 | 0.000 | 40.071 | 54.266 | |
| Group 1 | −3.174 | 2.164 | −0.176 | −1.467 | 0.144 | −7.438 | 1.089 | |
| Group 2 | 2.257 | 2.113 | 0.137 | 1.068 | 0.287 | −1.905 | 6.420 | |
| Group 3 | 0.896 | 2.132 | 0.051 | 0.420 | 0.675 | −3.303 | 5.095 | |
| Group 4 | 0.815 | 2.218 | 0.043 | 0.367 | 0.714 | −3.554 | 5.184 | |
| SNP5 rs803580 †† | (Constant) | 49.096 | 2.918 | 16.827 | 0.000 | 43.349 | 54.844 | |
| Group 1 | −4.981 | 1.951 | −0.259 | −2.553 | 0.011 | −8.825 | −1.138 | |
| Group 2 | 1.254 | 1.860 | 0.072 | 0.674 | 0.501 | −2.410 | 4.918 | |
| Group 3 | 0.113 | 2.049 | 0.006 | 0.055 | 0.956 | −3.924 | 4.150 | |
| Group 4 | −0.317 | 1.968 | −0.016 | −0.161 | 0.872 | −4.195 | 3.560 | |
| SNP6 rs1799983 ††† | (Constant) | 49.603 | 2.573 | 19.278 | 0.000 | 44.535 | 54.672 | |
| Group 1 | −6.357 | 2.258 | −0.261 | −2.815 | 0.005 | −10.806 | −1.909 | |
| Group 2 | 1.151 | 2.104 | 0.053 | 0.547 | 0.585 | −2.993 | 5.294 | |
| Group 3 | −0.717 | 2.264 | −0.030 | −0.317 | 0.752 | −5.177 | 3.744 | |
| Group 4 | −0.681 | 2.098 | −0.030 | −0.325 | 0.746 | −4.813 | 3.451 | |
| Variants | Unstandardized Coefficients | Standardized Coefficients | t | Sig. | 95.0% CI for B | ||
|---|---|---|---|---|---|---|---|
| B | Std. Error | Beta | Lower Bound | Upper Bound | |||
| (Constant) | 49.460 | 1.705 | 29.015 | 0.000 | 46.102 | 52.818 | |
| Atorva_Group1 | −10.566 | 2.779 | −0.274 | −3.802 | 0.000 | −16.040 | −5.091 |
| Atorva_Group2 | 1.659 | 2.695 | 0.045 | 0.615 | 0.539 | −3.651 | 6.968 |
| Atorva_Group3 | −0.808 | 2.624 | −0.023 | −0.308 | 0.758 | −5.977 | 4.361 |
| Simva_Group1 | −4.723 | 3.541 | −0.090 | −1.334 | 0.184 | −11.700 | 2.253 |
| Simva_Group2 | 0.078 | 3.154 | 0.002 | 0.025 | 0.980 | −6.135 | 6.291 |
| Simva_Group3 | 3.903 | 4.421 | 0.058 | 0.883 | 0.378 | −4.807 | 12.613 |
| Variants | Statin and Group | Unstandardized Coefficients | Standardized Coefficients | t | Sig. | 95.0% CI for B | ||
|---|---|---|---|---|---|---|---|---|
| B | Std. Error | Beta | Lower Bound | Upper Bound | ||||
| SNP1 rs4919686 * | (Constant) | 49.134 | 1.544 | 31.827 | 0.000 | 46.093 | 52.175 | |
| Atorva_Group1 | −8.585 | 2.397 | −0.249 | −3.581 | 0.000 | −13.308 | −3.862 | |
| Atorva_Group2 | 1.732 | 2.205 | 0.055 | 0.785 | 0.433 | −2.613 | 6.076 | |
| Atorva_Group3 | −0.637 | 2.167 | −0.021 | −0.294 | 0.769 | −4.906 | 3.633 | |
| Simva_Group1 | −3.353 | 3.013 | −0.074 | −1.113 | 0.267 | −9.288 | 2.582 | |
| Simva_Group2 | −0.158 | 2.520 | −0.004 | −0.063 | 0.950 | −5.123 | 4.806 | |
| Simva_Group3 | 3.250 | 3.503 | 0.060 | 0.928 | 0.354 | −3.650 | 10.151 | |
| SNP2 rs17703883 ** | (Constant) | 49.113 | 1.678 | 29.276 | 0.000 | 45.809 | 52.418 | |
| Atorva_Group1 | −4.611 | 1.275 | −0.258 | −3.617 | 0.000 | −7.123 | −2.100 | |
| Atorva_Group2 | 1.003 | 1.340 | 0.054 | 0.749 | 0.455 | −1.636 | 3.642 | |
| Atorva_Group3 | −0.231 | 1.304 | −0.013 | −0.177 | 0.860 | −2.799 | 2.338 | |
| Simva_Group1 | −2.005 | 1.708 | −0.079 | −1.174 | 0.241 | −5.369 | 1.359 | |
| Simva_Group2 | 0.213 | 1.571 | 0.009 | 0.135 | 0.892 | −2.881 | 3.307 | |
| Simva_Group3 | 2.125 | 2.207 | 0.063 | 0.963 | 0.337 | −2.222 | 6.473 | |
| SNP3 rs57989773 *** | (Constant) | 48.742 | 1.631 | 29.893 | 0.000 | 45.530 | 51.954 | |
| Atorva_Group1 | −8.084 | 2.358 | −0.242 | −3.428 | 0.001 | −12.729 | −3.438 | |
| Atorva_Group2 | 2.132 | 2.464 | 0.062 | 0.865 | 0.388 | −2.723 | 6.987 | |
| Atorva_Group3 | 0.227 | 2.491 | 0.007 | 0.091 | 0.928 | −4.681 | 5.134 | |
| Simva_Group1 | −4.005 | 3.515 | −0.077 | −1.139 | 0.256 | −10.930 | 2.919 | |
| Simva_Group2 | 0.796 | 3.122 | 0.018 | 0.255 | 0.799 | −5.354 | 6.946 | |
| Simva_Group3 | 4.621 | 4.406 | 0.068 | 1.049 | 0.295 | −4.058 | 13.300 | |
| SNP4 rs1042522 † | (Constant) | 49.072 | 1.583 | 31.007 | 0.000 | 45.955 | 52.190 | |
| Atorva_Group1 | −5.271 | 1.421 | −0.260 | −3.710 | 0.000 | −8.070 | −2.472 | |
| Atorva_Group2 | 0.831 | 1.434 | 0.041 | 0.579 | 0.563 | −1.994 | 3.656 | |
| Atorva_Group3 | −0.305 | 1.381 | −0.016 | −0.221 | 0.826 | −3.026 | 2.416 | |
| Simva_Group1 | −1.349 | 2.093 | −0.043 | −0.645 | 0.520 | −5.472 | 2.773 | |
| Simva_Group2 | 0.334 | 1.720 | 0.013 | 0.194 | 0.846 | −3.054 | 3.722 | |
| Simva_Group3 | 2.120 | 2.355 | 0.058 | 0.900 | 0.369 | −2.520 | 6.759 | |
| SNP5 rs803580 †† | (Constant) | 49.467 | 1.523 | 32.486 | 0.000 | 46.467 | 52.466 | |
| Atorva_Group1 | −6.163 | 1.568 | −0.270 | −3.930 | 0.000 | −9.253 | −3.074 | |
| Atorva_Group2 | 0.794 | 1.397 | 0.039 | 0.569 | 0.570 | −1.957 | 3.546 | |
| Atorva_Group3 | −0.376 | 1.556 | −0.017 | −0.241 | 0.809 | −3.442 | 2.690 | |
| Simva_Group1 | −3.395 | 1.963 | −0.114 | −1.730 | 0.085 | −7.263 | 0.472 | |
| Simva_Group2 | 0.120 | 1.909 | 0.004 | 0.063 | 0.950 | −3.641 | 3.880 | |
| Simva_Group3 | 1.581 | 2.538 | 0.040 | 0.623 | 0.534 | −3.418 | 6.580 | |
| SNP6 rs1799983 ††† | (Constant) | 49.611 | 1.483 | 33.450 | 0.000 | 46.689 | 52.532 | |
| Atorva_Group1 | −7.858 | 1.904 | −0.281 | −4.127 | 0.000 | −11.609 | −4.107 | |
| Atorva_Group2 | 1.231 | 1.844 | 0.046 | 0.668 | 0.505 | −2.402 | 4.864 | |
| Atorva_Group3 | −1.131 | 1.938 | −0.040 | −0.584 | 0.560 | −4.949 | 2.686 | |
| Simva_Group1 | −2.937 | 2.667 | −0.072 | −1.101 | 0.272 | −8.192 | 2.318 | |
| Simva_Group2 | −0.775 | 2.163 | −0.024 | −0.358 | 0.720 | −5.036 | 3.485 | |
| Simva_Group3 | 1.263 | 2.658 | 0.030 | 0.475 | 0.635 | −3.974 | 6.500 | |
| SNP | Re Restriction Enzyme | Incubation Temperature (°C) | Incubation Time (min) |
|---|---|---|---|
| Rs1799983 | Ban II | 37 | 60 |
| Rs1042522 | BstUI | 60 | 15 |
| Rs8023580 | HpyCH41V | 37 | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adeeb, N.B.A.; Al-Ani, H.A.; Daud, N.A.A.; Abu Bakar, R.; Ahmad, I.; Noor, D.A.M.; Bin Alwi, Z. Genetic Prediction of Erectile Dysfunction Caused by Statins Among Malaysian Cardiac Patients. Int. J. Mol. Sci. 2025, 26, 8447. https://doi.org/10.3390/ijms26178447
Adeeb NBA, Al-Ani HA, Daud NAA, Abu Bakar R, Ahmad I, Noor DAM, Bin Alwi Z. Genetic Prediction of Erectile Dysfunction Caused by Statins Among Malaysian Cardiac Patients. International Journal of Molecular Sciences. 2025; 26(17):8447. https://doi.org/10.3390/ijms26178447
Chicago/Turabian StyleAdeeb, Naam Bahjat Ahmed, Hadeer Akram Al-Ani, Nur Aizati Athirah Daud, Ruzilawati Abu Bakar, Imran Ahmad, Dzul Azri Mohamed Noor, and Zilfalil Bin Alwi. 2025. "Genetic Prediction of Erectile Dysfunction Caused by Statins Among Malaysian Cardiac Patients" International Journal of Molecular Sciences 26, no. 17: 8447. https://doi.org/10.3390/ijms26178447
APA StyleAdeeb, N. B. A., Al-Ani, H. A., Daud, N. A. A., Abu Bakar, R., Ahmad, I., Noor, D. A. M., & Bin Alwi, Z. (2025). Genetic Prediction of Erectile Dysfunction Caused by Statins Among Malaysian Cardiac Patients. International Journal of Molecular Sciences, 26(17), 8447. https://doi.org/10.3390/ijms26178447







