Cholesterol Regulates Airway Epithelial Cell Differentiation by Inhibiting p53 Nuclear Translocation
Abstract
1. Introduction
2. Results
2.1. MS/MS-Based Proteomic Analysis of Normal Differentiating phBECs Reveals Inhibition of the CHOL Biosynthesis Pathway
2.2. Chronic CHOL Exposure Impairs Nuclear Translocation of p53 In Vitro
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. Patient Material
4.3. Primary Human Bronchial Epithelial Cell (phBEC) Differentiation
4.4. Proteome Analysis from Normal Differentiating phBECs
4.5. Bioinformatic and Ingenuity Pathway Analysis
4.6. Chronic CHOL Exposure
4.7. Lactate Dehydrogenase (LDH) Assay
4.8. RNA Isolation and qRT-PCR Analysis
4.9. Immunofluorescence (IF) Analysis and Quantification
4.10. Primers and Antibodies
4.11. Trans Epithelial Electrical Resistance (TEER) Measurement
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| phBECs | Primary human bronchial epithelial cells |
| CHOL | Cholesterol |
| IPA | Ingenuity Pathway Analysis |
| ALI | Air Liquid Interface |
References
- Montoro, D.T.; Haber, A.L.; Biton, M.; Vinarsky, V.; Lin, B.; Birket, S.E.; Yuan, F.; Chen, S.; Leung, H.M.; Villoria, J.; et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018, 560, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Mastalerz, M.; Ansari, M.; Schiller, H.B.; Staab-Weijnitz, C.A. Emerging Roles of Airway Epithelial Cells in Idiopathic Pulmonary Fibrosis. Cells 2022, 11, 1050. [Google Scholar] [CrossRef]
- Mastalerz, M.; Dick, E.; Chakraborty, A.; Hennen, E.; Schamberger, A.C.; Schröppel, A.; Lindner, M.; Hatz, R.; Behr, J.; Hilgendorff, A.; et al. Validation of in vitro models for smoke exposure of primary human bronchial epithelial cells. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2022, 322, L129–L148. [Google Scholar] [CrossRef]
- Schamberger, A.C.; Staab-Weijnitz, C.A.; Mise-Racek, N.; Eickelberg, O. Cigarette smoke alters primary human bronchial epithelial cell differentiation at the air-liquid interface. Sci. Rep. 2015, 5, 8163. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, I.E.; Eickelberg, O. New cellular and molecular mechanisms of lung injury and fibrosis in idiopathic pulmonary fibrosis. Lancet 2012, 380, 680–688. [Google Scholar] [CrossRef]
- Shaykhiev, R. Airway Basal Cells in Chronic Obstructive Pulmonary Disease: A Continuum or a Dead End? Am. J. Respir. Cell Mol. Biol. 2021, 65, 10–12. [Google Scholar] [CrossRef]
- Ross, A.J.; Dailey, L.A.; Brighton, L.E.; Devlin, R.B. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2007, 37, 169–185. [Google Scholar] [CrossRef]
- Djidrovski, I.; Georgiou, M.; Tasinato, E.; Leonard, M.O.; Bor, J.V.D.; Lako, M.; Armstrong, L. Direct transcriptomic comparison of xenobiotic metabolism and toxicity pathway induction of airway epithelium models at an air–liquid interface generated from induced pluripotent stem cells and primary bronchial epithelial cells. Cell Biol. Toxicol. 2023, 39, 1–18. [Google Scholar] [CrossRef]
- Wang, J.; Ma, Z.; Carr, S.A.; Mertins, P.; Zhang, H.; Zhang, Z.; Chan, D.W.; Ellis, M.J.; Townsend, R.R.; Smith, R.D.; et al. Proteome profiling outperforms transcriptome profiling for coexpression based gene function prediction. Mol. Cell. Proteom. 2017, 16, 121–134. [Google Scholar] [CrossRef]
- Gowdy, K.M.; Fessler, M.B. Emerging roles for cholesterol and lipoproteins in lung disease. Pulm. Pharmacol. Ther. 2013, 26, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, Y.; Liu, X.; Zheng, N.; Gu, Y.; Song, Y.; Wang, X. Regulatory roles of external cholesterol in human airway epithelial mitochondrial function through STARD3 signalling. Clin. Transl. Med. 2022, 12, e902. [Google Scholar] [CrossRef]
- Jing, D.; Yu, J.-K.; Chen, H.-P.; Dong, L.-L.; Li, W.; Li, Z.-Y.; Zhou, J.-S. Cholesterol Accumulation Enhances Cigarette Smoke-Induced Airway Epithelial Inflammation. Int. J. Chronic Obstr. Pulm. Dis. 2025, 20, 411–423. [Google Scholar] [CrossRef]
- McCrae, C.; Dzgoev, A.; Ståhlman, M.; Horndahl, J.; Svärd, R.; Große, A.; Großkopf, T.; Skujat, M.-A.; Williams, N.; Schubert, S.; et al. Lanosterol Synthase Regulates Human Rhinovirus Replication in Human Bronchial Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2018, 59, 713–722. [Google Scholar] [CrossRef]
- Pérez-Gil, J. Structure of pulmonary surfactant membranes and films: The role of proteins and lipid–Protein interactions. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 1676–1695. [Google Scholar] [CrossRef]
- Andersson, J.M.; Grey, C.; Larsson, M.; Ferreira, T.M.; Sparr, E. Correction for: Effect of cholesterol on the molecular structure and transitions in a clinical-grade lung surfactant extract. Proc. Natl. Acad. Sci. USA 2017, 114, E3592–E3601. [Google Scholar] [CrossRef]
- Sallese, A.; Suzuki, T.; McCarthy, C.; Bridges, J.; Filuta, A.; Arumugam, P.; Shima, K.; Ma, Y.; Wessendarp, M.; Black, D.; et al. Targeting cholesterol homeostasis in lung diseases. Sci. Rep. 2017, 7, 10211. [Google Scholar] [CrossRef]
- Basset-Léobon, C.; Lacoste-Collin, L.; Aziza, J.; Bes, J.C.; Jozan, S.; Courtade-Saïdi, M. Cut-off values and significance of Oil Red O-positive cells in bronchoalveolar lavage fluid. Cytopathology 2010, 21, 245–250. [Google Scholar] [CrossRef]
- Wilson, A.M.; Nair, P.; Hargreave, F.E.; Efthimiadis, A.E.; Anvari, M.; Allen, C.J.; ELVIS Research Study Group. Lipid and smoker’s inclusions in sputum macrophages in patients with airway diseases. Respir. Med. 2011, 105, 1691–1695. [Google Scholar] [CrossRef]
- Xuan, L.; Han, F.; Gong, L.; Lv, Y.; Wan, Z.; Liu, H.; Zhang, D.; Jia, Y.; Yang, S.; Ren, L.; et al. Association between chronic obstructive pulmonary disease and serum lipid levels: A meta-analysis. Lipids Health. Dis. 2018, 17, 263. [Google Scholar] [CrossRef]
- Fessler, M.B. A new frontier in immunometabolism. cholesterol in lung health and disease. Ann. Am. Thorac. Soc. 2017, 14, S399–S405. [Google Scholar] [CrossRef]
- Jia, J.; Conlon, T.M.; Sarker, R.S.; Taşdemir, D.; Smirnova, N.F.; Srivastava, B.; Verleden, S.E.; Güneş, G.; Wu, X.; Prehn, C.; et al. Cholesterol metabolism promotes B-cell positioning during immune pathogenesis of chronic obstructive pulmonary disease. EMBO Mol. Med. 2018, 10, e8349. [Google Scholar] [CrossRef]
- White, N.M.; Jiang, D.; Burgess, J.D.; Bederman, I.R.; Previs, S.F.; Kelley, T.J. Altered cholesterol homeostasis in cultured and in vivo models of cystic fibrosis. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2007, 292, L476–L486. [Google Scholar] [CrossRef]
- Cui, G.; Cottrill, K.A.; Strickland, K.M.; Mashburn, S.A.; Koval, M.; McCarty, N.A. Alteration of Membrane Cholesterol Content Plays a Key Role in Regulation of Cystic Fibrosis Transmembrane Conductance Regulator Channel Activity. Front. Physiol. 2021, 12, 652513. [Google Scholar] [CrossRef]
- McCarthy, C.; Lee, E.; Bridges, J.P.; Sallese, A.; Suzuki, T.; Woods, J.C.; Bartholmai, B.J.; Wang, T.; Chalk, C.; Carey, B.C.; et al. Statin as a novel pharmacotherapy of pulmonary alveolar proteinosis. Nat. Commun. 2018, 9, 3127. [Google Scholar] [CrossRef]
- Nardacci, R.; Colavita, F.; Castilletti, C.; Lapa, D.; Matusali, G.; Meschi, S.; Del Nonno, F.; Colombo, D.; Capobianchi, M.R.; Zumla, A.; et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021, 12, 263. [Google Scholar] [CrossRef]
- Chen, F.; Matsuda, A.; Budinger, G.R.S.; Sporn, P.H.S.; Casalino-Matsuda, S.M. Hypercapnia increases ACE2 expression and pseudo-SARS-CoV-2 entry in bronchial epithelial cells by augmenting cellular cholesterol. Front. Immunol. 2023, 14, 1251120. [Google Scholar] [CrossRef]
- Shim, A.; Song, J.-H.; Kwon, B.-E.; Lee, J.-J.; Ahn, J.-H.; Kim, Y.-J.; Rhee, K.-J.; Chang, S.-Y.; Cha, Y.; Lee, Y.-S.; et al. Therapeutic and prophylactic activity of itraconazole against human rhinovirus infection in a murine model. Sci. Rep. 2016, 6, 23110. [Google Scholar] [CrossRef]
- McConnell, A.M.; Yao, C.; Yeckes, A.R.; Wang, Y.; Selvaggio, A.S.; Tang, J.; Kirsch, D.G.; Stripp, B.R. p53 Regulates Progenitor Cell Quiescence and Differentiation in the Airway. Cell Rep. 2016, 17, 2173–2182. [Google Scholar] [CrossRef]
- Chakraborty, A.; Giraldo-Arias, J.; Merl-Pham, J.; Dick, E.; Mastalerz, M.; Zöller, M.; Hatz, R.A.; Behr, J.; Hilgendorff, A.; Hauck, S.M.; et al. Cholesterol Modulates Airway Epithelial Cell Differentiation Via Inhibition of P53 Nuclear Translocation. Am. J. Respir. Crit. Care Med. 2025, 211, A2452. [Google Scholar] [CrossRef]
- Chakraborty, A.; Zöller, M.; Sardogan, A.; Klotz, M.; Mastalerz, M.; Marchi, H.; Meixner, R.; A Hatz, R.; Behr, J.; Hilgendorff, A.; et al. Development of a Polidocanol-based Human In vitro Model to Explore Airway Epithelial Repair. Am. J. Respir. Cell Mol. Biol. 2025. [Google Scholar] [CrossRef]
- Gomi, K.; Arbelaez, V.; Crystal, R.G.; Walters, M.S. Activation of NOTCH1 or NOTCH3 signaling skews human airway basal cell differentiation toward a secretory pathway. PLoS ONE 2015, 10, e0116507. [Google Scholar] [CrossRef]
- Rock, J.R.; Gao, X.; Xue, Y.; Randell, S.H.; Kong, Y.-Y.; Hogan, B.L.M. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 2011, 8, 639–648. [Google Scholar] [CrossRef]
- Xing, Y.; Li, A.; Borok, Z.; Li, C.; Minoo, P. NOTCH1 is required for regeneration of clara cells during repair of airway injury. Stem Cells 2012, 30, 946–955. [Google Scholar] [CrossRef]
- Xu, K.; Moghal, N.; Egan, S.E. Notch signaling in lung development and disease. Notch Signal. Embryol. Cancer 2012, 727, 89–98. [Google Scholar]
- Briot, A.; Civelek, M.; Seki, A.; Hoi, K.; Mack, J.J.; Lee, S.D.; Kim, J.; Hong, C.; Yu, J.; Fishbein, G.; et al. Endothelial NOTCH1 is suppressed by circulating lipids and antagonizes inflammation during atherosclerosis. J. Exp. Med. 2015, 212, 2147–2163. [Google Scholar] [CrossRef]
- Aquila, G.; Fortini, C.; Pannuti, A.; Delbue, S.; Pannella, M.; Morelli, M.B.; Caliceti, C.; Castriota, F.; de Mattei, M.; Ongaro, A.; et al. Distinct gene expression profiles associated with Notch ligands Delta-like 4 and Jagged1 in plaque material from peripheral artery disease patients: A pilot study. J. Transl. Med. 2017, 15, 98. [Google Scholar] [CrossRef]
- Dotto, G.P. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat. Rev. Cancer 2009, 9, 587–595. [Google Scholar] [CrossRef]
- Mogi, A.; Kuwano, H. TP53 mutations in nonsmall cell lung cancer. J. Biomed. Biotechnol. 2011, 2011, 583929. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Zhang, L.; Qian, W.; Shen, S.; Wang, J.; Wu, S.; Xu, W.; Chen, B.; Lin, M.; et al. Single-cell transcriptomics highlights immunological dysregulations of monocytes in the pathobiology of COPD. Respir. Res. 2022, 23, 367. [Google Scholar] [CrossRef]
- Expert Panel on Detection, Evaluation; Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 2001, 285, 2486–2497. [Google Scholar] [CrossRef]
- Estronca, L.M.B.B.; Filipe, H.A.L.; Salvador, A.; Moreno, M.J.; Vaz, W.L.C. Homeostasis of free cholesterol in the blood: A preliminary evaluation and modeling of its passive transport. J. Lipid Res. 2014, 55, 1033–1043. [Google Scholar] [CrossRef]
- Rearick, J.I.; Jetten, A.M. Accumulation of cholesterol 3-sulfate during in vitro squamous differentiation of rabbit tracheal epithelial cells and its regulation by retinoids. J. Biol. Chem. 1986, 261, 13898–13904. [Google Scholar] [CrossRef]
- Ogden, J.; Sellers, R.; Sahoo, S.; Oojageer, A.; Chaturvedi, A.; Dive, C.; Lopez-Garcia, C. A human model to deconvolve genotype-phenotype causations in lung squamous cell carcinoma. Nat. Commun. 2025, 16, 3215. [Google Scholar] [CrossRef]
- Zhang, Y.; Black, K.E.; Phung, T.-K.N.; Thundivalappil, S.R.; Lin, T.; Wang, W.; Xu, J.; Zhang, C.; Hariri, L.P.; Lapey, A.; et al. Human Airway Basal Cells Undergo Reversible Squamous Differentiation and Reshape Innate Immunity. Am. J. Respir. Cell Mol. Biol. 2023, 68, 664–678. [Google Scholar] [CrossRef]
- Marin, L.; Traini, D.; Bebawy, M.; Colombo, P.; Buttini, F.; Haghi, M.; Ong, H.X.; Young, P. Multiple dosing of simvastatin inhibits airway mucus production of epithelial cells: Implications in the treatment of chronic obstructive airway pathologies. Eur. J. Pharm. Biopharm. 2013, 84, 566–572. [Google Scholar] [CrossRef]
- Lee, E.J.; Song, K.J.; Kwon, J.H.; Park, A.Y.; Jo, K.-H.; Kim, K.-S. Chronic cholesterol depletion by lovastatin suppresses MUC5AC gene expression in human airway epithelial cells. Am. J. Rhinol. Allergy 2014, 28, e125–e129. [Google Scholar] [CrossRef]
- Varshney, P.; Yadav, V.; Saini, N. Lipid rafts in immune signalling: Current progress and future perspective. Immunology 2016, 149, 13–24. [Google Scholar] [CrossRef]
- Maja, M.; Tyteca, D. Alteration of cholesterol distribution at the plasma membrane of cancer cells: From evidence to pathophysiological implication and promising therapy strategy. Front. Physiol. 2022, 13, 999883. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Jung, S.; Kim, D.H.; Choi, Y.J.; Kim, S.Y.; Park, H.; Lee, H.; Choi, C.-M.; Sung, Y.H.; Lee, J.C.; Rho, J.K. Contribution of p53 in sensitivity to EGFR tyrosine kinase inhibitors in non-small cell lung cancer. Sci. Rep. 2021, 11, 19667. [Google Scholar] [CrossRef]
- Jackson, N.D.; Everman, J.L.; Chioccioli, M.; Feriani, L.; Goldfarbmuren, K.C.; Sajuthi, S.P.; Rios, C.L.; Powell, R.; Armstrong, M.; Gomez, J.; et al. Single-Cell and Population Transcriptomics Reveal Pan-epithelial Remodeling in Type 2-High Asthma. Cell Rep. 2020, 32, 107872. [Google Scholar] [CrossRef] [PubMed]
- Duclos, G.E.; Teixeira, V.H.; Autissier, P.; Gesthalter, Y.B.; Reinders-Luinge, M.A.; Terrano, R.; Dumas, Y.M.; Liu, G.; Mazzilli, S.A.; Brandsma, C.-A.; et al. Characterizing smoking-induced transcriptional heterogeneity in the human bronchial epithelium at single-cell resolution. Sci. Adv. 2019, 5, eaaw3413. [Google Scholar] [CrossRef]
- Collin, A.M.; Lecocq, M.; Detry, B.; Carlier, F.M.; Bouzin, C.; de Sany, P.; Hoton, D.; Verleden, S.; Froidure, A.; Pilette, C.; et al. Loss of ciliated cells and altered airway epithelial integrity in cystic fibrosis. J. Cyst. Fibros. 2021, 20, e129–e139. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Y.; Shi, M.; Gao, F.; Huang, L.; Wang, W.; Wei, D.; Shi, C.; Yu, Y.; Xia, X.; et al. The novel molecular mechanism of pulmonary fibrosis: Insight into lipid metabolism from reanalysis of single-cell RNA-seq databases. Lipids Health Dis. 2024, 23, 98. [Google Scholar] [CrossRef]
- Merl-Pham, J.; Basak, T.; Knüppel, L.; Ramanujam, D.; Athanason, M.; Behr, J.; Engelhardt, S.; Eickelberg, O.; Hauck, S.M.; Vanacore, R.; et al. Quantitative proteomic profiling of extracellular matrix and site-specific collagen post-translational modifications in an in vitro model of lung fibrosis. Matrix Biol. Plus 2019, 1, 100005. [Google Scholar] [CrossRef]
- Grosche, A.; Hauser, A.; Lepper, M.F.; Mayo, R.; von Toerne, C.; Merl-Pham, J.; Hauck, S.M. The proteome of native adult müller Glial cells from murine retina. Mol. Cell. Proteom. 2016, 15, 462–480. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Merl, J.; Ueffing, M.; Hauck, S.M.; von Toerne, C. Direct comparison of MS-based label-free and SILAC quantitative proteome profiling strategies in primary retinal Müller cells. Proteomics 2012, 12, 1902–1911. [Google Scholar] [CrossRef] [PubMed]
- Hauck, S.M.; Dietter, J.; Kramer, R.L.; Hofmaier, F.; Zipplies, J.K.; Amann, B.; Feuchtinger, A.; Deeg, C.A.; Ueffing, M. Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol. Cell. Proteom. 2010, 9, 2292–2305. [Google Scholar] [CrossRef] [PubMed]
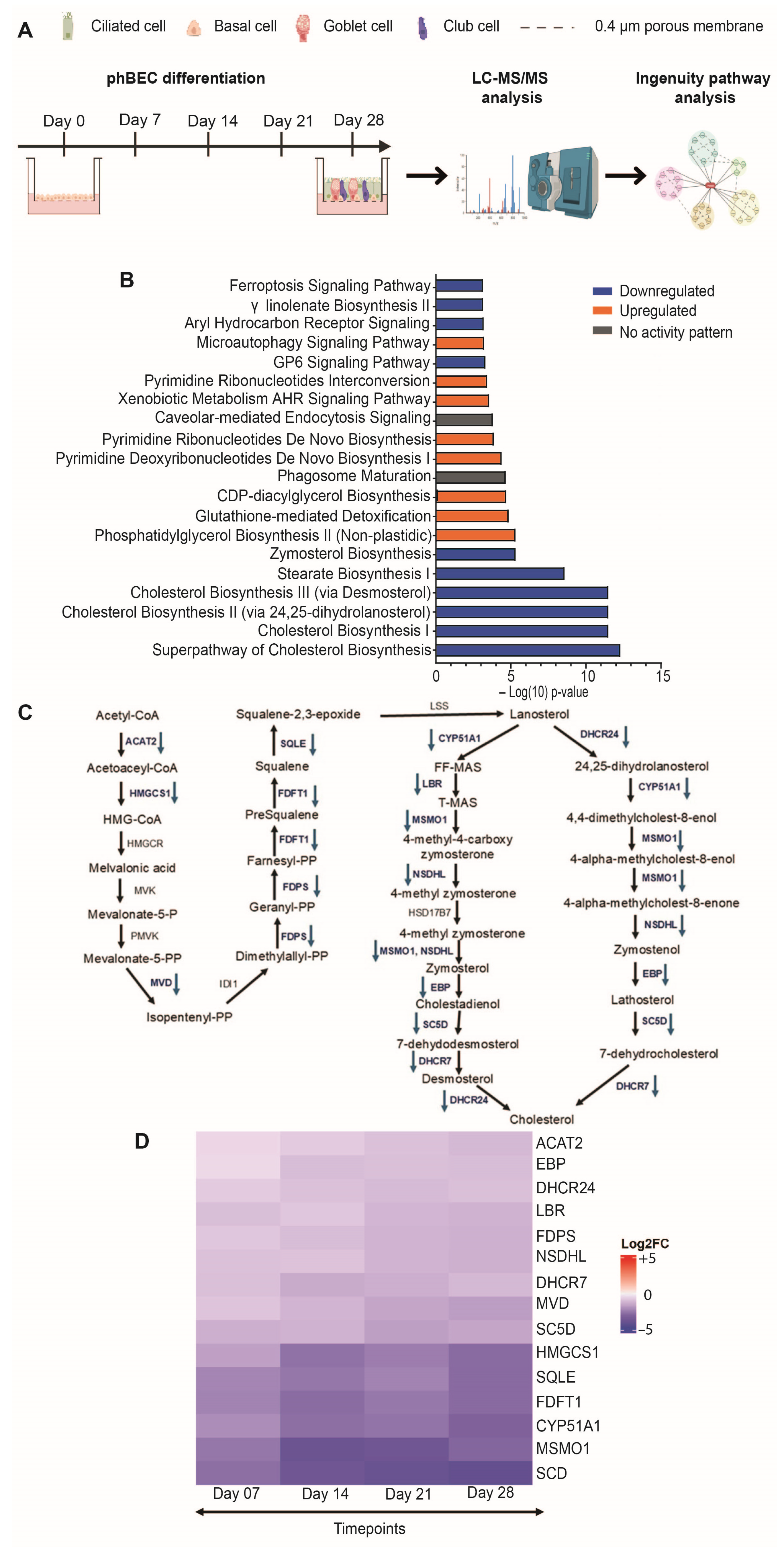
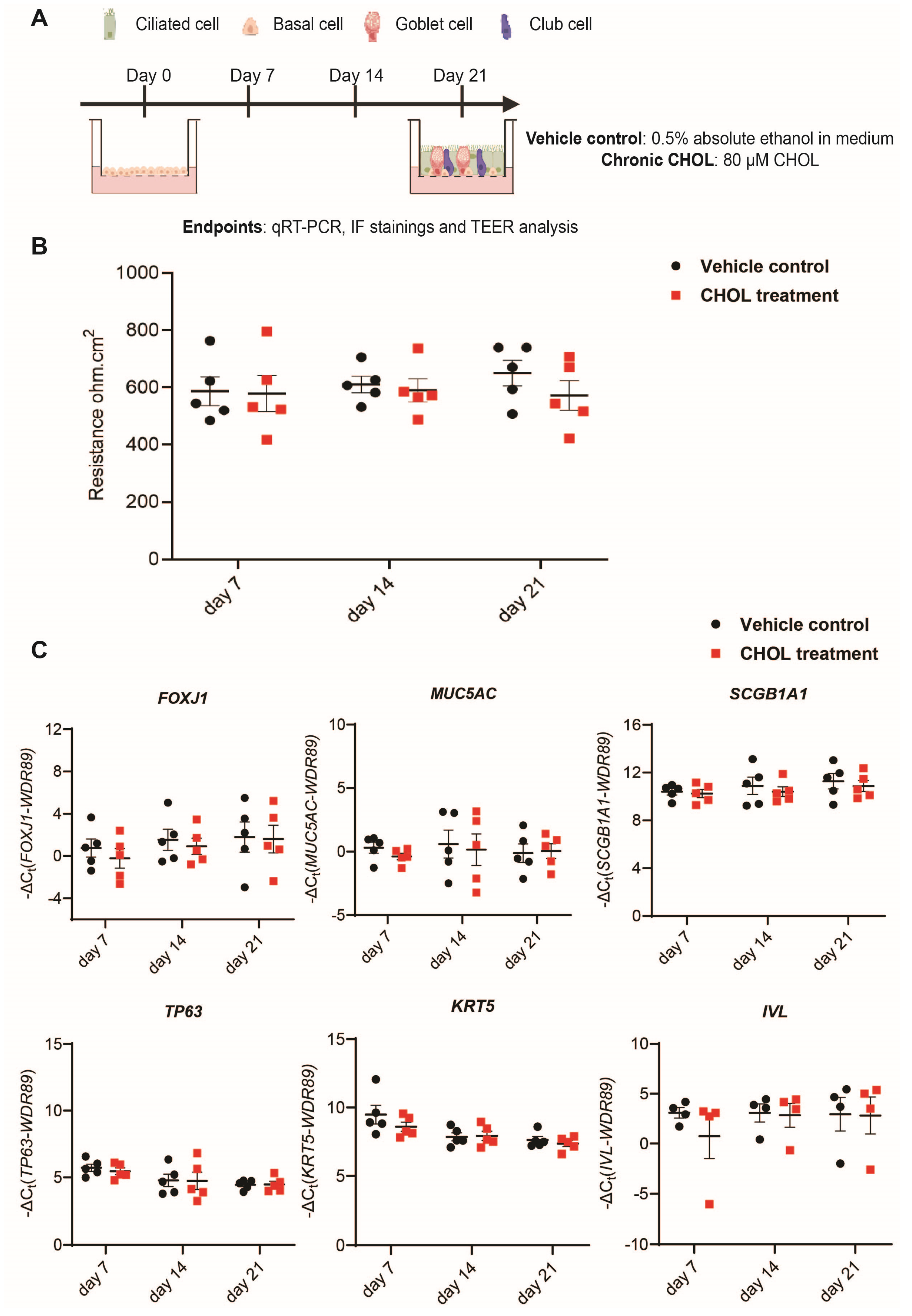
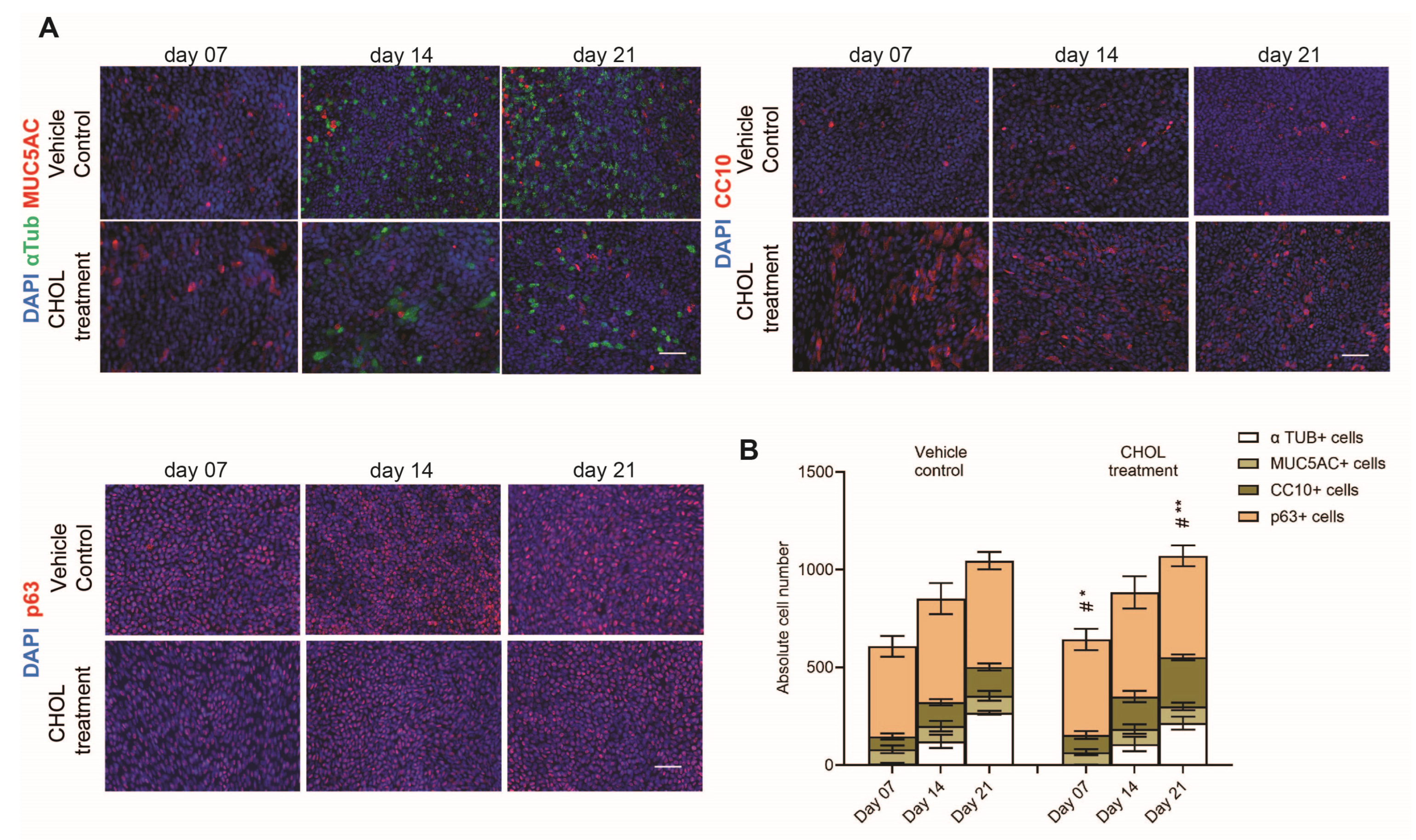
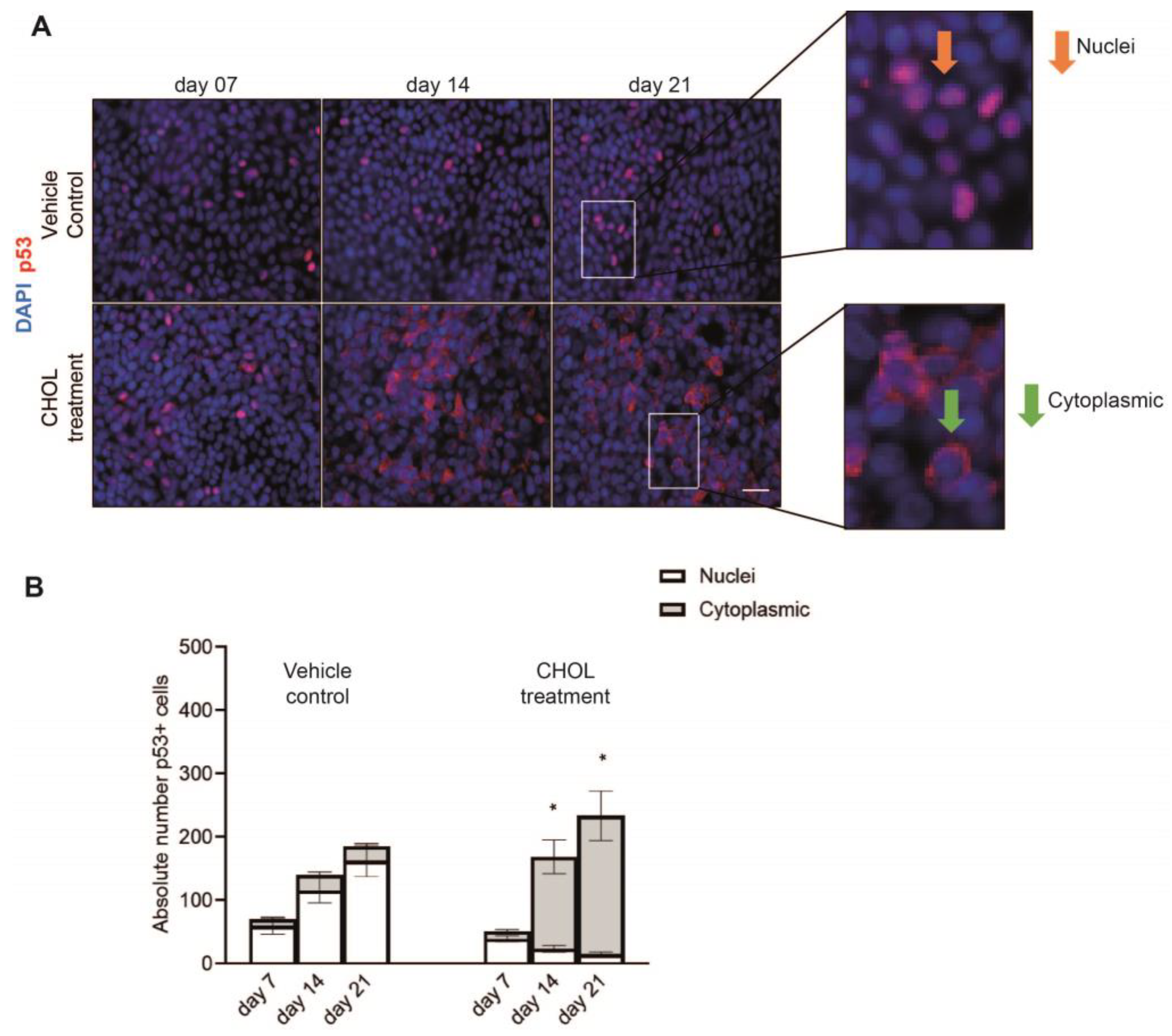
| Accession | Peptide Count | Unique Peptides | Confidence Score | Gene Symbol | Description |
|---|---|---|---|---|---|
| Q9BWD1 | 13 | 10 | 407 | ACAT2 | Q9BWD1|THIC_HUMAN Acetyl-CoA acetyltransferase, cytosolic OS = Homo sapiens GN = ACAT2 PE = 1 SV = 2 |
| P37268 | 12 | 10 | 409 | FDFT1 | P37268|FDFT_HUMAN Squalene synthase OS = Homo sapiens GN = FDFT1 PE = 1 SV = 1 |
| Q15738 | 11 | 10 | 287 | NSDHL | Q15738|NSDHL_HUMAN Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating OS = Homo sapiens GN = NSDHL PE = 1 SV = 2 |
| Q16850 | 14 | 8 | 411 | CYP51A1 | Q16850|CP51A_HUMAN Lanosterol 14-alpha demethylase OS = Homo sapiens GN = CYP51A1 PE = 1 SV = 3 |
| Q01581 | 10 | 8 | 341 | HMGCS1 | Q01581|HMCS1_HUMAN Hydroxymethylglutaryl-CoA synthase, cytoplasmic OS = Homo sapiens GN = HMGCS1 PE = 1 SV = 2 |
| Q15392 | 8 | 7 | 268 | DHCR24 | Q15392|DHC24_HUMAN Delta (24)-sterol reductase OS = Homo sapiens GN = DHCR24 PE = 1 SV = 2 |
| P14324 | 6 | 6 | 275 | FDPS | P14324|FPPS_HUMAN Farnesyl pyrophosphate synthase OS = Homo sapiens GN = FDPS PE = 1 SV = 4 |
| P53602 | 7 | 6 | 209 | MVD | P53602|MVD1_HUMAN Diphosphomevalonate decarboxylase OS = Homo sapiens GN = MVD PE = 1 SV = 1 |
| O00767 | 7 | 6 | 267 | SCD | O00767|ACOD_HUMAN Acyl-CoA desaturase OS = Homo sapiens GN = SCD PE = 1 SV = 2 |
| Q15800 | 4 | 4 | 141 | MSMO1 | Q15800|MSMO1_HUMAN Methylsterol monooxygenase 1 OS = Homo sapiens GN = MSMO1 PE = 1 SV = 1 |
| Q9UBM7 | 3 | 3 | 68 | DHCR7 | Q9UBM7|DHCR7_HUMAN 7-dehydrocholesterol reductase OS = Homo sapiens GN = DHCR7 PE = 1 SV = 1 |
| Q14534 | 2 | 2 | 54 | SQLE | Q14534|ERG1_HUMAN Squalene monooxygenase OS = Homo sapiens GN = SQLE PE = 1 SV = 3 |
| Q15125 | 2 | 1 | 89 | EBP | Q15125|EBP_HUMAN 3-beta-hydroxysteroid-Delta (8), Delta (7)-isomerase OS = Homo sapiens GN = EBP PE = 1 SV = 3 |
| Q14739 | 2 | 1 | 51 | LBR | Q14739|LBR_HUMAN Lamin-B receptor OS = Homo sapiens GN = LBR PE = 1 SV = 2 |
| O75845 | 2 | 1 | 47 | SC5D | O75845|SC5D_HUMAN Lathosterol oxidase OS = Homo sapiens GN = SC5D PE = 1 SV = 2 |
| Upstream Regulator | Molecule Type | Prediction Activation State | Activation Z-Score | p-Value of Overlap | Target Molecules in Dataset |
|---|---|---|---|---|---|
| TP53 | transcription regulator | Activated | 3.079 | 0.000000116 | ACAA1, ACL, ACOX1, ACSL3, ADA, AK1, ALDH1A1, ALDH1A3, ARHGEF2, ARL6IP1, ARVCF, BIRC3, CCN2, CD276, CD44, CES2, CLU, COL18A1, COL1A1, COL3A1, COL7A1, CRYAB, CTSD, CYP51A1, DHCR7, DKK1, DUT, ECM1, EGFR, FAM§C, FASN, FDFT1, FDPS, FDXR, FEN1, FGFBP1, FOSL1, FTH1, GDF15, GLB1, HMGCS1, IFI30, IFI35, IFIT1, IGFBP7, IL1B, IL1RN, ITGB4, LAMC2, LBR, LGALS3, MAD2L1, MCM2, MCM3, MCM6, MCM7, ME1, MET, MPZL2, MTHFD2, MVD, NDRG1, NFKB2, NOTCH1, NUCKS1, OMA1, PFKP, PLAU, POLD1, PPP5C, PPT1, PRKCB, PRODH, PROM1, PUM3, RBM34, S100A9, SCD, SEL1L, SLC16A1, SLC2A1, SMC2, SMC4, SOD2, SPATA18, SQLE, STX6, TFRC, THBD, TIGAR, TLR3, TNFRSF10B, TUBB, VCAN |
| CST5 | other | Activated | 2.623 | 0.0000034 | ACAT2, AHCTF1, ANK3, ANXA3, ARHGEF2, C17orf49, CD44, CNBP, CORO1A, CTSB, DDX21, DRAP1, EBNA1BP2, EEF1B2, EIF5B, FBX = 2, GALNT5, GDA, GPRC5A, INTS1, ITPRID2, LYAR, MSN, MYO5B, NIFK, NT5E, PITRM1, RBM28, RPF2, RRS1, RSL1D1, SSRP1, SURF6, TOMM40, VASP, VCAN, WDR3, WDR36 |
| OGA | enzyme | Activated | 2.558 | 0.0247 | ACTG1, ALDOC, CDK5, CELSR1, CEP43, DHCR7, FDFT1, FDPS, FGFBP1, FLNA, FSCN1, GALK2, HMGCS1, LAMB1, MSMO1, NSDHL, PLAU, PROM1, S100A2, TNC, TP53BP1, TRADD |
| IFT88 | other | Activated | 2.449 | 0.000111 | ACAT2, ACLY, CYP51A1, FDPS, MVD, NSDHL |
| KIF3A | enzyme | Activated | 2.449 | 0.0000379 | ACAT2, ACLY, CYP51A1, FDPS, MVD, NSDHL |
| ILF3 | transcription regulator | Activated | 2.132 | 0.0000013 | ACAT2, ACTG1, CCDC170, CFH, DHCR7, EBP, EGFR, FADS2, FDPS, IL1B, IL1RN, LDLR, LGALS3, MCM5, NSDHL, PLAU, SCD, SLC3A2, STEAP4, TLR3, TNC |
| SREBF1 | transcription regulator | Inhibited | −2.207 | 0.0000261 | ACADS, ACLA, AK4, ALDOA, CAP2, CYP51A1, FASN, FDPS, GLB1, IFI30, LDLR, LGALS3, MDK, NPC1, OAT, P4HA2, PLEKHA4, PPAT, RNASET2, SCD, SERPINA3, TF |
| NUPR1 | transcription regulator | Inhibited | −2.795 | 0.142 | ACSS1, ALDH5A1, ALDOC, ANK3, BRI3BP, CCN1, CEBPB, COL3A1, DHCR24, ETV6, GDF15, GSTA4, HSPA2, LARS2, LBR, MKI67, NDRG1, NEK9, NUCKS1, NUP50, P4HA2, PARP9, PRR12, RAB32, RIBC2, RNF19B, SAMHD1, SHROOM3, SLC2A1, STK38, SUOX, TCTN2, TMPO, TNFAIP8L1, UPP1, ZNF512 |
| THEM6 | other | Inhibited | −3.302 | 0.000385 | ACAA1, ASNS; CYP51A1, DHCR7, FDFT1, FDPS, HMGCS1, MSMO1, MVD, NSDHL, SQLE |
| Gene | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| FOXJ1 | TCGTATGCCACGCTCATCTG | CTTGTAGATGGCCGACAGGG |
| IVL | GGAGGTCCCATCAAAGCAAGA | GCTCCTTCTGCTGTGCTCA |
| KRT5 | GAGATCGCCACTTACCGCAA | TGCTTGTGACAACAGAGATGT |
| MUC5AC | AGCAGGGTCCTCATGAAGGTGGAT | AATGAGGACCCCAGACTGGCTGAA |
| SCGB1A1 | TTCAGCGTGTCATCGAAACCC | ACAGTGAGCTTTGGGCTATTTTT |
| TP63 | CCCGTTTCGTCAGAACACAC | CATAAGTCTCACGGCCCCTC |
| WDR89 | AGTACGTTCCATCCCAGCAATCC | AGGCCATCAGATGAACCTGAGACT |
| Target | Host | Catalog Number | Provider | Dilution |
|---|---|---|---|---|
| αTUB | Rabbit | ab 179484 | Abcam | 1:500 |
| CC10 | Mouse | sc 365992 | Santa Cruz | 1:300 |
| MUC5AC | Mouse | ab 3649 | Abcam | 1:250 |
| p53 | Mouse | sc126 | Santa Cruz | 1:100 |
| p63 | Mouse | ab 735 | Abcam | 1:125 |
| Donkey anti-mouse (red 568) | Mouse | A10037 | Thermofisher | 1:500 |
| Goat anti-rabbit (green 488) | Rabbit | A32731 | Thermofisher | 1:500 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, A.; Giraldo-Arias, J.; Merl-Pham, J.; Dick, E.; Mastalerz, M.; Zöller, M.; Marchi, H.; Le Gleut, R.; Hatz, R.A.; Behr, J.; et al. Cholesterol Regulates Airway Epithelial Cell Differentiation by Inhibiting p53 Nuclear Translocation. Int. J. Mol. Sci. 2025, 26, 8324. https://doi.org/10.3390/ijms26178324
Chakraborty A, Giraldo-Arias J, Merl-Pham J, Dick E, Mastalerz M, Zöller M, Marchi H, Le Gleut R, Hatz RA, Behr J, et al. Cholesterol Regulates Airway Epithelial Cell Differentiation by Inhibiting p53 Nuclear Translocation. International Journal of Molecular Sciences. 2025; 26(17):8324. https://doi.org/10.3390/ijms26178324
Chicago/Turabian StyleChakraborty, Ashesh, Juliana Giraldo-Arias, Juliane Merl-Pham, Elisabeth Dick, Michal Mastalerz, Marie Zöller, Hannah Marchi, Ronan Le Gleut, Rudolf A. Hatz, Jürgen Behr, and et al. 2025. "Cholesterol Regulates Airway Epithelial Cell Differentiation by Inhibiting p53 Nuclear Translocation" International Journal of Molecular Sciences 26, no. 17: 8324. https://doi.org/10.3390/ijms26178324
APA StyleChakraborty, A., Giraldo-Arias, J., Merl-Pham, J., Dick, E., Mastalerz, M., Zöller, M., Marchi, H., Le Gleut, R., Hatz, R. A., Behr, J., Hilgendorff, A., Hauck, S. M., & Staab-Weijnitz, C. A. (2025). Cholesterol Regulates Airway Epithelial Cell Differentiation by Inhibiting p53 Nuclear Translocation. International Journal of Molecular Sciences, 26(17), 8324. https://doi.org/10.3390/ijms26178324








