The Complete Mitochondrial Genome of the Deep-Dwelling Goby Suruga fundicola (Teleostei, Gobiidae) Reveals Evidence of Recombination in the Control Region
Abstract
1. Introduction
2. Results and Discussion
2.1. General Features of S. fundicola Mitogenome
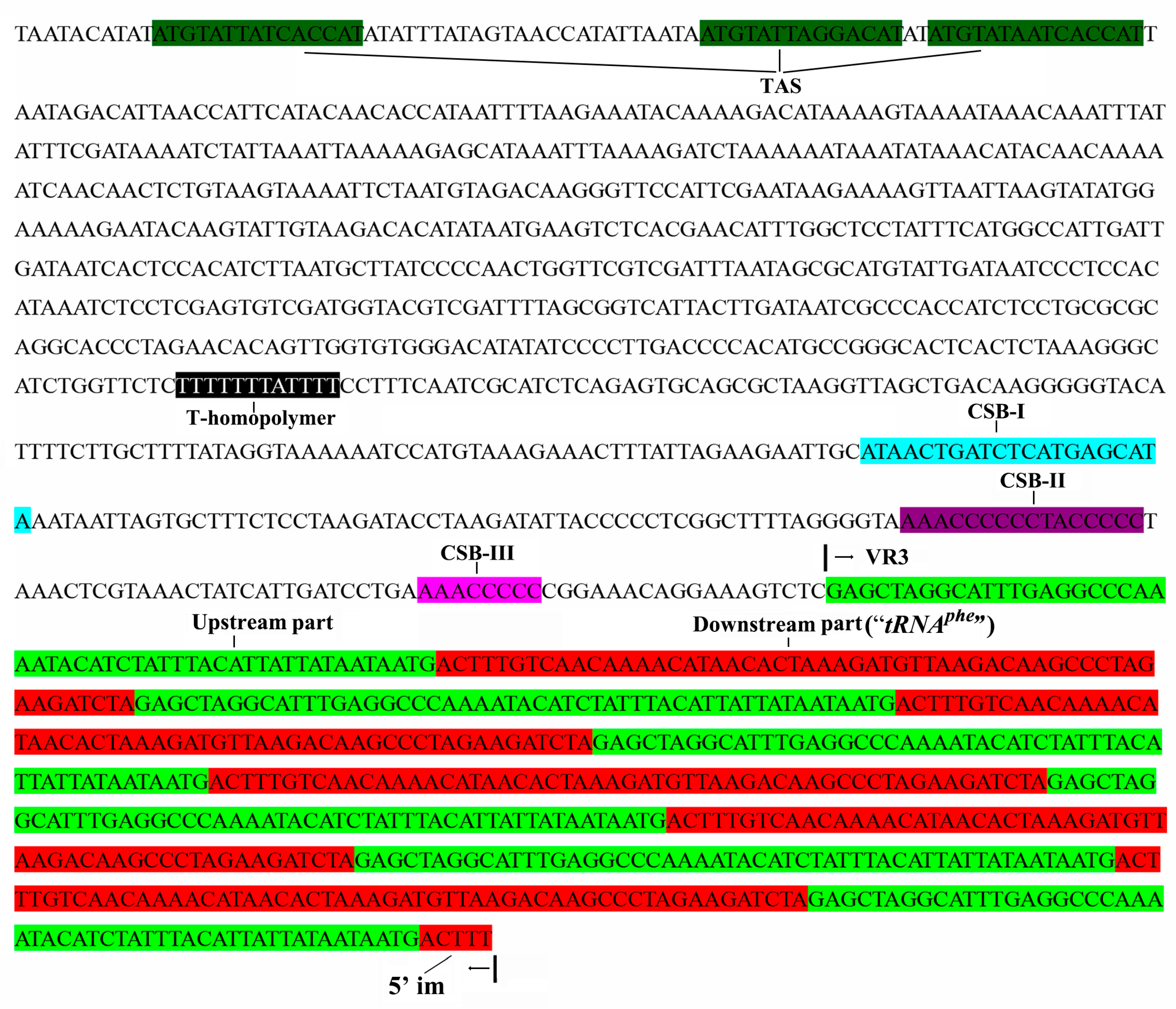
2.2. Variation in the Long Terminal Repeat
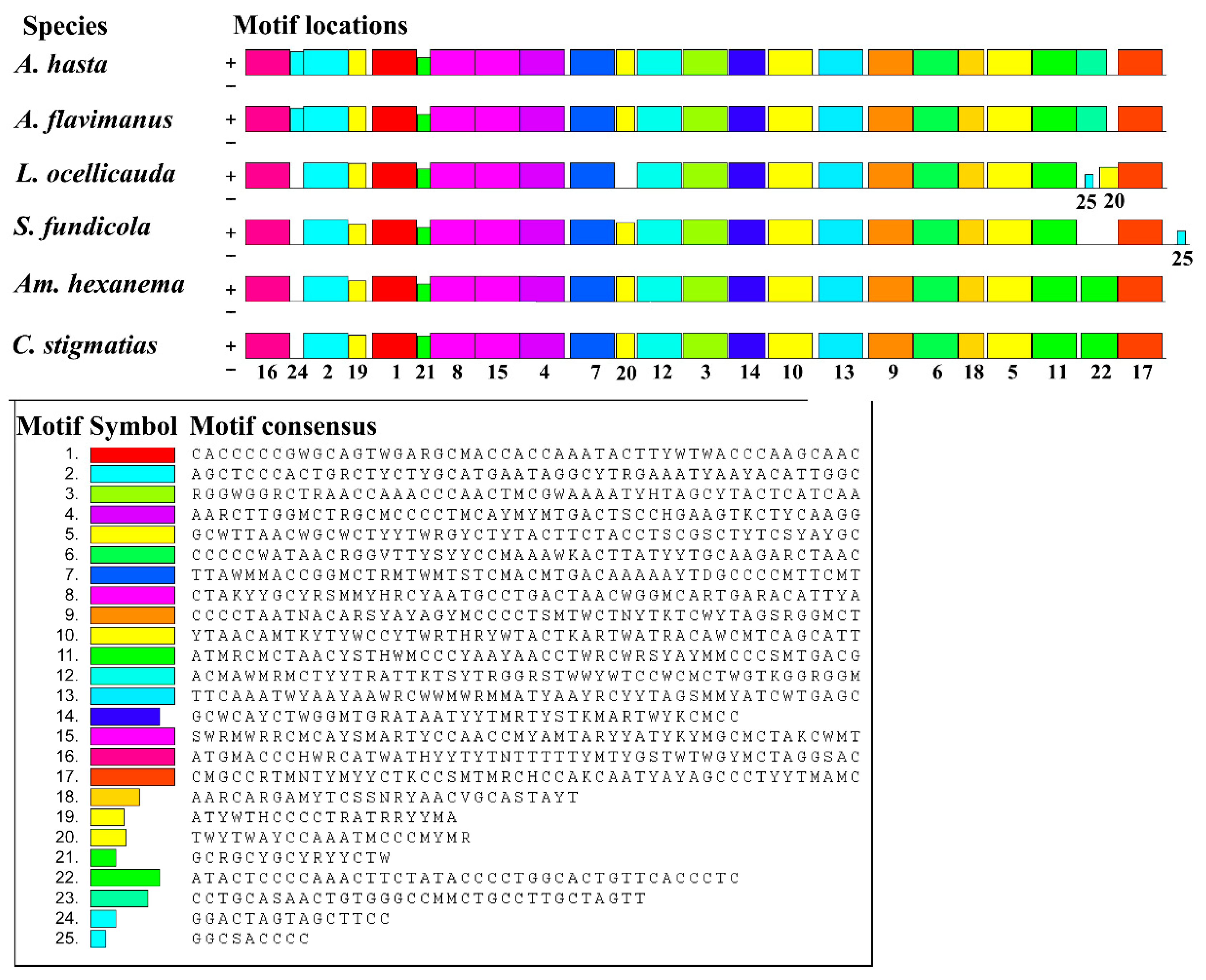
2.3. Comparative Analysis
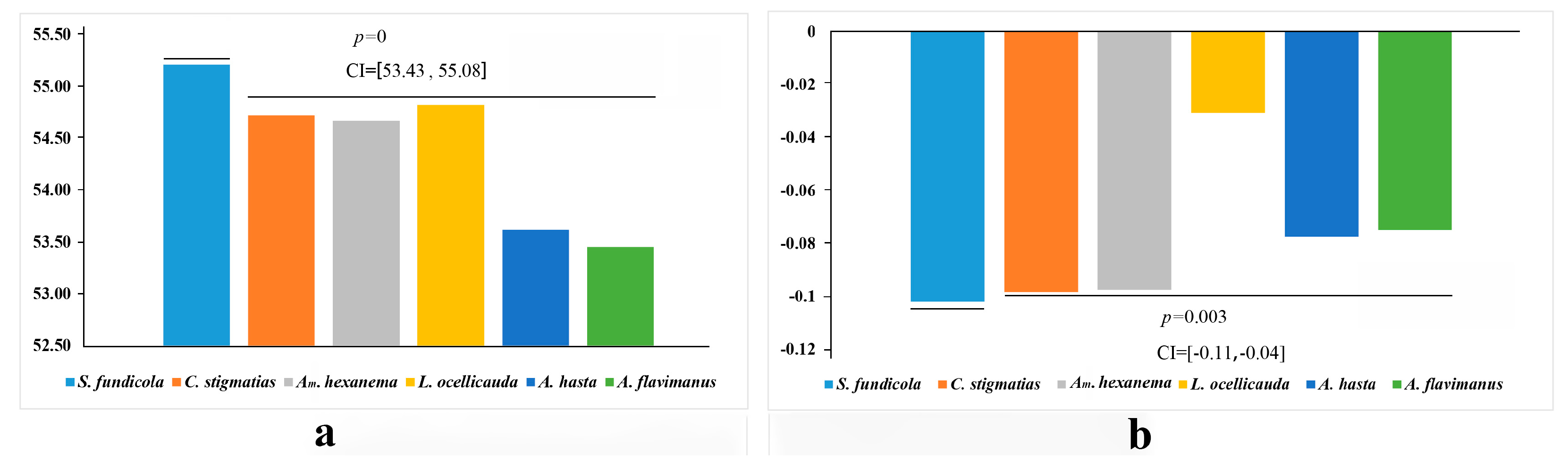
2.3.1. A + T% and AT-Skew of the Acanthogobius Group Mitogenomes PCGs
2.3.2. Transfer RNA of the Acanthogobius Group
2.3.3. ND2 Gene of the Acanthogobius Group Mitogenomes
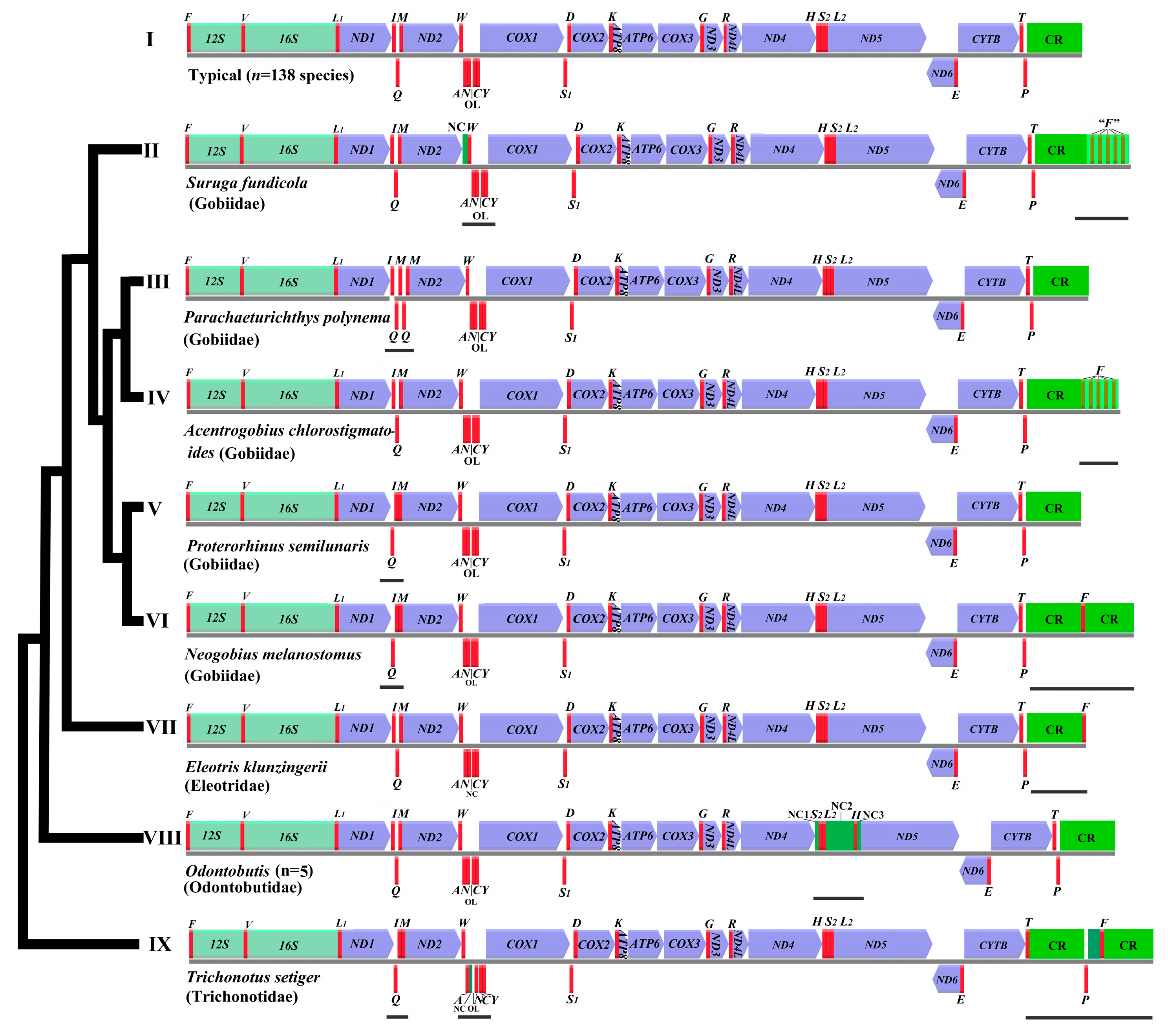
2.3.4. Gene Arrangement Analysis of Goby Fish Mitogenomes
2.3.5. Start and Stop Codons of the Mitogenomes of Gobies
2.4. Phylogenetic Inferences of Gobies
2.5. Selective Pressure Analysis
3. Materials and Methods
3.1. Sample Collection and DNA Extraction
3.2. Library Construction and DNA Sequencing
3.3. Sequence Assembly, Annotation, and Analysis of S. fundicola
3.4. Comparative Analysis of the Goby Mitogenomes
3.5. Molecular Phylogenetic Analysis
3.6. Selective Pressure Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hampl, V.; Epika, I.; Eliá, M. Was the mitochondrion necessary to start Eukaryogenesis? Trends Microbiol. 2019, 27, 96–104. [Google Scholar] [CrossRef]
- Burzyński, A.; Zbawicka, M.; Skibinski, D.O.F.; Wenne, R. Evidence for Recombination of mtDNA in the marine mussel Mytilus trossulus from the Baltic. Mol. Biol. Evol. 2003, 20, 388–392. [Google Scholar] [CrossRef]
- Ladoukakis, E.D.; Eleftherios, Z. Direct Evidence for Homologous Recombination in Mussel (Mytilus galloprovincialis) Mitochondrial DNA. Mol. Biol. Evol. 2001, 18, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Arnason, E.; Rand, D.M. Heteroplasmy of short tandem repeats in mitochondrial DNA of Atlantic cod, Gadus morhua. Genetics 1992, 132, 211–220. [Google Scholar] [CrossRef]
- Hoarau, G.; Holla, S.; Lescasse, R.; Stam, W.T.; Olsen, J.L. Heteroplasmy and Evidence for Recombination in the Mitochondrial Control Region of the Flatfish Platichthys flesus. Mol. Biol. Evol. 2002, 19, 2261–2264. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Liu, N.; Zhang, H.-L.; Yang, X.-J.; Huang, Y.; Lei, F. Extreme variation in patterns of tandem repeats in mitochondrial control region of yellow-browed tits (Sylviparus modestus, Paridae). Sci. Rep. 2015, 5, 13227. [Google Scholar] [CrossRef]
- Patterson, E.C.; Lall, G.M.; Neumann, R.; Ottolini, B.; Batini, C.; Sacchini, F.; Foster, A.P.; Wetton, J.H.; Jobling, M.A. Mitogenome sequences of domestic cats demonstrate lineage expansions and dynamic mutation processes in a mitochondrial minisatellite. BMC Genom. 2023, 24, 690. [Google Scholar] [CrossRef]
- Marilena, D.A.; Gajewski, C.D.; Lin, M.-T.; Mauck, W.M.; Shao, L.-Z.; Giorgio, L.; Moraes, C.T.; Giovanni, M. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 2004, 13, 3171–3179. [Google Scholar] [CrossRef]
- Strakova, A.; Leathlobhair, M.N.; Wang, G.D.; Yin, T.-T.; Airikkalaotter, I.; Allen, J.L.; Allum, K.M.; Bansseissa, L.; Bisson, J.L.; Castillo, D.A. Mitochondrial genetic diversity, selection and recombination in a canine transmissible cancer. eLife 2016, 5, e14552. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Cui, X.-F.; Chen, S.-H.; Li, Y.; Zhang, S.-Y.; Yang, Y.-F.; Li, Y.-Y.; Guo, Y.-S.; Wang, Z.-D.; Liao, J. The First Complete Mitogenome Characterization and Phylogenetic Implications of Elops machnata (Teleostei: Elopiformes: Elopidae). Biology 2025, 14, 739. [Google Scholar] [CrossRef]
- Bibb, M.J.; Van Etten, R.A.; Wright, C.T.; Walberg, M.W.; Clayton, D.A. Sequence and gene organization of mouse mitochondrial DNA. Cell 1981, 26, 167–180. [Google Scholar] [CrossRef]
- Bohnsack, J.A.; Sutherland, D.L. Artificial Reef Research: A Review with Recommendations for Future Priorities. Bull. Mar. Sci. -Miami-. 1985, 37, 11–39. [Google Scholar]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef]
- Serrano, M.J.; Goudet, J.; Cumer, T. Characterization of the diversity of barn owl’s mitochondrial genome reveals high copy number variations in the control region. PLoS ONE 2024, 19, e0295595. [Google Scholar] [CrossRef] [PubMed]
- Tapanainen, R.; Aasumets, K.; Fekete, Z.; Goffart, S.; Dufour, E.; Pohjoismäki, J.L.O. Species-specific variation in mitochondrial genome tandem repeat polymorphisms in hares (Lepus spp., Lagomorpha, Leporidae) provides insight into their evolution. Gene 2024, 926, 148644. [Google Scholar] [CrossRef]
- Adrian-Kalchhauser, I.; Svensson, O.; Kutschera, V.E.; Alm Rosenblad, M.; Pippel, M.; Winkler, S.; Schloissnig, S.; Blomberg, A.; Burkhardt-Holm, P. The mitochondrial genome sequences of the round goby and the sand goby reveal patterns of recent evolution in gobiid fish. BMC Genom. 2017, 18, 177. [Google Scholar] [CrossRef] [PubMed]
- Dadkhah, K.; Mianji, G.R.; Barzegar, A.; Farhadi, A. Characterization of the mitochondrial Huso huso genome and new aspects of its organization in the presence of tandem repeats in 12S rRNA. BMC Ecol. Evol. 2023, 23, 55. [Google Scholar] [CrossRef]
- Wilkinson, G.S.; Mayer, F.; Kerth, G.; Petri, B. Evolution of repeated sequence arrays in the D-loop region of bat mitochondrial DNA. Genetics 1997, 146, 1035–1048. [Google Scholar] [CrossRef]
- Birdsong, R.S.; Murdy, E.O.; Pezold, F.L. A Study of the Vertebral Column and Median Fin Osteology in Gobioid Fishes with Comments on Gobioid Relationships. Bull. Mar. Sci. 1988, 42, 174–214. [Google Scholar]
- Shibukawa, K.; Iwata, A. Review of the East Asian gobiid genus Chaeturichthys (Teleostei: Perciformes: Gobioidei), with description of a new species. Bull. Natl. Mus. Nat. Sci. 2013, 4, 31–51. [Google Scholar]
- Agorreta, A.; San Mauro, D.; Schliewen, U.; Van Tassell, J.L.; Kovačić, M.; Zardoya, R.; Rüber, L. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Mol. Phylogenetics Evol. 2013, 69, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Thacker, C.E. Phylogenetic placement of the European sand gobies in Gobionellidae and characterization of gobionellid lineages (Gobiiformes: Gobioidei). Zootaxa 2013, 3619, 369–382. [Google Scholar] [CrossRef] [PubMed]
- McCraney, W.T.; Thacker, C.E.; Alfaro, M.E. Supermatrix phylogeny resolves goby lineages and reveals unstable root of Gobiaria. Mol. Phylogenetics Evol. 2020, 151, 106862. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, B.-J.; Gao, T.-X.; Song, N. The mitochondrial genome of Chaeturichthys stigmatias provides novel insight into the interspecific difference with Amblychaeturichthys hexanema. Acta Oceanol. Sin. 2021, 40, 74–81. [Google Scholar] [CrossRef]
- An, C.-T.; Li, A.; Wang, H.; Li, B.-S.; Liu, K.-Y.; Sun, H.-Y.; Liu, S.; Zhuang, Z.-M.; van der Laan, R. Identification of the rare deep-dwelling goby Suruga fundicola Jordan & Snyder, 1901 (Gobiiformes, Gobiidae) from the Yellow Sea. Zoosyst. Evol. 2023, 99, 489–501. [Google Scholar]
- Akihito; Ikeda, Y.; Aizawa, M. Suborder Gobioidei. In Fishes of Japan with Pictorial Keys to the Species, 3rd ed.; Nakabo, T., Ed.; Tokai University Press: Tokyo, Japan, 2013; pp. 1347–1608 + 2109–2211. ISBN 978-4-4860-1804-9. [Google Scholar]
- Akihito; Hayashi, M.; Yoshino, T.; Shimada, K.; Yamamoto, T. Suborder Gobioidei. In The fishes of the Japanese Archipelago; Masuda, H., Amaoka, K., Araga, C., Uyeno, T., Yoshino, T., Eds.; Tokai University Press: Tokyo, Japan, 1984; pp. 236–289, pls. 235–258, 353–355. [Google Scholar]
- Choi, Y.; Lee, H.-H. First Record of the Goby, Suruga fundicola (Perciformes: Gobiidae) from Tongyeong, Korea. Korean J. Ichthyol. 2019, 31, 255–258. [Google Scholar] [CrossRef]
- Warrant, E. The eyes of deep-sea fishes and the changing nature of visual scenes with depth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1155–1159. [Google Scholar] [CrossRef]
- Shen, Y.-Y.; Liang, L.; Zhu, Z.-H.; Zhou, W.-P.; Irwin, D.M.; Zhang, Y.-P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. USA 2010, 107, 8666–8671. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Cao, J.-H.; Xiang, H.-M.; Liu, Z.-X.; Jiang, W.-S. Comparative mitogenomic analysis of Chinese cavefish Triplophysa (Cypriniformes: Nemacheilidae): Novel gene tandem duplication and evolutionary implications. BMC Genom. 2025, 26, 293. [Google Scholar] [CrossRef]
- Kim, I.C.; Kweon, H.S.; Kim, Y.J.; Kim, C.B.; Gye, M.C.; Lee, W.O.; Lee, Y.S.; Lee, J.S. The complete mitochondrial genome of the javeline goby Acanthogobius hasta (Perciformes, Gobiidae) and phylogenetic considerations. Gene 2004, 336, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.-Q.; Jin, X.-X.; Sun, Y.-N. The complete mitochondrial genome of Lophiogobius ocellicauda (Perciformes, Gobiidae). Mitochondrial DNA 2014, 25, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, H.; Yang, L.; Liu, S.; Zhuang, Z. Complete Mitochondrial Genome of Pseudocaranx dentex (Carangidae, Perciformes) Provides Insight into Phylogenetic and Evolutionary Relationship among Carangidae Family. Genes 2021, 12, 1234. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.-Q.; Wang, M.-H.; Li, D.-M.; Tang, S.-K.; Zhang, T.-Q.; Bian, W.-J.; Chen, X.-H. Complete mitochondrial genome of Odontobutis haifengensis (Perciformes, Odontobutiae): A unique rearrangement of tRNAs and additional non-coding regions identified in the genus. Odontobutis Genom. 2018, 110, 382–388. [Google Scholar]
- Hernández, M.Á.; Campos, F.; Gutiérrez-corchero, F.; Amezcua, A. Identification of Lanius species and subspecies using tandem repeats in the mitochondrial DNA control region. IBIS 2003, 146, 227–230. [Google Scholar] [CrossRef]
- Keogh, S.M.; Johnson, N.A.; Smith, C.H.; Sietman, B.E.; Garner, J.T.; Randklev, C.R.; Simons, A.M. Secondary contact erodes Pleistocene diversification in a wide-ranging freshwater mussel (Quadrula). Mol. Ecol. 2025, 34, e17572. [Google Scholar] [CrossRef]
- Padhi, A. Geographic variation within a tandemly repeated mitochondrial DNA D-loop region of a North American freshwater fish, Pylodictis olivaris. Gene 2014, 538, 63–68. [Google Scholar] [CrossRef]
- Mundy, N.I.; Winchell, C.S.; Woodruff, D.S. Tandem repeats and heteroplasmy in the mitochondrial DNA control region of the loggerhead shrike (Lanius ludovicianus). J. Hered. 1996, 87, 21–26. [Google Scholar] [CrossRef]
- Françoso, E.; Zuntini, A.R.; Ricardo, P.C.; Araujo, N.S.; Silva, J.P.N.; Brown, M.J.F.; Arias, M.C. The complete mitochondrial genome of Trigonisca nataliae (Hymenoptera, Apidae) assemblage reveals heteroplasmy in the control region. Gene 2023, 881, 147621. [Google Scholar] [CrossRef]
- Cerasale, D.J.; Dor, R.; Winkler, D.W.; Lovette, I.J. Phylogeny of the Tachycineta genus of New World swallows: Insights from complete mitochondrial genomes. Mol. Phylogenetics Evol. 2012, 63, 64–71. [Google Scholar] [CrossRef]
- Slack, K.E.; Delsuc, F.; McLenachan, P.A.; Arnason, U.; Penny, D. Resolving the root of the avian mitogenomic tree by breaking up long branches. Mol. Phylogenet. Evol. 2007, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhou, L.; Yang, W.T.; Miao, L.J.; Li, Z.; Zhang, X.J.; Wang, Y.; Gui, J.F. Comparative mitogenome analyses uncover mitogenome features and phylogenetic implications of the subfamily Cobitinae. BMC Genom. 2021, 22, 50. [Google Scholar] [CrossRef]
- Tang, Q.; Liu, Y.; Li, C.H.; Zhao, J.F.; Wang, T. Comparative Mitogenome Analyses Uncover Mitogenome Features and Phylogenetic Implications of the Reef Fish Family Holocentridae (Holocentriformes). Biology 2023, 12, 1273. [Google Scholar] [CrossRef]
- Ting, K.; Luke, T.; Jing Yan, L.; Jia Mei, J.; Prosanta, C.; Sparks, J.S.; Naylor, G.J.P.; Chen Hong, L. Phylogenomic analysis on the exceptionally diverse fish clade Gobioidei (Actinopterygii: Gobiiformes) and data-filtering based on molecular clocklikeness. Mol. Phylogenet. Evol. 2018, 128, 192–202. [Google Scholar]
- Sha, Z.; Xiao, N. The first two complete mitogenomes of the order Apodida from deep-sea chemoautotrophic environments: New insights into the gene rearrangement, origin and evolution of the deep-sea sea cucumbers. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2021, 39, 100839. [Google Scholar]
- Zhao, B.; Gao, S.; Zhao, M.; Lv, H.; Song, J.; Wang, H.; Zeng, Q.; Liu, J. Mitochondrial genomic analyses provide new insights into the “missing” atp8 and adaptive evolution of Mytilidae. BMC Genom. 2022, 23, 738. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Freyer, C.; Elson, J.L.; Larsson, N.G.R. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat. Rev. Genet. 2008, 9, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Kuhle, B.; Chihade, J.; Schimmel, P. Relaxed sequence constraints favor mutational freedom in idiosyncratic metazoan mitochondrial tRNAs. Nat. Commun. 2020, 11, 969. [Google Scholar] [CrossRef]
- Wallace, D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet. 2005, 39, 359–407. [Google Scholar] [CrossRef]
- Shen, X.-J.; Pu, Z.-Q.; Chen, X.; Murphy, R.W.; Shen, Y.-Y. Convergent Evolution of Mitochondrial Genes in Deep-Sea Fishes. Front. Genet. 2019, 10, 925. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, J.; Xu, T.; Qiu, J.-W.; Qian, P.-Y. Phylogenetic Relationships and Adaptation in Deep-Sea Mussels: Insights from Mitochondrial Genomes. Int. J. Mol. Sci. 2021, 22, 1900. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, Y.-Y.; Wang, X.; Jiang, W.; Yin, J.-P.; Lin, Q. Comparative analysis of mitochondrial genome of a deep-sea crab Chaceon granulates reveals positive selection and novel genetic features. J. Ocean. Limnol. 2020, 38, 427–437. [Google Scholar] [CrossRef]
- Gutiérrez, E.G.; Ortega, J.; Savoie, A.; Baeza, J.A. The mitochondrial genome of the mountain wooly tapir, Tapirus pinchaque and a formal test of the effect of altitude on the adaptive evolution of mitochondrial protein coding genes in odd-toed ungulates. BMC Genom. 2023, 24, 527. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.-P.; Lin, B.-Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3374–3376. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 5, 484–489. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Jemt, E.; Persson, Ö.; Shi, Y.; Mehmedovic, M.; Uhler, J.P.; López, M.D.; Freyer, C.; Gustafsson, C.M.; Samuelsson, T.; Falkenberg, M. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015, 43, 9262–9275. [Google Scholar] [CrossRef]
- Nicholls, T.J.; Minczuk, M. In D-loop: 40 years of mitochondrial 7S DNA. Exp. Gerontol. 2014, 56, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Sbisà, E.; Tanzariello, F.; Reyes, A.; Pesole, G.; Saccone, C. Mammalian mitochondrial D-loop region structural analysis: Identification of new conserved sequences and their functional and evolutionary implications. Gene 1997, 205, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.; Van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References; California Academy of Sciences: San Francisco, CA, USA; Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 10 August 2025).
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Kainer, D.; Mayer, C.; Stamatakis, A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014, 14, 82. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Anisimova, M.; Gil, M.; Dufayard, J.-F.; Dessimoz, C.; Gascuel, O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree-Version 1.4.3, A Graphical Viewer of Phylogenetic Trees. Computer Program Distributed by the Author. 2017. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 5 June 2024).
- Wang, D.-P.; Zhang, Y.-B.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Gao, F.-L.; Chen, C.-J.; Arab, D.A.; Du, Z.-G.; He, Y.-H.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Bielawski, J.P.; Yang, Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 2001, 18, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-Z.; Nielsen, R.; Yang, Z.-H. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
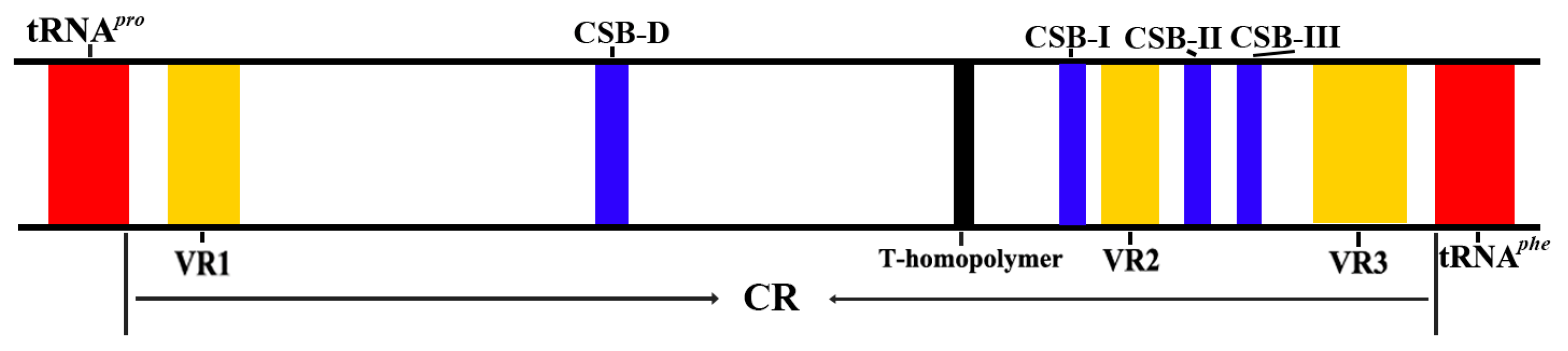
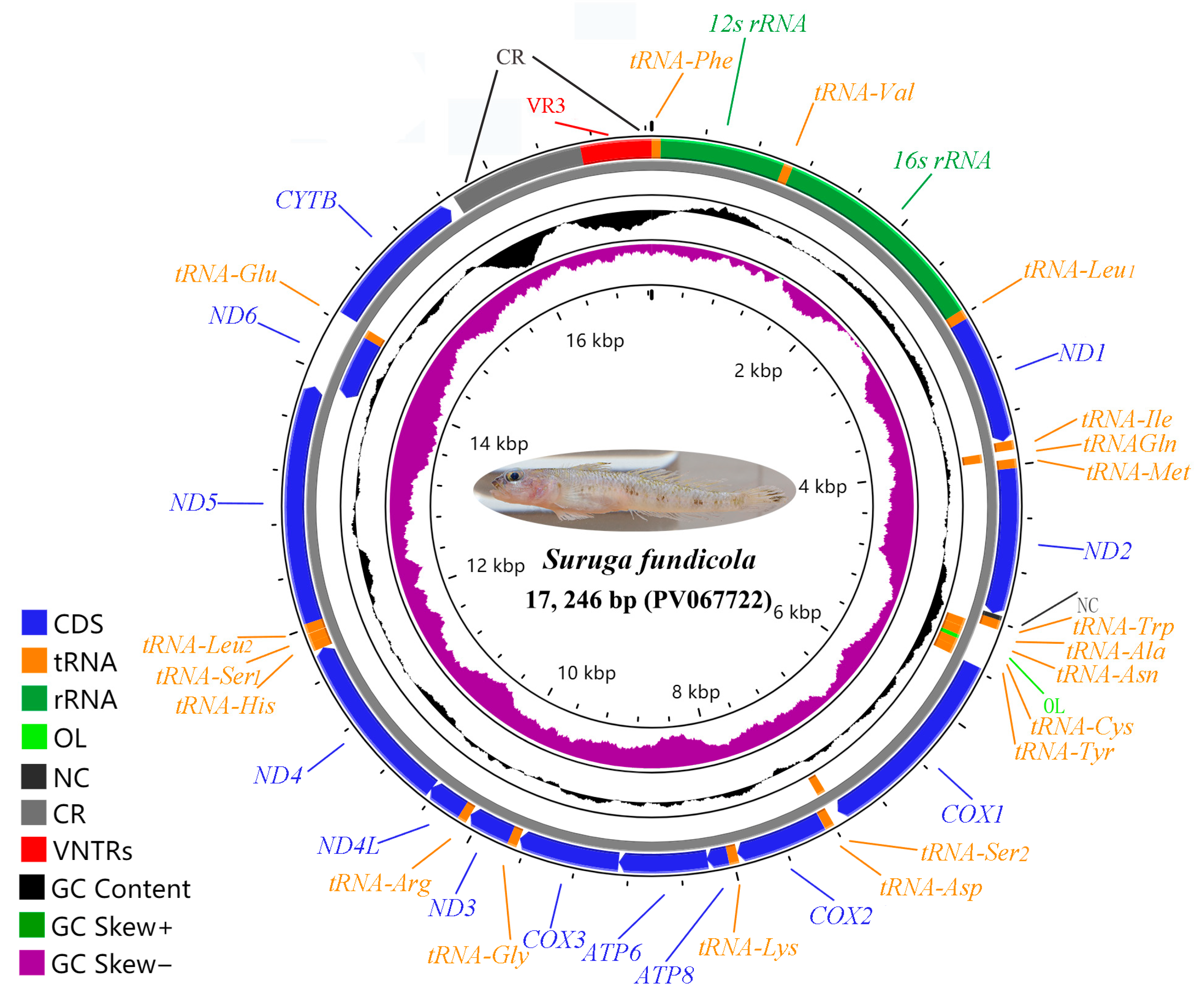
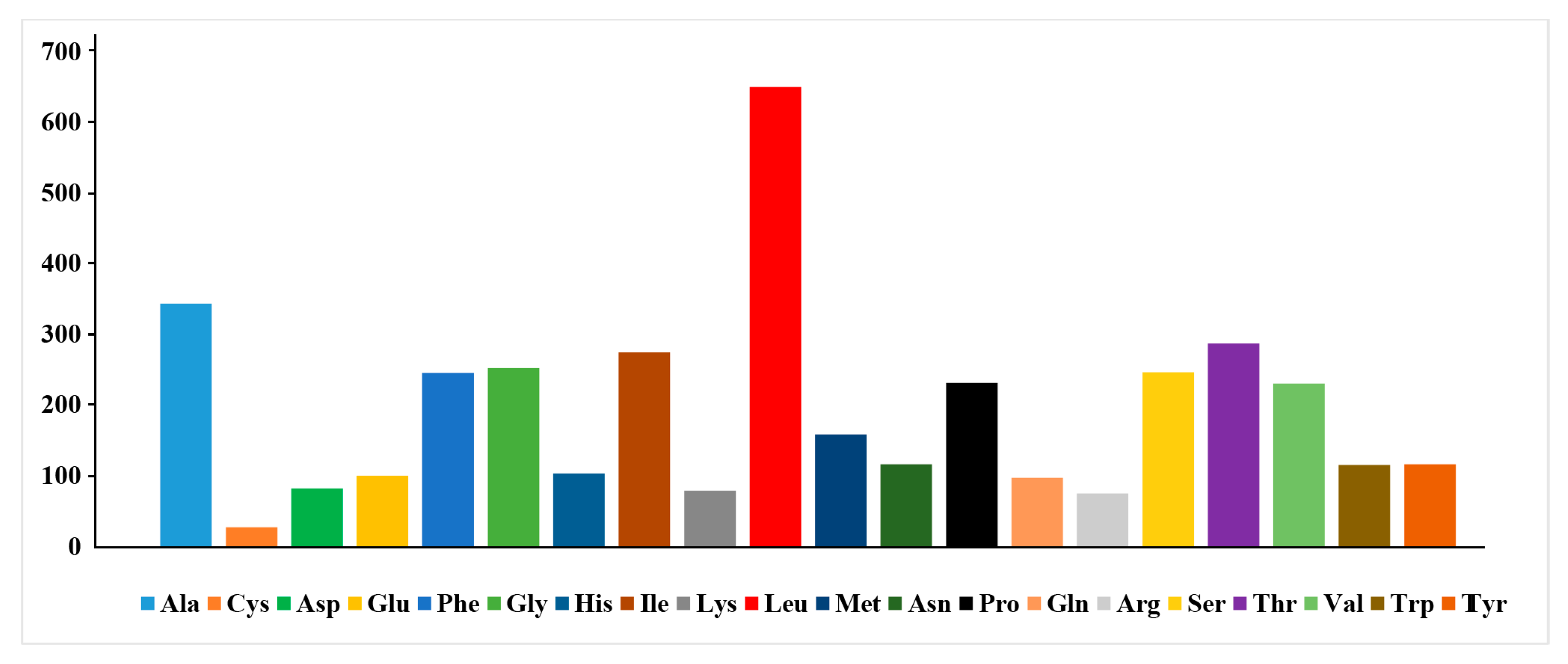
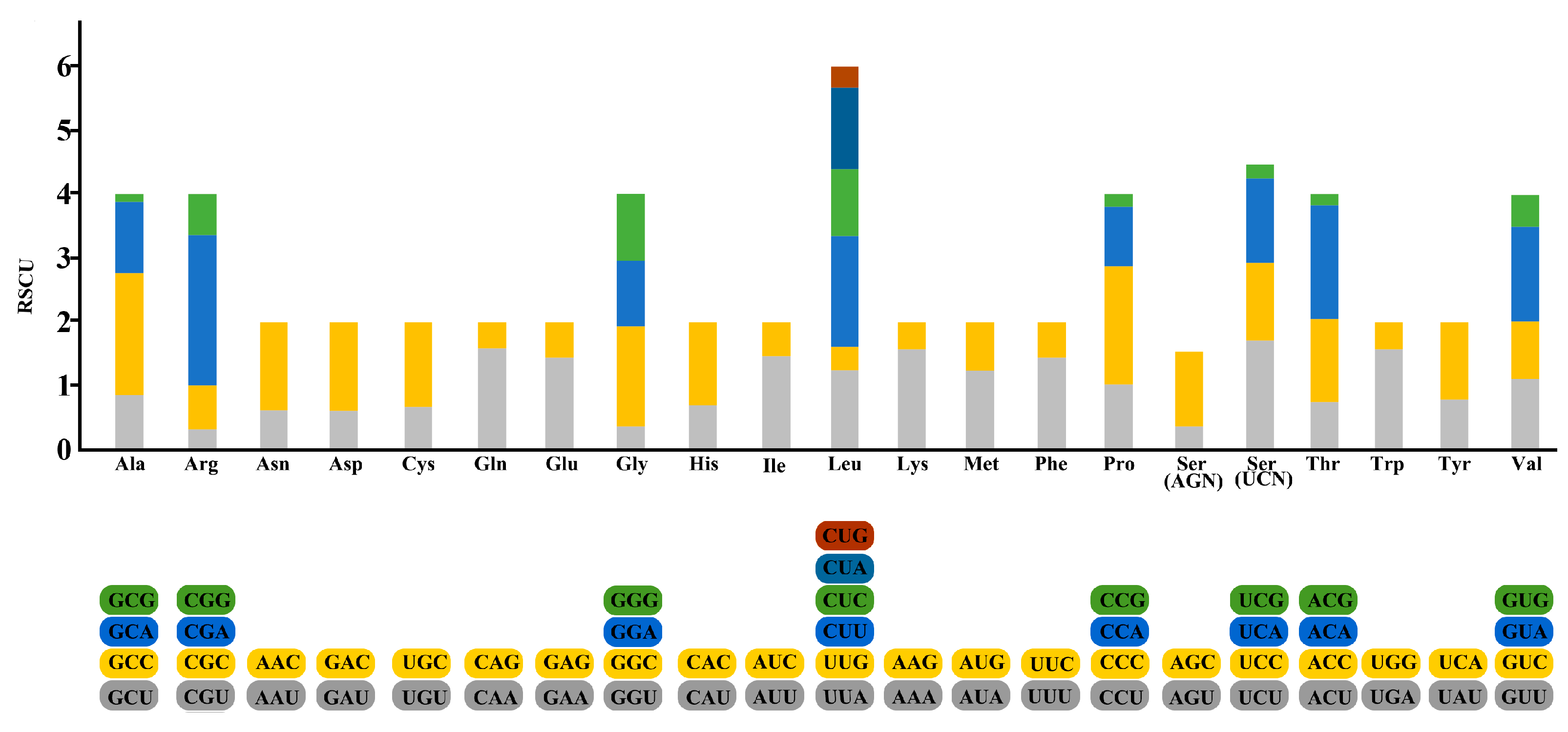


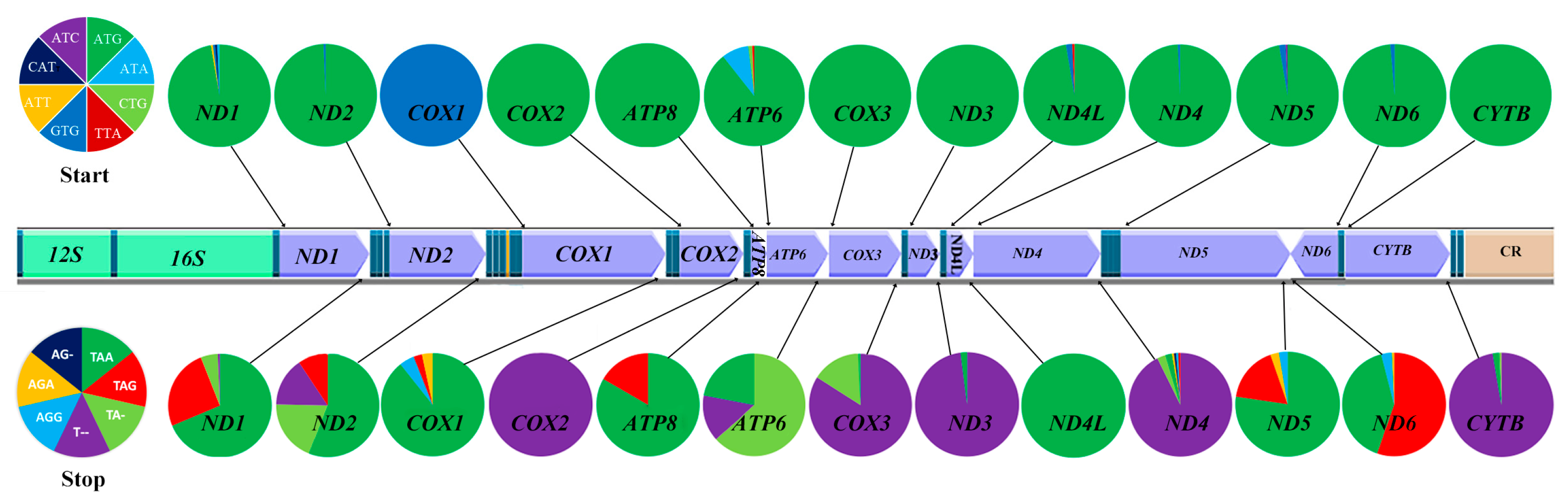
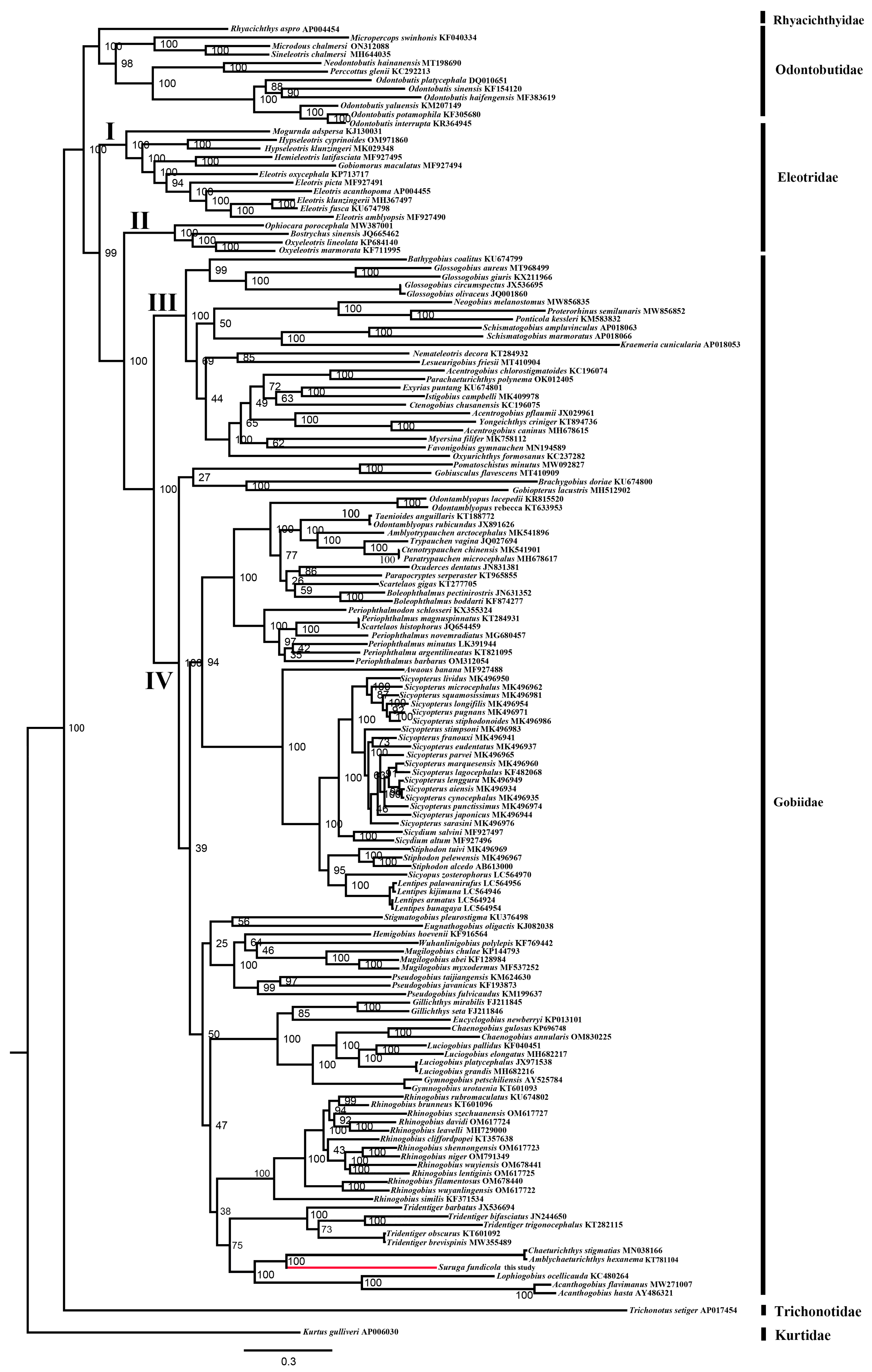
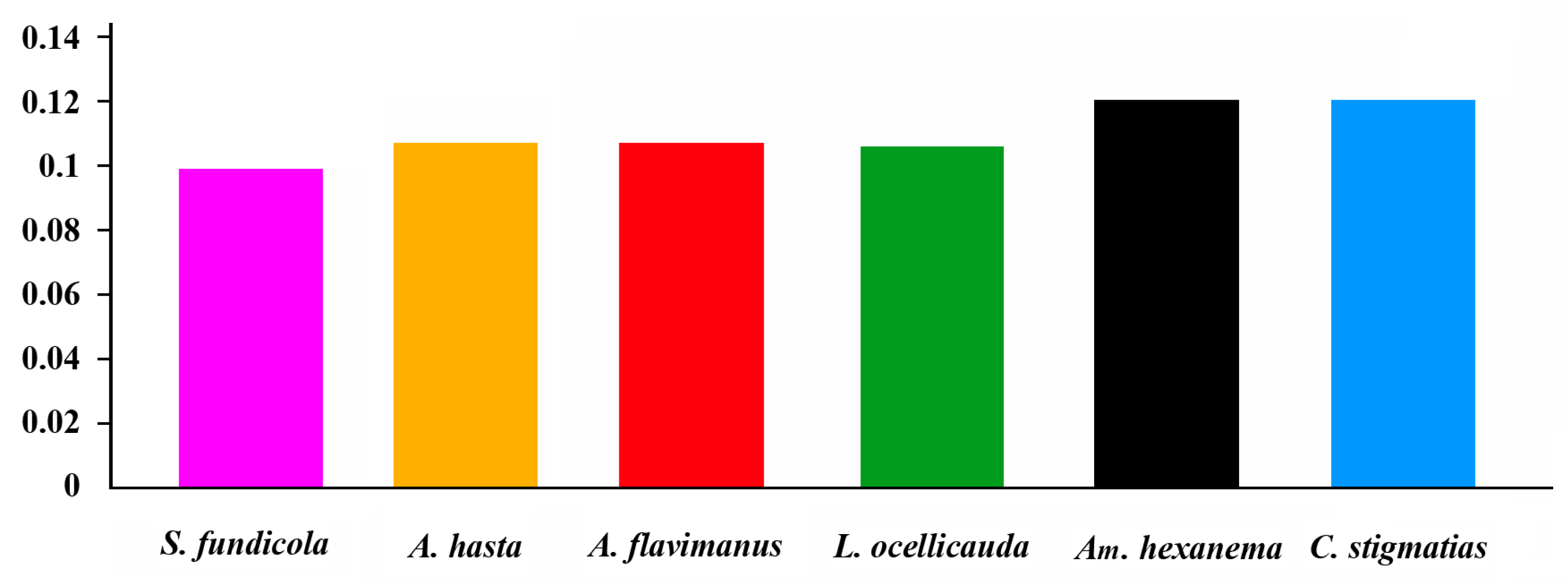
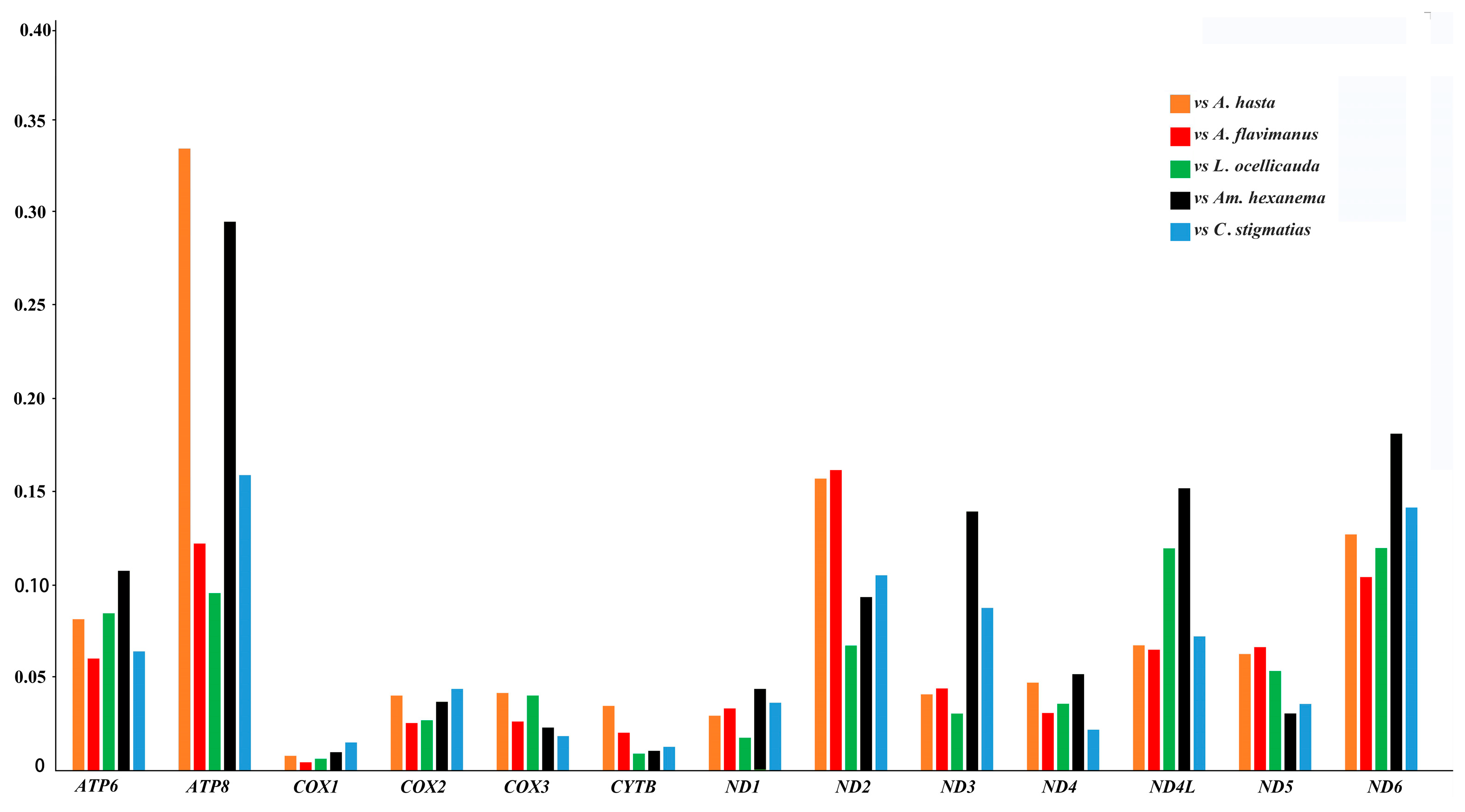
| Gene | Strand | Location | Size (bp) | Anticodon | Start Codon | Stop Codon | Intergenics Length |
|---|---|---|---|---|---|---|---|
| tRNAPhe (S) | + | 1–68 | 68 | GAA | 0 | ||
| 12srRNA | + | 69–1012 | 944 | 0 | |||
| tRNAVal (V) | + | 1013–1083 | 71 | TAC | 0 | ||
| 16srRNA | + | 1084–2761 | 1678 | 0 | |||
| tRNALeu(UUR) (L1) | + | 2762–2836 | 75 | TAA | 0 | ||
| ND1 | + | 2837–3811 | 975 | ATG | TAG | 3 | |
| tRNAIle (I) | + | 3815–3884 | 70 | GAT | −1 | ||
| tRNAGln (Q) | − | 3884–3954 | 71 | TTG | −1 | ||
| tRNAMet (M) | + | 3954–4023 | 70 | CAT | 1 | ||
| ND2 | + | 4025–5146 | 1122 | ATG | TAA | 0 | |
| NC | + | 5147–5181 | 35 | ||||
| tRNATrp (W) | + | 5183–5253 | 71 | TCA | 2 | ||
| tRNAAla (A) | − | 5256–5324 | 69 | TGC | 1 | ||
| tRNAAsn (N) | − | 5326–5398 | 73 | GTT | 33 | ||
| OL | − | 5399–5431 | |||||
| tRNACys (C) | − | 5432–5496 | 65 | GCA | 0 | ||
| tRNATyr (Y) | − | 5496–5566 | 71 | GTA | 1 | ||
| COX1 | + | 5569–7122 | 1554 | GTG | TAA | 0 | |
| tRNASer (S2) | − | 7123–7193 | 71 | TGA | 3 | ||
| tRNAAsp (D) | + | 7197–7268 | 72 | GTC | 3 | ||
| COX2 | + | 7272–7962 | 691 | ATG | T– | 0 | |
| tRNALys (K) | + | 7963–8036 | 74 | TTT | 1 | ||
| ATP8 | + | 8038–8202 | 165 | ATG | TAA | −5 | |
| ATP6 | + | 8196–8878 | 683 | ATG | TA– | 0 | |
| COX3 | + | 8879–9662 | 784 | ATG | T– | 0 | |
| tRNAGly (G) | + | 9663–9734 | 72 | TCC | 0 | ||
| ND3 | + | 9735–10,083 | 349 | ATG | T– | 0 | |
| tRNAArg (R) | + | 10,084–10,152 | 69 | TCG | 0 | ||
| ND4L | + | 10,153–10,449 | 297 | ATG | TAA | −5 | |
| ND4 | + | 10,443–11,823 | 1381 | ATG | T– | 0 | |
| tRNAHis (H) | + | 11,824–11,891 | 68 | GTG | 0 | ||
| tRNASerr(AGY) (S1) | + | 11,892–11,959 | 68 | GCT | 3 | ||
| tRNALeu(CUN) (L2) | + | 11,963–12,035 | 73 | TAG | 0 | ||
| ND5 | + | 12,036–13,874 | 1839 | ATG | TAA | −2 | |
| ND6 | − | 13,871–14,392 | 522 | ATG | TAG | 0 | |
| tRNAGlu (E) | − | 14,393–14,461 | 69 | TTC | 5 | ||
| CYTB | + | 14,467–15,610 | 1144 | ATG | T– | 0 | |
| tRNAThr (T) | + | 15,611–15,682 | 72 | TGT | −1 | ||
| tRNAPro (P) | − | 15,682–15,751 | 70 | TGG | 0 | ||
| Control region | + | 15,752–17,246 | 1495 | – |
| Length | T% | C% | A% | G% | AT% | AT- Skew% | GC- Skew% | |
|---|---|---|---|---|---|---|---|---|
| PCGs | 11,223 | 30.4 | 28.5 | 24.8 | 16.3 | 55.2 | −0.10 | −0.27 |
| tRNA | 1552 | 27.0 | 20.9 | 28.6 | 23.5 | 55.6 | 0.03 | 0.06 |
| rRNA | 2622 | 20.9 | 25.3 | 33.2 | 20.6 | 54.1 | 0.23 | −0.10 |
| CR | 1495 | 28.8 | 19.3 | 37.7 | 14.2 | 66.5 | 0.13 | −0.15 |
| Genome | 17,246 | 27.6 | 27.0 | 28.6 | 16.8 | 56.2 | 0.02 | −0.23 |
| Model | Ln L | Model Compared | LRT p-Value | Positive Sites | |
|---|---|---|---|---|---|
| ATP6 | MA | −2587.829 | MA vs. MA 0 | 0.266 | 7 D 0.960 * |
| MA 0 | −2588.449 | Not Allowed | |||
| ATP8 | MA | −726.169 | MA vs. MA 0 | 1.000 | |
| MA 0 | −726.169 | Not Allowed | |||
| COX1 | MA | −4856.932 | MA vs. MA 0 | 1.000 | |
| MA 0 | −4856.932 | Not Allowed | |||
| COX2 | MA | −2035.809 | MA vs. MA 0 | 0.670 | |
| MA 0 | −2035.718 | Not Allowed | |||
| COX3 | MA | −2355.767 | MA vs. MA 0 | 1.000 | |
| MA 0 | −2355.767 | Not Allowed | |||
| CYTB | MA | −3942.845 | MA vs. MA 0 | 1.000 | |
| MA 0 | −3942.845 | Not Allowed | |||
| ND1 | MA | −3522.005 | MA vs. MA 0 | 1.000 | |
| MA 0 | −3522.005 | Not Allowed | |||
| ND2 | MA | −4337.265 | MA vs. MA 0 | 0.385 | 2 N 0.987 *, 18 G 0.980 *, 21 A 0.980 *, 62 T 0.976 *, 240 T 0.967 *, 329 T 0.953 *, 345 L 0.982 * |
| MA 0 | −4337.643 | Not Allowed | |||
| ND3 | MA | −1202.254 | MA vs. MA 0 | 0.346 | 88 T 0.952 * |
| MA 0 | −1202.698 | Not Allowed | |||
| ND4 | MA | −5157.592 | MA vs. MA 0 | 1.000 | 351 S 0.957 * |
| MA 0 | −5157.592 | Not Allowed | |||
| ND4L | MA | −1051.586 | MA vs. MA 0 | 0.804 | |
| MA 0 | −1051.617 | Not Allowed | |||
| ND5 | MA | −6756.556 | MA vs. MA 0 | 0.472 | |
| MA 0 | −6756.815 | Not Allowed | |||
| ND6 | MA | −2199.261 | MA vs. MA 0 | 1.000 | |
| MA 0 | −2199.261 | Not Allowed |
| Region | Initial Denaturation | Denaturation | Annealing | Extension | Cycles | Volume |
|---|---|---|---|---|---|---|
| ND2 | 94 °C, 4 min | 94 °C, 50 s | 57 °C, 30 S | 50 S | 35 | 25 μL |
| VR3 | 50 °C, 30 S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, C.; Li, A.; Wang, H.; Che, S.; van der Laan, R.; Liu, S.; Zhuang, Z. The Complete Mitochondrial Genome of the Deep-Dwelling Goby Suruga fundicola (Teleostei, Gobiidae) Reveals Evidence of Recombination in the Control Region. Int. J. Mol. Sci. 2025, 26, 8317. https://doi.org/10.3390/ijms26178317
An C, Li A, Wang H, Che S, van der Laan R, Liu S, Zhuang Z. The Complete Mitochondrial Genome of the Deep-Dwelling Goby Suruga fundicola (Teleostei, Gobiidae) Reveals Evidence of Recombination in the Control Region. International Journal of Molecular Sciences. 2025; 26(17):8317. https://doi.org/10.3390/ijms26178317
Chicago/Turabian StyleAn, Changting, Ang Li, Huan Wang, Shuai Che, Richard van der Laan, Shufang Liu, and Zhimeng Zhuang. 2025. "The Complete Mitochondrial Genome of the Deep-Dwelling Goby Suruga fundicola (Teleostei, Gobiidae) Reveals Evidence of Recombination in the Control Region" International Journal of Molecular Sciences 26, no. 17: 8317. https://doi.org/10.3390/ijms26178317
APA StyleAn, C., Li, A., Wang, H., Che, S., van der Laan, R., Liu, S., & Zhuang, Z. (2025). The Complete Mitochondrial Genome of the Deep-Dwelling Goby Suruga fundicola (Teleostei, Gobiidae) Reveals Evidence of Recombination in the Control Region. International Journal of Molecular Sciences, 26(17), 8317. https://doi.org/10.3390/ijms26178317






