Stabilizing the Baseline: Reference Gene Evaluation in Three Invasive Reynoutria Species
Abstract
1. Introduction
2. Results
2.1. Primer Specificity and Amplification Efficiency
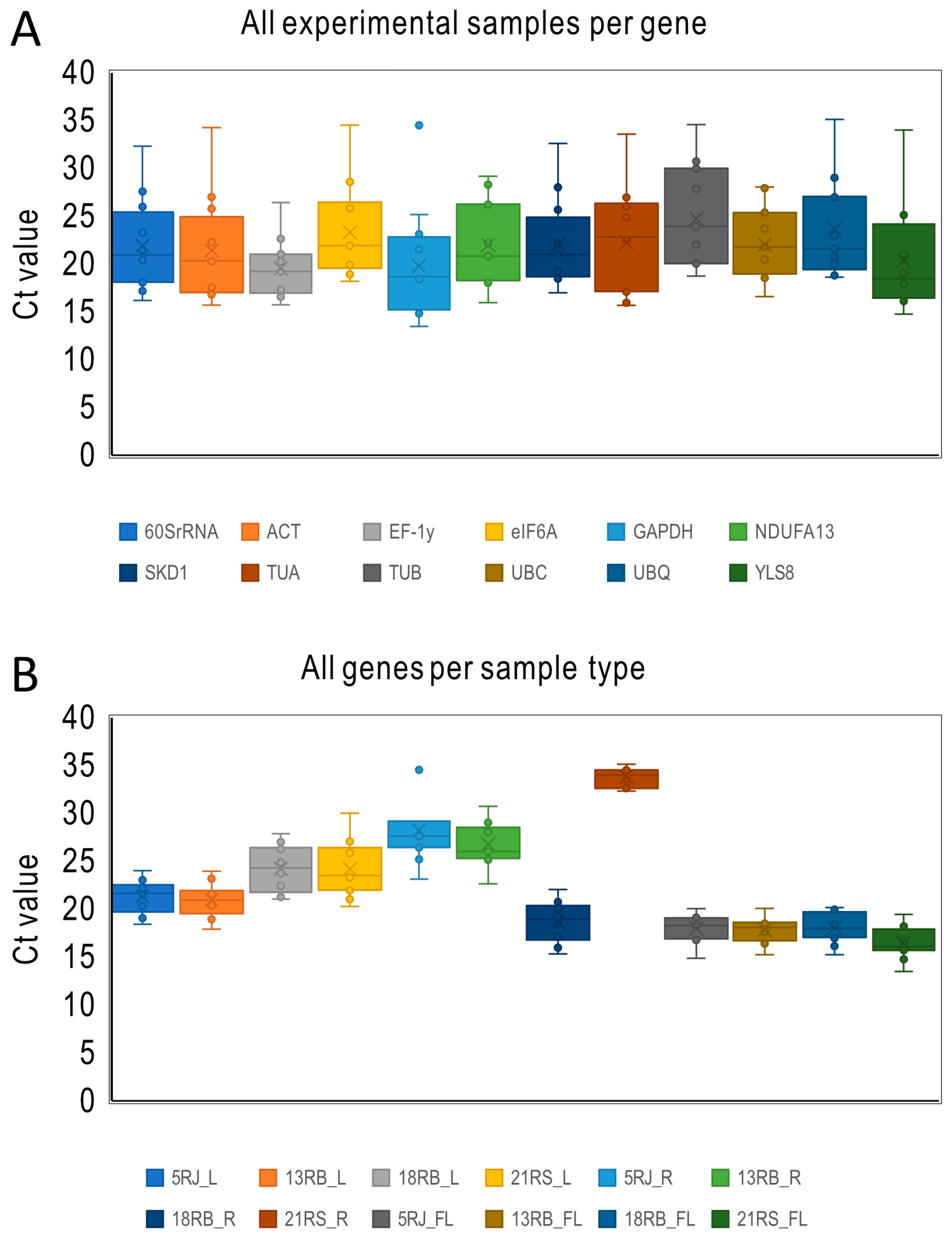
2.2. Expression Profiles of Candidate Reference Genes
2.3. Expression Stability Assessment
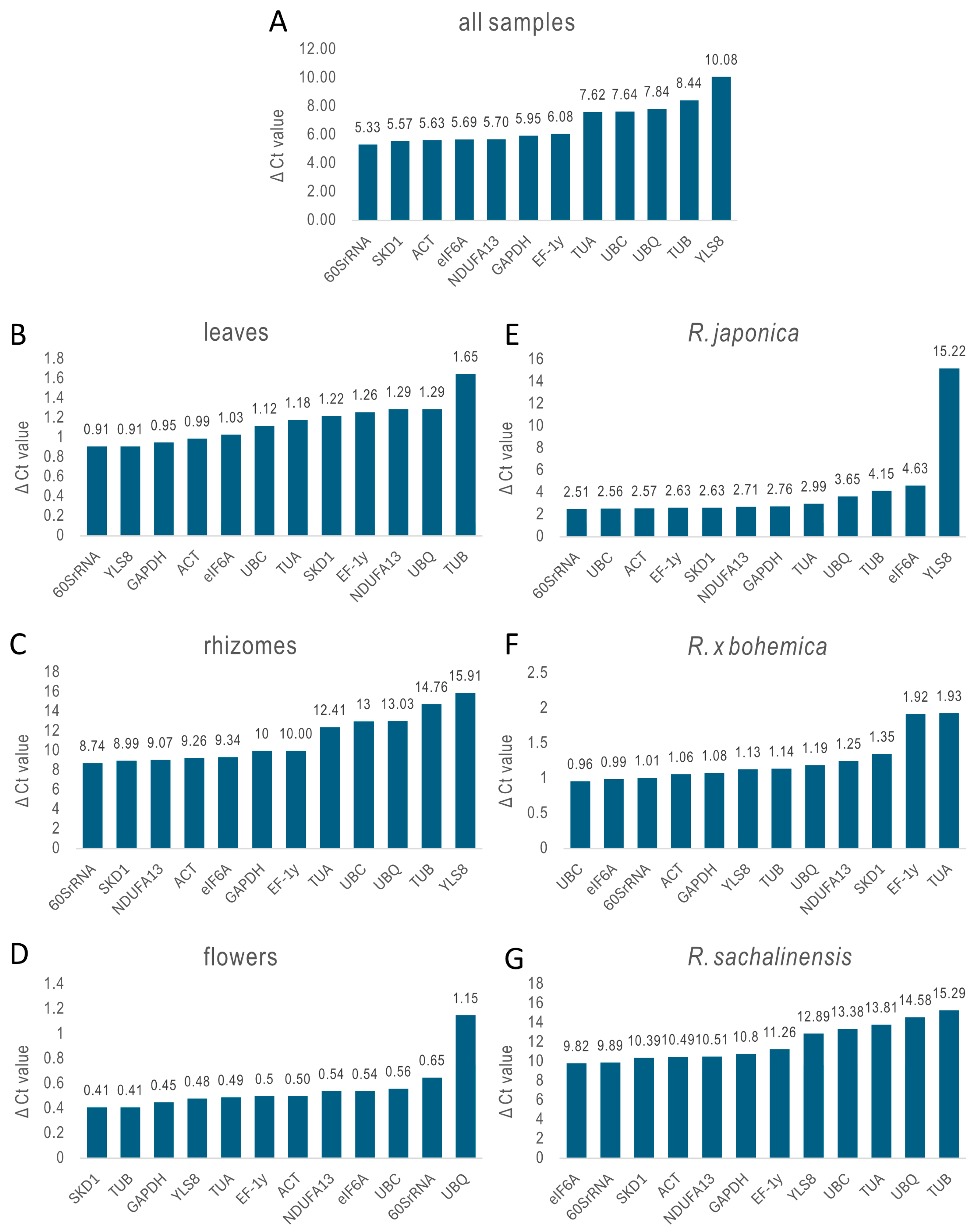
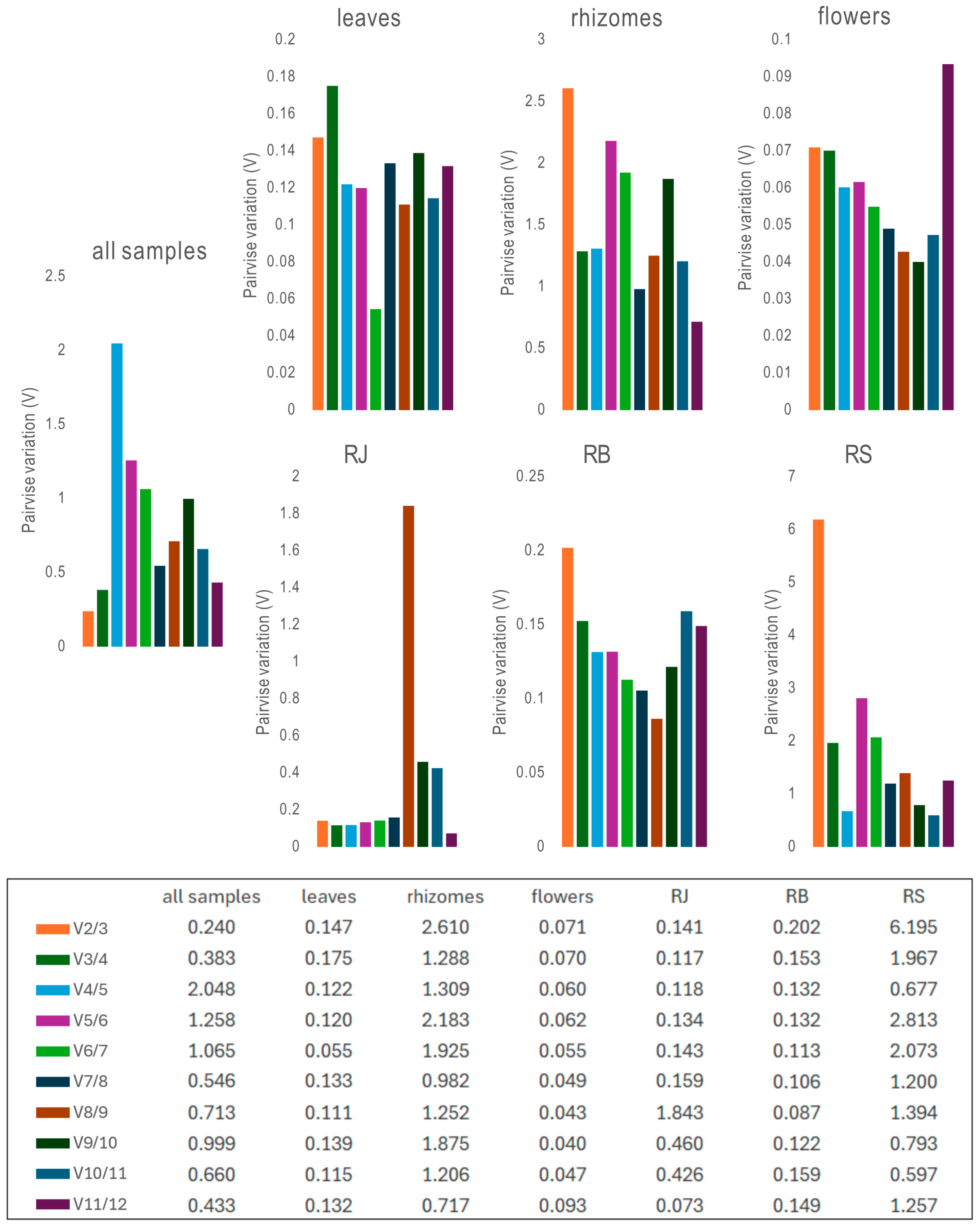
2.3.1. ΔCt Method
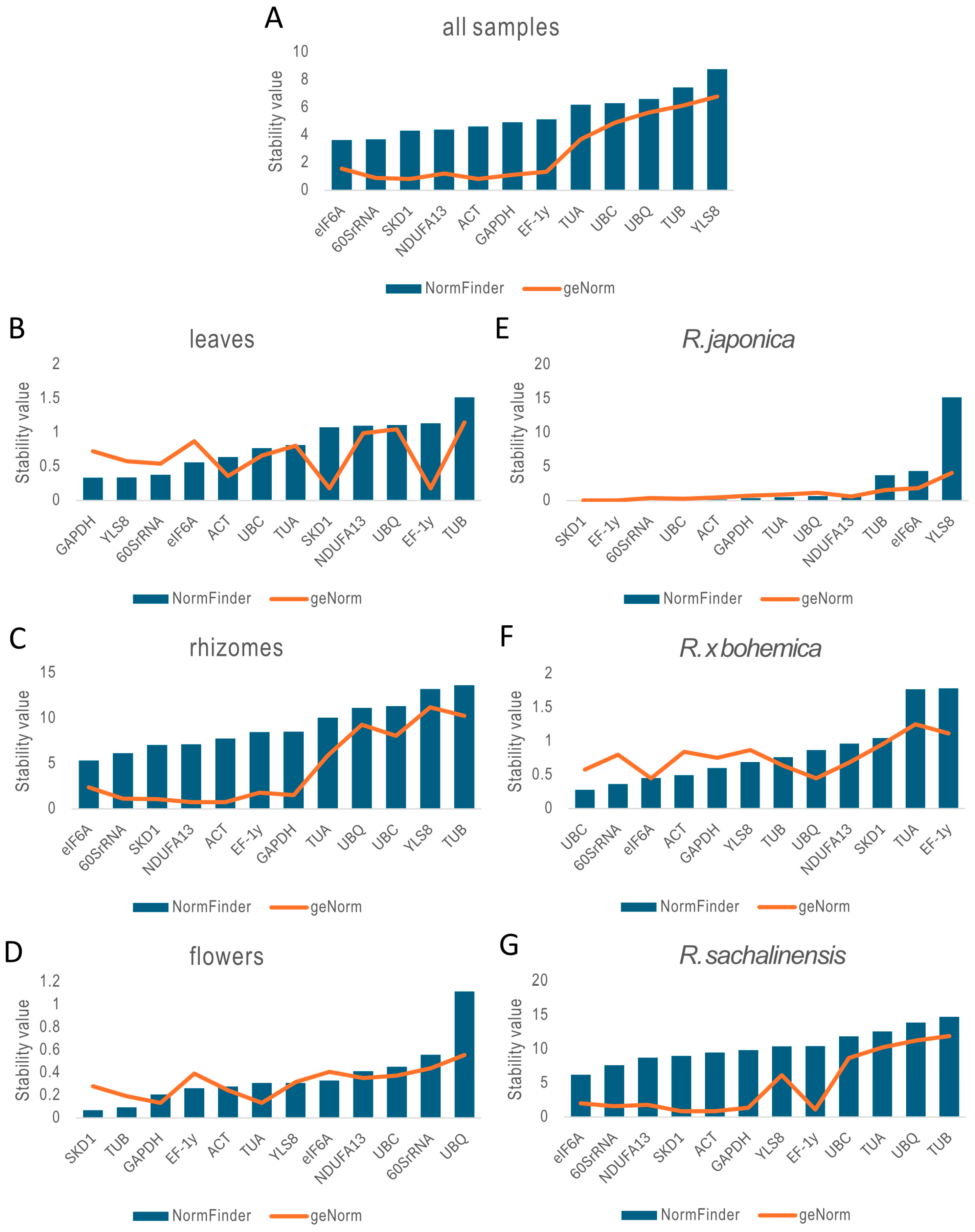
2.3.2. geNorm Analysis
2.3.3. NormFinder Analysis
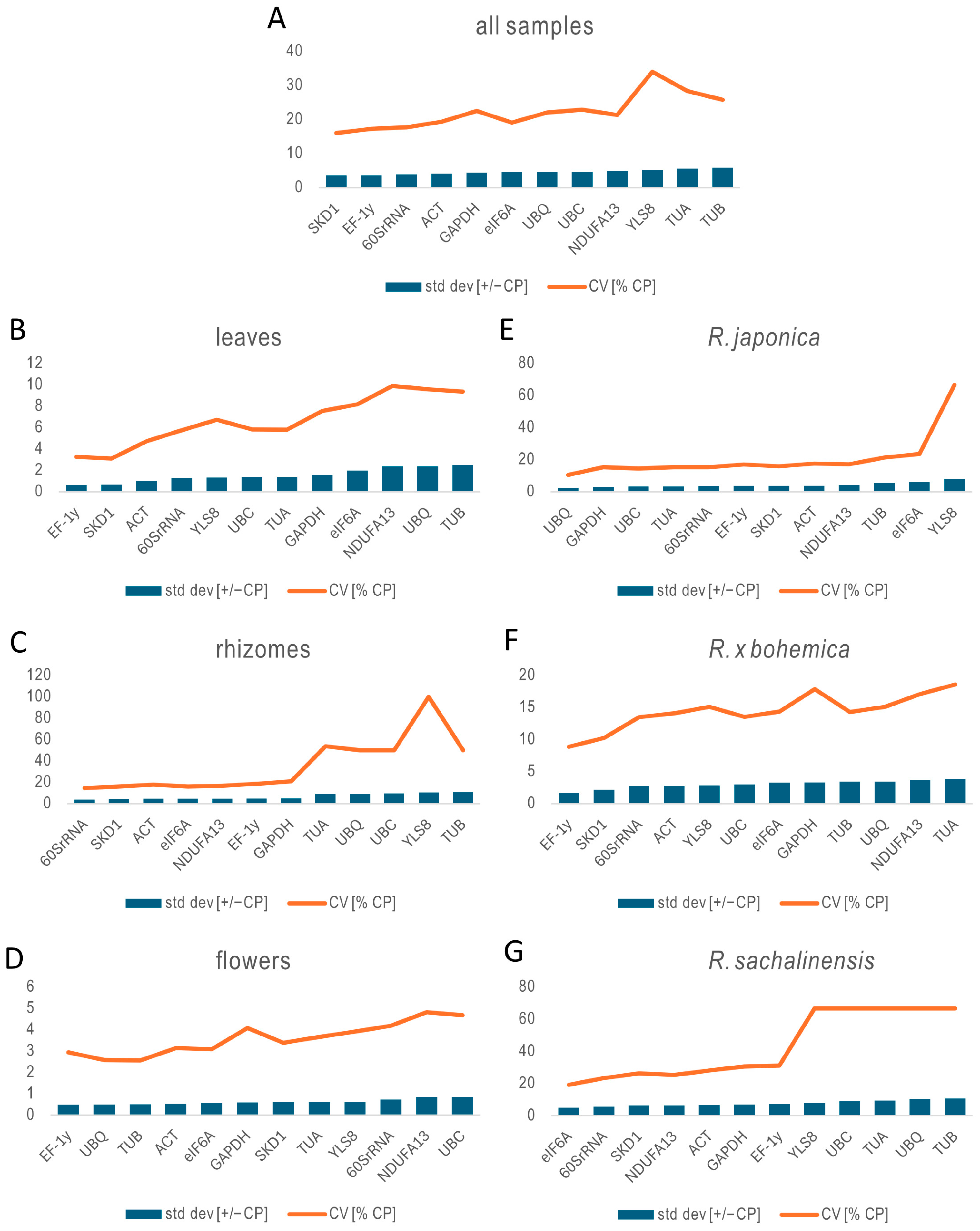
2.3.4. BestKeeper Analysis
2.3.5. Comprehensive Stability Ranking
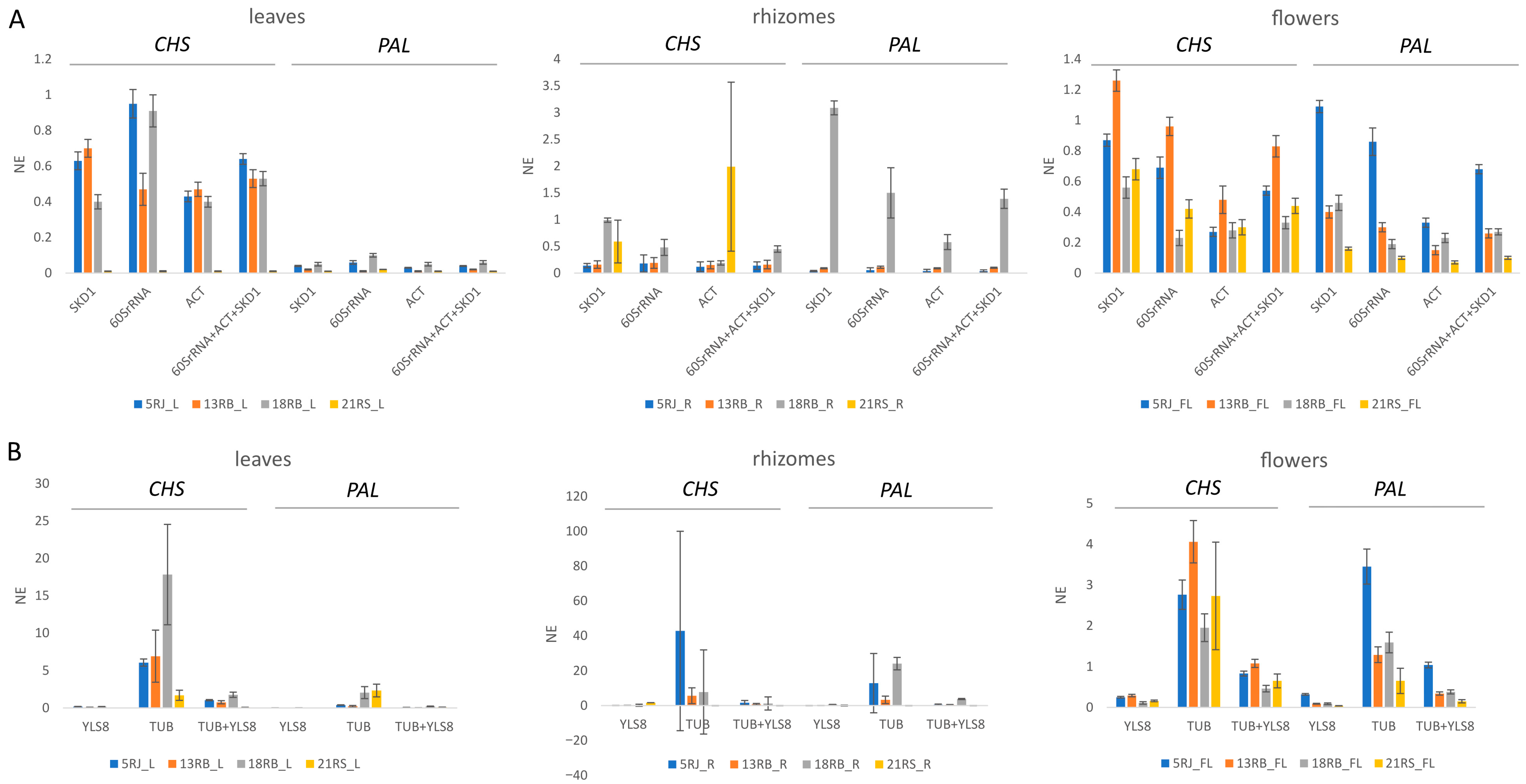
2.4. Evaluation of Candidate Reference Genes for Expression Analysis in Reynoutria
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. RNA Extraction and cDNA Synthesis
4.3. Selection of Candidate Reference Genes and Primer Design
4.4. Quantitative Real-Time PCR
4.5. Statistical Data Analysis
4.6. Validation of Selected Reference Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 60SrRNA | 60S ribosomal RNA |
| ACT | actin |
| EF-1γ | elongation factor 1 gamma |
| eIF6A | eukaryotic translation initiation factor 6A |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| NDUFA13 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-A |
| SKD1 | suppressor of K+ transport defect1 |
| TUA | α-tubulin |
| TUB | β-tubulin |
| UBC | ubiquitin-conjugating enzyme |
| UBQ | ubiquitin domain–containing protein |
| YLS8 | thioredoxin-like protein |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| PP2A | protein phosphatase 2A |
| UBQ14 | polyubiquitin 14 |
| UBQ4-1 | polyubiquitin 4 |
| SAMS | S-adenosylmethionine synthase |
| Ct | cycle threshold |
| PAL | phenylalanine ammonia-lyase |
| CHS | chalcone synthase |
| RJ | Reynoutria japonica |
| RB | Reynoutria × bohemica |
| RS | Reynoutria sachalinensis |
| ANOVA | analysis of variance |
| SD | standard deviation |
| CV | coefficient of variation |
| L | leaves |
| R | rhizomes |
| FL | flowers |
References
- Bustin, S.A. Quantification of MRNA Using Real-Time Reverse Transcription PCR (RT-PCR): Trends and Problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Gachon, C.; Mingam, A.; Charrier, B. Real-Time PCR: What Relevance to Plant Studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef]
- Shi, L.; Cai, Y.; Yao, J.; Zhang, Q.; He, B.; Lin, S. Reference Genes Selection and Validation for Cinnamomum burmanni by Real-Time Quantitative Polymerase Chain Reaction. Int. J. Mol. Sci. 2024, 25, 3500. [Google Scholar] [CrossRef]
- Xing, L.; Zhang, Y.; Xing, Y.; Ge, M.; Miao, H.; Huo, X. Reference Genes Selection and Validation in Yam by Real-Time Quantitative Polymerase Chain Reaction. Sci. Rep. 2025, 15, 19947. [Google Scholar] [CrossRef]
- Wong, M.L.; Medrano, J.F. Real-time PCR. Encycl. Ref. Genom. Proteom. Mol. Med. 2005, 39, 1603. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to Do Successful Gene Expression Analysis Using Real-Time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef]
- Hao, X.; Horvath, D.P.; Chao, W.S.; Yang, Y.; Wang, X.; Xiao, B. Identification and Evaluation of Reliable Reference Genes for Quantitative Real-Time PCR Analysis in Tea Plant (Camellia sinensis (L.) O. Kuntze). Int. J. Mol. Sci. 2014, 15, 22155–22172. [Google Scholar] [CrossRef]
- Schmidt, G.W.; Delaney, S.K. Stable Internal Reference Genes for Normalization of Real-Time RT-PCR in Tobacco (Nicotiana tabacum) during Development and Abiotic Stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef]
- Zhao, M.; Fan, H.; Tu, Z.; Cai, G.; Zhang, L.; Li, A.; Xu, M. Stable Reference Gene Selection for Quantitative Real-Time PCR Normalization in Passion Fruit (Passiflora edulis Sims.). Mol. Biol. Rep. 2022, 49, 5985–5995. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of Housekeeping Genes as Internal Control for Studying Gene Expression in Rice by Quantitative Real-Time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Souček, P.; Pavlů, J.; Medveďová, Z.; Reinöhl, V.; Brzobohatý, B. Stability of Housekeeping Gene Expression in Arabidopsis thaliana Seedlings under Differing Macronutrient and Hormonal Conditions. J. Plant Biochem. Biotechnol. 2017, 26, 415–424. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference Genes in Real-Time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Reddy, D.S.; Bhatnagar-Mathur, P.; Cindhuri, K.S.; Sharma, K.K. Evaluation and Validation of Reference Genes for Normalization of Quantitative Real-Time PCR Based Gene Expression Studies in Peanut. PLoS ONE 2013, 8, e78555. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Li, Y.; Sun, H. Selection of Reference Genes for RT-QPCR Normalization in Blueberry (Vaccinium corymbosum × angustifolium) under Various Abiotic Stresses. FEBS Open Bio 2020, 10, 1418–1435. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.J.; Ke, W.; Drangowska-Way, A.; O’Rourke, E.J.; Lewis, N.E. What Are Housekeeping Genes? PLoS Comput. Biol. 2022, 18, e1010295. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Liu, Q.; Tong, H.; Zhang, T.; Gu, C.; Liu, L.; Huang, S.; Yuan, H. Selection and Validation of Appropriate Reference Genes for RT-QPCR Analysis of Flowering Stages and Different Genotypes of Iris germanica L. Sci. Rep. 2021, 11, 9901. [Google Scholar] [CrossRef]
- Mane, V.P.; Heuer, M.A.; Hillyer, P.; Navarro, M.B.; Rabin, R.L. Systematic Method for Determining an Ideal Housekeeping Gene for Real-Time PCR Analysis. J. Biomol. Tech. 2008, 19, 342. [Google Scholar]
- Alberternst, B.; Böhmer, H.J. NOBANIS—Invasive Alien Species Fact Sheet Fallopia japonica. From: Online Database of the European Network on Invasive Alien Species—NOBANIS. Available online: https://www.nonnativespecies.org/assets/Invasive_Alien_Species_Fact_Sheet_-_Fallopia_japonica_-_NOBANIS_-_downloaded_from_website_March_2014.pdf (accessed on 16 April 2013).
- Tokarska-Guzik, B. The Establishment and Spread of Alien Plant Species (Kenophytes) in the Flora of Poland; Wydawnictwo Uniwersytetu Śląskiego: Katowice, Poland, 2005; ISBN 8322614853. [Google Scholar]
- Desjardins, S.D.; Pashley, C.H.; Bailey, J.P. A Taxonomic, Cytological and Genetic Survey of Japanese Knotweed s.l. in New Zealand Indicates Multiple Secondary Introductions from Europe and a Direct Introduction from Japan. N. Z. J. Bot. 2023, 61, 49–66. [Google Scholar] [CrossRef]
- Jugieau, E.; Talmot, V.; Staentzel, C.; Noir, S.; Hardion, L. A Knot of Hybrids: Differentiating Asian Knotweeds in North-Eastern France Using Genetic, Cytological, and Morphological Data. J. Syst. Evol. 2024, 62, 1218–1226. [Google Scholar] [CrossRef]
- Ke, J.; Li, M.T.; Xu, S.; Ma, J.; Liu, M.Y.; Han, Y. Advances for Pharmacological Activities of Polygonum cuspidatum—A Review. Pharm. Biol. 2023, 61, 177–188. [Google Scholar] [CrossRef]
- Fan, P.; Terrier, L.; Hay, A.E.; Marston, A.; Hostettmann, K. Antioxidant and Enzyme Inhibition Activities and Chemical Profiles of Polygonum sachalinensis F. Schmidt ex Maxim (Polygonaceae). Fitoterapia 2010, 81, 124–131. [Google Scholar] [CrossRef]
- Strašil, Z.; Kára, J. Study of Knotweed (Reynoutria) as Possible Phytomass Resource for Energy and Industrial Utilization. Res. Agric. Eng. 2010, 56, 85–91. [Google Scholar] [CrossRef]
- Nawrot-Hadzik, I.; Slusarczyk, S.; Granica, S.; Hadzik, J.; Matkowski, A. Phytochemical Diversity in Rhizomes of Three Reynoutria Species and Their Antioxidant Activity Correlations Elucidated by LC-ESI-MS/MS Analysis. Molecules 2019, 24, 1136. [Google Scholar] [CrossRef]
- Stafiniak, M.; Bielecka, M.; Kujawa, K.; Jezierska-Domaradzka, A.; Pencakowski, B.; Basiak, A.; Matkowski, A.; Nawrot-Hadzik, I. Integrative Morphological, Phytochemical, and Molecular Identification of Three Invasive and Medicinal Reynoutria Species. Sci. Rep. 2025, 15, 6001. [Google Scholar] [CrossRef]
- Candassamy, S.V.; Wu, J.J.; Zhou, X.; Wang, R.H.; Qi, Z.C.; Yan, X.L. The Complete Chloroplast Genome of Medicinal Herb Reynoutria japonica Houtt. (Polygonaceae). Mitochondrial DNA B Resour. 2020, 5, 1983–1985. [Google Scholar] [CrossRef]
- Chen, J.; Huang, T.; Fan, H.; Lin, F.; Ma, H.; Cao, J.; Chai, T.; Ma, L.; Wang, H. A Comparative Phylogenetic Analysis on the Chloroplast Genome in Different Reynoutria japonica Populations. Genes 2022, 13, 1979. [Google Scholar] [CrossRef]
- Chen, J.; Ma, H.; Fan, H.; Lin, F.; Chai, T.; Wang, H. De Novo Assembly and Comparative Analysis of the Mitochondrial Genome of Reynoutria japonica. Front. Genet. 2023, 14, 1289811. [Google Scholar] [CrossRef]
- Tippery, N.P.; Olson, A.L.; Wendtlandt, J.L. Using the Nuclear LEAFY Gene to Reconstruct Phylogenetic Relationships among Invasive Knotweed (Reynoutria, Polygonaceae) Populations. Invasive Plant Sci. Manag. 2021, 14, 92–100. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Z.; Bao, W.; Hu, H.; Chen, M.; Chai, T.; Wang, H. Identification and Evaluation of Reference Genes for Quantitative Real-Time PCR Analysis in Polygonum cuspidatum Based on Transcriptome Data. BMC Plant Biol. 2019, 19, 498. [Google Scholar] [CrossRef]
- Løvdal, T.; Lillo, C. Reference Gene Selection for Quantitative Real-Time PCR Normalization in Tomato Subjected to Nitrogen, Cold, and Light Stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef]
- Wang, H.; Cai, Q.-Z.; Liu, L.; Yang, Q.; Zhou, L.-Y. Reference Gene Screening for Real-Time Quantitative PCR in Polygonum multiflorum. Zhongguo Zhong Yao Za Zhi 2021, 46, 80–85. [Google Scholar] [CrossRef]
- Liu, H.; Wu, W.; Hou, K.; Chen, J.; Zhao, Z. Deep Sequencing Reveals Transcriptome Re-Programming of Polygonum multiflorum Thunb. Roots to the Elicitation with Methyl Jasmonate. Mol. Genet. Genom. 2016, 291, 337–348. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T. Determination of Most Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A MiRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A Web-Based Tool for Comprehensively Analyzing and Identifying Reference Genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.M.; Yakovlev, I.A.; Strauss, S.H. Validating Internal Controls for Quantitative Plant Gene Expression Studies. BMC Plant Biol. 2004, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-Time RT-PCR Normalisation; Strategies and Considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Panina, Y.; Germond, A.; Masui, S.; Watanabe, T.M. Validation of Common Housekeeping Genes as Reference for QPCR Gene Expression Analysis During IPS Reprogramming Process. Sci. Rep. 2018, 8, 8716. [Google Scholar] [CrossRef]
- Dundas, J.; Ling, M. Reference Genes for Measuring MRNA Expression. Theory Biosci. 2012, 131, 215–223. [Google Scholar] [CrossRef]
- Arukwe, A. Toxicological Housekeeping Genes: Do They Really Keep the House? Environ. Sci. Technol. 2006, 40, 7944–7949. [Google Scholar] [CrossRef]
- Bonefeld, B.E.; Elfving, B.; Wegener, G. Reference Genes for Normalization: A Study of Rat Brain Tissue. Synapse 2008, 62, 302–309. [Google Scholar] [CrossRef]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of Internal Control for Gene Expression Study in Soybean by Quantitative Real-Time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, N.; Si, H.; Calderón-Urrea, A. Selection and Validation of Reference Genes for RT-QPCR Analysis in Potato under Abiotic Stress. Plant Methods 2017, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference Gene Validation for RT-QPCR, a Note on Different Available Software Packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef]
- Yin, X.; He, T.; Yi, K.; Zhao, Y.; Hu, Y.; Liu, J.; Zhang, X.; Meng, L.; Wang, L.; Liu, H.; et al. Comprehensive Evaluation of Candidate Reference Genes for Quantitative Real-Time PCR-Based Analysis in Caucasian Clover. Sci. Rep. 2021, 11, 3269. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, C.; Yang, S.; Zhong, Q.; Tian, J. Screening and Verification of Reference Genes for Analysis of Gene Expression in Garlic (Allium sativum L.) under Cold and Drought Stress. Plants 2023, 12, 763. [Google Scholar] [CrossRef]
- Yuan, Y.; Liu, C.; Bao, J.; Li, F. Selection and Validation of Appropriate Reference Genes for QRT-PCR Analysis of Iris Germanica L. Under Various Abiotic Stresses. Food Sci. Nutr. 2024, 13, e4765. [Google Scholar] [CrossRef]
- Zhou, L.; Shi, Q.; Wang, Y.; Li, K.; Zheng, B.; Miao, K. Evaluation of Candidate Reference Genes for Quantitative Gene Expression Studies in Tree Peony. J. Am. Soc. Hortic. Sci. 2016, 141, 99–111. [Google Scholar] [CrossRef]
- Wang, X.; Hu, H.; Wu, Z.; Fan, H.; Wang, G.; Chai, T.; Wang, H. Tissue-Specific Transcriptome Analyses Reveal Candidate Genes for Stilbene, Flavonoid and Anthraquinone Biosynthesis in the Medicinal Plant Polygonum cuspidatum. BMC Genom. 2021, 22, 353. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Noge, K.; Asano, Y. Cytochrome P450 CYP71AT96 Catalyses the Final Step of Herbivore-Induced Phenylacetonitrile Biosynthesis in the Giant Knotweed, Fallopia sachalinensis. Plant Mol. Biol. 2016, 91, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Abueg, L.A.L.; Afgan, E.; Allart, O.; Awan, A.H.; Bacon, W.A.; Baker, D.; Bassetti, M.; Batut, B.; Bernt, M.; Blankenberg, D.; et al. The Galaxy Platform for Accessible, Reproducible, and Collaborative Data Analyses: 2024 Update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification Efficiency: Linking Baseline and Bias in the Analysis of Quantitative PCR Data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Li, J.; Han, J.; Hu, Y.; Yang, J. Selection of Reference Genes for Quantitative Real-Time Pcr during Flower Development in Tree Peony (Paeonia suffruticosa Andr.). Front. Plant Sci. 2016, 7, 516. [Google Scholar] [CrossRef]
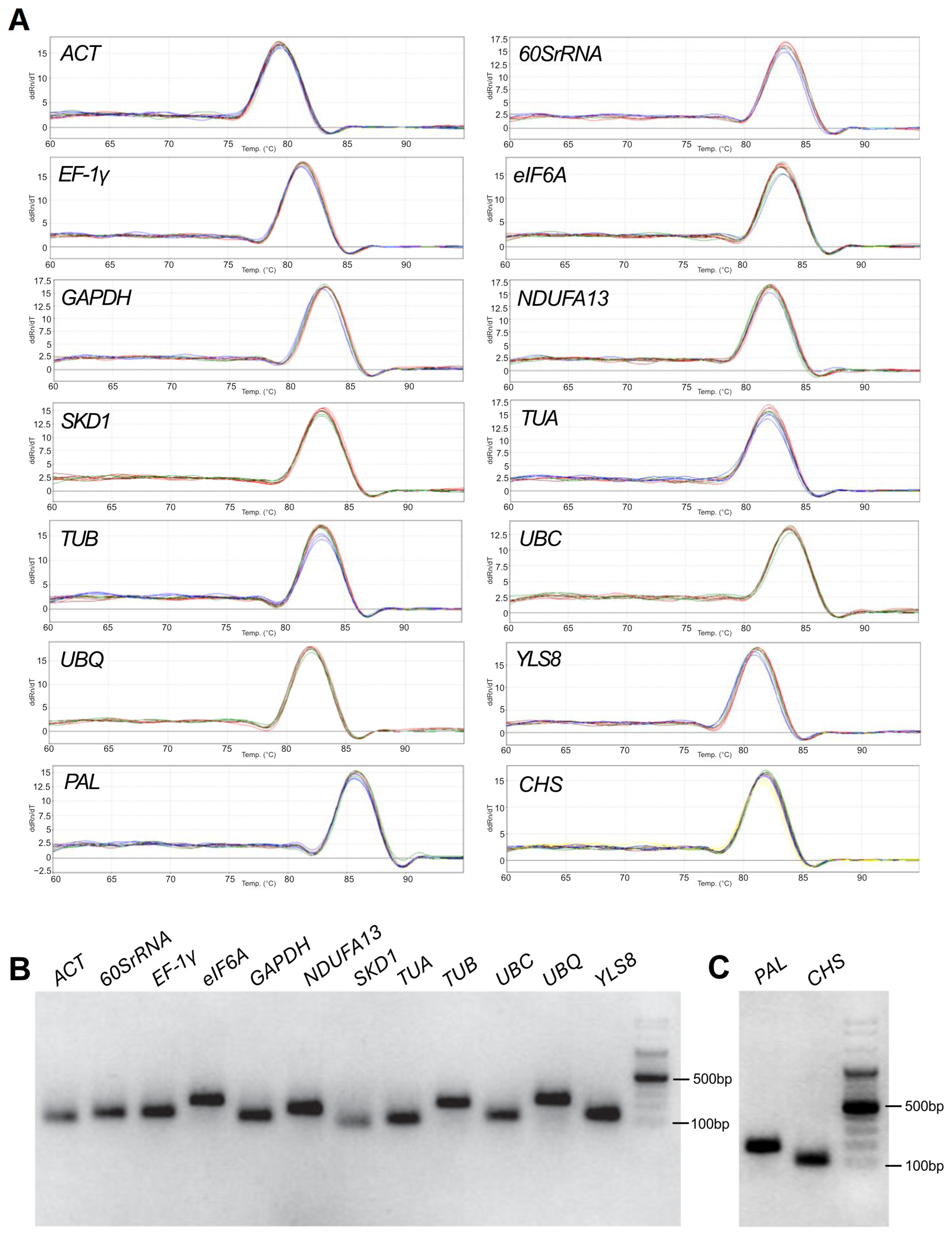
| Primer Name | Primer Sequence 5′ —> 3′ | Target Based on Alignment Analysis | Product Length [bp] | Efficiency [%] | R2 | Reference | Accession Number |
|---|---|---|---|---|---|---|---|
| Actin 7 (ACT) | |||||||
| actF1 | GCCGTCTATGATTGGAATGG | Reynoutria spp. | 99 | 104 | 0.998 | [31] | MK288156 |
| actR1 | TACCGTACAAGTCCTTCCTAA | Reynoutria spp. | [31] | ||||
| Tubulin-alpha 6 (TUA) | |||||||
| tuaF2 | AGGGCCGTTTGCATGATTTC | Reynoutria spp. | 101 | 101 | 0.996 | This study | MK288157 |
| tuaR2 | TGAACAAAGGCACGCTTAGC | Reynoutria spp. | This study | ||||
| Tubulin-beta 2 (TUB) | |||||||
| tubF1 | ATCCGACACTGTTGTTGAGC | R. japonica/Reynoutria spp. | 211 | 102 | 0.999 | [31] | MK288158 |
| tubR2 | TTGGCCAGGGAAACGTAAAC | Reynoutria spp. | This study | ||||
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | |||||||
| gapdhF2 | TGACCACAGTTCACGCAATG | Reynoutria spp. | 108 | 101 | 0.999 | This study | MK288159 |
| gapdhR2 | TGCTGCTGGGAATGATGTTG | Reynoutria spp. | This study | ||||
| Elongation factor 1-gamma (EF-1γ) | |||||||
| ef1γF2 | AGCCGCATCATGACCAAAAC | Reynoutria spp. | 131 | 101 | 0.998 | This study | MK288160 |
| ef1γR2 | ATTGCTGGAACGGATGTTGC | Reynoutria spp. | This study | ||||
| Ubiquitin domain-containing protein (UBQ) | |||||||
| ubqF2 | ATTGGAGCAGATGCAGCAAC | Reynoutria spp. | 240 | 100 | 0.999 | This study | MK288161 |
| ubqR2 | ATTTCACGCATGAGCTCTGG | Reynoutria spp. | This study | ||||
| Ubiquitin-conjugating enzyme (UBC) | |||||||
| ubcF1 | ATTTGATGGCGTGGAGTTGC | R. japonica | 119 | 101 | 0.998 | [31] | MK288162 |
| ubcR2 | TTTACACTTTGGGGGCTTGC | Reynoutria spp. | This study | ||||
| 60S ribosomal RNA (60SrRNA) | |||||||
| 60SF | ACTGTGATTTCGCAGACGCA | R. japonica | 124 | 101 | 0.997 | [31] | MK288163 |
| 60SR | CCTGGTGCTTGGTGAGACGG | R. japonica | [31] | ||||
| Eukaryotic translation initiation factor 6A (eIF6A) | |||||||
| elf6aF2 | TGGTTGCAATTGGTGGATCC | Reynoutria spp. | 220 | 100 | 0.99 | This study | MK288164 |
| elf6aR2 | TCAATGCGCTGGACAACAAC | Reynoutria spp. | This study | ||||
| Suppressor of K+ transport growth defect 1 (SKD1) | |||||||
| skdF2 | AGAAGCCGAATGTGAAGTGG | Reynoutria spp. | 75 | 102 | 0.994 | This study | MK288165 |
| skdR2 | ATATGACCGCTTCCTGCAAC | Reynoutria spp. | This study | ||||
| Thioredoxin-like protein YLS8 (YLS8) | |||||||
| ylsF2 | TGCCCGACTTCAACACAATG | Reynoutria spp. | 138 | 102 | 0.999 | This study | MK288166 |
| ylsR2 | ACTCCTGCTTGTCCTTTAGAGC | Reynoutria spp. | This study | ||||
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-A (NDUFA13) | |||||||
| ndufaF1 | ATGTACCAGGTCGGCGTAGG | R. japonica/Reynoutria spp. | 161 | 104 | 0.999 | [31] | MK288167 |
| ndufaR1 | TCCTTCATAATTCTGGCTTCC | R. japonica/Reynoutria spp. | [31] | ||||
| Phenylalanine ammonia lyase (PAL) | |||||||
| palF | TATTGTCTGTCGGCGTCAAC | Reynoutria spp. | 187 | 104 | 0.997 | This study | MK288155 |
| palR | TTCTCCTTGTCGCCGTTTTC | Reynoutria spp. | This study | ||||
| Chalcone synthase (CHS) | |||||||
| chsF | AAACATGTCGAGTGCGTGTG | Reynoutria spp. | 108 | 105 | 0.999 | This study | MT415958 |
| chsR | AACAAAACGCCCCACTCAAG | Reynoutria spp. | This study | ||||
| Method | Ranking Order (from the Most Stable to the Least Stable Gene) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| all samples | ||||||||||||
| Delta CT | 60SrRNA | SKD1 | ACT | eIF6A | NDUFA13 | GAPDH | EF-1γ | TUA | UBC | UBQ | TUB | YLS8 |
| BestKeeper | SKD1 | EF-1γ | 60SrRNA | ACT | GAPDH | eIF6A | UBQ | UBC | NDUFA13 | YLS8 | TUA | TUB |
| NormFinder | eIF6A | 60SrRNA | SKD1 | NDUFA13 | ACT | GAPDH | EF-1γ | TUA | UBC | UBQ | TUB | YLS8 |
| geNorm | ACT|SKD1 | 60SrRNA | GAPDH | NDUFA13 | EF-1γ | eIF6A | TUA | UBC | UBQ | TUB | YLS8 | |
| Comprehensive ranking | SKD1 | 60SrRNA | ACT | eIF6A | EF-1γ | GAPDH | NDUFA13 | TUA | UBC | UBQ | TUB | YLS8 |
| leaves | ||||||||||||
| Delta CT | 60SrRNA | YLS8 | GAPDH | ACT | eIF6A | UBC | TUA | SKD1 | EF-1γ | NDUFA13 | UBQ | TUB |
| BestKeeper | EF-1γ | SKD1 | ACT | 60SrRNA | YLS8 | UBC | TUA | GAPDH | eIF6A | UBQ | NDUFA13 | TUB |
| NormFinder | GAPDH | YLS8 | 60SrRNA | eIF6A | ACT | UBC | TUA | SKD1 | NDUFA13 | UBQ | EF-1γ | TUB |
| geNorm | EF-1γ|SKD1 | ACT | 60SrRNA | YLS8 | UBC | GAPDH | TUA | eIF6A | NDUFA13 | UBQ | TUB | |
| Comprehensive ranking | 60SrRNA | EF-1γ | YLS8 | SKD1 | GAPDH | ACT | UBC | eIF6A | TUA | NDUFA13 | UBQ | TUB |
| rhizomes | ||||||||||||
| Delta CT | 60SrRNA | SKD1 | NDUFA13 | ACT | eIF6A | GAPDH | EF-1γ | TUA | UBC | UBQ | TUB | YLS8 |
| BestKeeper | 60SrRNA | SKD1 | ACT | eIF6A | NDUFA13 | EF-1γ | GAPDH | TUA | UBQ | UBC | YLS8 | TUB |
| NormFinder | eIF6A | 60SrRNA | SKD1 | NDUFA13 | ACT | EF-1γ | GAPDH | TUA | UBQ | UBC | YLS8 | TUB |
| geNorm | ACT|NDUFA13 | SKD1 | 60SrRNA | GAPDH | EF-1γ | eIF6A | TUA | UBC | UBQ | TUB | YLS8 | |
| Comprehensive ranking | 60SrRNA | SKD1 | NDUFA13 | ACT | eIF6A | GAPDH | EF-1γ | TUA | UBC | UBQ | TUB | YLS8 |
| flowers | ||||||||||||
| Delta CT | SKD1 | TUB | GAPDH | YLS8 | TUA | EF-1γ | ACT | NDUFA13 | eIF6A | UBC | 60SrRNA | UBQ |
| BestKeeper | EF-1γ | UBQ | TUB | ACT | eIF6A | GAPDH | SKD1 | TUA | YLS8 | 60SrRNA | NDUFA13 | UBC |
| NormFinder | SKD1 | TUB | GAPDH | EF-1γ | ACT | TUA | YLS8 | eIF6A | NDUFA13 | UBC | 60SrRNA | UBQ |
| geNorm | GAPDH|TUA | TUB | ACT | SKD1 | YLS8 | NDUFA13 | UBC | EF-1γ | eIF6A | 60SrRNA | UBQ | |
| Comprehensive ranking | SKD1 | TUB | GAPDH | EF-1γ | TUA | ACT | YLS8 | UBQ | eIF6A | NDUFA13 | UBC | 60SrRNA |
| R. japonica | ||||||||||||
| Delta CT | 60SrRNA | UBC | ACT | EF-1γ | SKD1 | NDUFA13 | GAPDH | TUA | UBQ | TUB | eIF6A | YLS8 |
| BestKeeper | UBQ | GAPDH | UBC | TUA | 60SrRNA | EF-1γ | SKD1 | ACT | NDUFA13 | TUB | eIF6A | YLS8 |
| NormFinder | SKD1 | EF-1γ | 60SrRNA | UBC | ACT | GAPDH | TUA | UBQ | NDUFA13 | TUB | eIF6A | YLS8 |
| geNorm | EF-1γ|SKD1 | UBC | 60SrRNA | ACT | NDUFA13 | GAPDH | TUA | UBQ | TUB | eIF6A | YLS8 | |
| Comprehensive ranking | SKD1 | EF-1γ | 60SrRNA | UBC | GAPDH | ACT | UBQ | TUA | NDUFA13 | TUB | eIF6A | YLS8 |
| R. × bohemica | ||||||||||||
| Delta CT | UBC | eIF6A | 60SrRNA | ACT | GAPDH | YLS8 | TUB | UBQ | NDUFA13 | SKD1 | EF-1γ | TUA |
| BestKeeper | EF-1γ | SKD1 | 60SrRNA | ACT | YLS8 | UBC | eIF6A | GAPDH | TUB | UBQ | NDUFA13 | TUA |
| NormFinder | UBC | 60SrRNA | eIF6A | ACT | GAPDH | YLS8 | TUB | UBQ | NDUFA13 | SKD1 | TUA | EF-1γ |
| geNorm | eIF6A|UBQ | UBC | TUB | NDUFA13 | GAPDH | 60SrRNA | ACT | YLS8 | SKD1 | EF-1γ | TUA | |
| Comprehensive ranking | UBC | eIF6A | 60SrRNA | ACT | UBQ | GAPDH | EF-1γ | YLS8 | TUB | SKD1 | NDUFA13 | TUA |
| R. sachalinensis | ||||||||||||
| Delta CT | eIF6A | 60SrRNA | SKD1 | ACT | NDUFA13 | GAPDH | EF-1γ | YLS8 | UBC | TUA | UBQ | TUB |
| BestKeeper | eIF6A | 60SrRNA | SKD1 | NDUFA13 | ACT | GAPDH | EF-1γ | YLS8 | UBC | TUA | UBQ | TUB |
| NormFinder | eIF6A | 60SrRNA | NDUFA13 | SKD1 | ACT | GAPDH | YLS8 | EF-1γ | UBC | TUA | UBQ | TUB |
| geNorm | ACT|SKD1 | EF-1γ | GAPDH | 60SrRNA | NDUFA13 | eIF6A | YLS8 | UBC | TUA | UBQ | TUB | |
| Comprehensive ranking | eIF6A | SKD1 | 60SrRNA | ACT | NDUFA13 | GAPDH | EF-1γ | YLS8 | UBC | TUA | UBQ | TUB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stafiniak, M.; Makowski, W.; Matkowski, A.; Bielecka, M. Stabilizing the Baseline: Reference Gene Evaluation in Three Invasive Reynoutria Species. Int. J. Mol. Sci. 2025, 26, 8265. https://doi.org/10.3390/ijms26178265
Stafiniak M, Makowski W, Matkowski A, Bielecka M. Stabilizing the Baseline: Reference Gene Evaluation in Three Invasive Reynoutria Species. International Journal of Molecular Sciences. 2025; 26(17):8265. https://doi.org/10.3390/ijms26178265
Chicago/Turabian StyleStafiniak, Marta, Wojciech Makowski, Adam Matkowski, and Monika Bielecka. 2025. "Stabilizing the Baseline: Reference Gene Evaluation in Three Invasive Reynoutria Species" International Journal of Molecular Sciences 26, no. 17: 8265. https://doi.org/10.3390/ijms26178265
APA StyleStafiniak, M., Makowski, W., Matkowski, A., & Bielecka, M. (2025). Stabilizing the Baseline: Reference Gene Evaluation in Three Invasive Reynoutria Species. International Journal of Molecular Sciences, 26(17), 8265. https://doi.org/10.3390/ijms26178265







