Analysis of the Entire Mitogenome of the Threatened Freshwater Stingray Potamotrygon leopoldi (Myliobatiformes: Potamotrygonidae) and Comprehensive Phylogenetic Assessment in the Xingu River, Brazilian Amazon
Abstract
1. Introduction
2. Result
2.1. Mitochondrial Genome Structure
2.2. Protein-Coding Genes (PCGs)
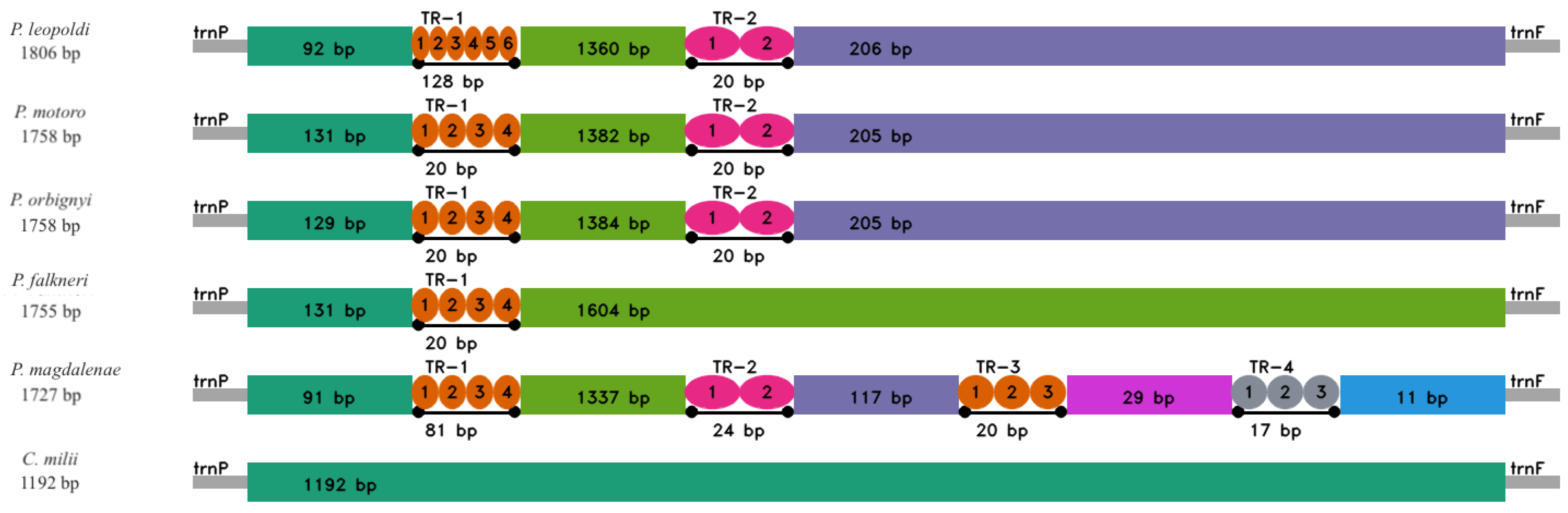
2.3. Control Region
2.4. Adaptive Molecular Evolution
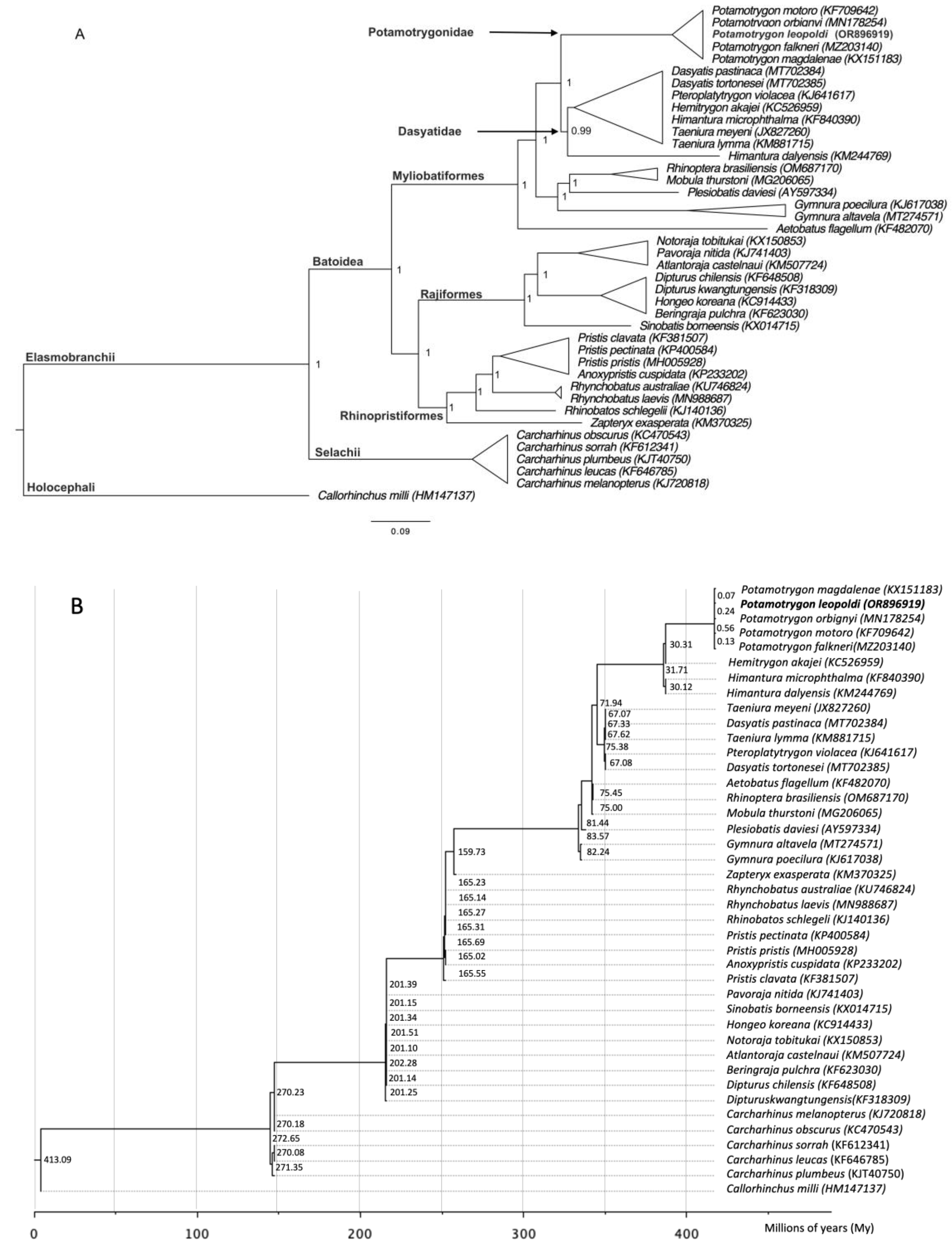
2.5. Phylogenetic Analyses
3. Discussion
3.1. Mitochondrial Genome Characterization
3.2. Protein-Coding Genes
3.2.1. Control Region (CR)
3.2.2. Adaptive Molecular Evolution
3.3. Phylogenetic Analyses
4. Materials and Methods
4.1. Sample Collection and DNA Extraction
4.2. Assembly, Annotation, and Bioinformatic Analysis of the Complete Mitogenome
4.3. Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stewart, J.B.; Freyer, C.; Elson, J.L.; Wredenberg, A.; Cansu, Z.; Trifunovic, A.; Larsson, N.-G. Strong Purifying Selection in Transmission of Mammalian Mitochondrial DNA. PLoS Biol. 2008, 6, e10. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.M.; George, M., Jr.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Tu, P.; Xu, P.; Sun, Y.; Yu, F.; Tu, N.; Guo, L.; Yang, Y. The Mitochondrial Response to DNA Damage. Front. Cell Dev. Biol. 2021, 9, 669379. [Google Scholar] [CrossRef] [PubMed]
- COMEXSTAT. Exportação e Importação Geral do Brasil. 2023. Available online: http://comexstat.mdic.gov.br/pt/home (accessed on 2 December 2024).
- Carvalho, M.R.d.; Ragno, M.P. An unusual, dwarf new species of Neotropical freshwater stingray, Plesiotrygon nana sp. nov., from the upper and mid Amazon basin: The second species of Plesiotrygon (Chondrichthyes: Potamotrygonidae). Papéis Avulsos Zool. 2011, 51, 101–138. [Google Scholar] [CrossRef]
- Charvet-Almeida, P.; GÛes, M.L.C. Neotropical Freshwater Stingrays: Diversity and conservation status. Shark News 2008, 14, 47–51. [Google Scholar]
- De Carvalho, M.R.; Lovejoy, N.R. Morphology and phylogenetic relationships of a remarkable new genus and two new species of Neotropical freshwater stingrays from the Amazon basin (Chondrichthyes: Potamotrygonidae). Zootaxa 2011, 2776, 13–48. [Google Scholar] [CrossRef]
- Torres, Y.; Faria, V.V.; Charvet, P. Current status and future perspectives of Neotropical freshwater stingrays (Potamotrygoninae, Myliobatiformes) genetics. Environ. Biol. Fishes 2022, 105, 1111–1127. [Google Scholar] [CrossRef]
- Lasso, C.; Rosa, R.; Morales, M.; Garrone Neto, D.; Carvalho, M. Rayas de Agua Dulce (Potamotrygonidae) de Suramérica. Parte II: Colombia, Brasil, Perú, Bolivia, Paraguay, Uruguay y Argentina; Instituto de Investigación de Recursos Biológicos Alexander von Humboldt: Bogotá, Colombia, 2016. [Google Scholar]
- CITES. Convention on International Trade in Endangered Species of Wild Fauna and Flora, Apendix III. 2023. Available online: https://cites.org/eng/app/index.php (accessed on 2 December 2024).
- IUCN. The IUCN Red List of Threatened Species. Version 2021-1. Available online: https://www.iucnredlist.org (accessed on 1 November 2024).
- Ibama. Instrução Normativa 7, de 30 de Abril de 2015 Instrução Normativa. 2015, 55. Available online: https://www.ibama.gov.br/component/legislacao/?view=legislacao&legislacao=135756#:~:text=Institui%20e%20normatiza%20as%20categorias,autorizativos%20para%20as%20categorias%20estabelecidas (accessed on 2 December 2024).
- Fontenelle, J.P.; Portella Luna Marques, F.; Kolmann, M.A.; Lovejoy, N.R. Biogeography of the neotropical freshwater stingrays (Myliobatiformes: Potamotrygoninae) reveals effects of continent-scale paleogeographic change and drainage evolution. J. Biogeogr. 2021, 48, 1406–1419. [Google Scholar] [CrossRef]
- Sanches, D.; Martins, T.; Lutz, Í.; Veneza, I.; Silva, R.D.; AraÚJo, F.; Muriel-Cunha, J.; Sampaio, I.; Garcia, M.; Sousa, L.; et al. Mitochondrial DNA suggests Hybridization in Freshwater Stingrays Potamotrygon (POTAMOTRYGONIDAE: MYLIOBATIFORMES) from the Xingu river, Amazonia and reveals speciation in Paratrygon aireba. An. Acad. Bras. Ciências 2021, 93, e20191325. [Google Scholar] [CrossRef]
- Dziedzic, E.; Sidlauskas, B.; Cronn, R.; Anthony, J.; Cornwell, T.; Friesen, T.A.; Konstantinidis, P.; Penaluna, B.E.; Stein, S.; Levi, T. Creating, curating and evaluating a mitogenomic reference database to improve regional species identification using environmental DNA. Mol. Ecol. Resour. 2023, 23, 1880–1904. [Google Scholar] [CrossRef]
- Hoban, M.L.; Whitney, J.; Collins, A.G.; Meyer, C.; Murphy, K.R.; Reft, A.J.; Bemis, K.E. Skimming for barcodes: Rapid production of mitochondrial genome and nuclear ribosomal repeat reference markers through shallow shotgun sequencing. PeerJ 2022, 10, e13790. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, A.; He, F.; Zhang, Y.; Shen, L.; Yu, J.; Zhang, X. Draft Genome of White-blotched River Stingray Provides Novel Clues for Niche Adaptation and Skeleton Formation. Genom. Proteom. Bioinform. 2022, 21, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef] [PubMed]
- Maduna, S.N.; Vivian-Smith, A.; Jónsdóttir, Ó.D.B.; Imsland, A.K.D.; Klütsch, C.F.C.; Nyman, T.; Eiken, H.G.; Hagen, S.B. Mitogenomics of the suborder Cottoidei (Teleostei: Perciformes): Improved assemblies, mitogenome features, phylogeny, and ecological implications. Genomics 2022, 114, 110297. [Google Scholar] [CrossRef] [PubMed]
- Ory, D.; Cuenot, Y.; Vigouroux, R.; Covain, R.; Brosse, S.; Murienne, J. Complete mitochondrial genome of the river stingray Potamotrygon orbignyi (Myliobatiformes: Potamotrygonidae). Mitochondrial DNA Part B 2019, 4, 3153–3154. [Google Scholar] [CrossRef]
- Reyes, A.; Gissi, C.; Pesole, G.; Saccone, C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol. Biol. Evol. 1998, 15, 957–966. [Google Scholar] [CrossRef]
- Krishnan, N.M.; Seligmann, H.; Raina, S.Z.; Pollock, D.D. Detecting gradients of asymmetry in site-specific substitutions in mitochondrial genomes. DNA Cell Biol. 2004, 23, 707–714. [Google Scholar] [CrossRef]
- Hassanin, A.; Léger, N.; Deutsch, J. Evidence for Multiple Reversals of Asymmetric Mutational Constraints during the Evolution of the Mitochondrial Genome of Metazoa, and Consequences for Phylogenetic Inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Swire, J.; Judson, O.P.; Burt, A. Mitochondrial genetic codes evolve to match amino acid requirements of proteins. J. Mol. Evol. 2005, 60, 128–139. [Google Scholar] [CrossRef]
- Grande, C.; Templado, J.; Zardoya, R. Evolution of gastropod mitochondrial genome arrangements. BMC Evol. Biol. 2008, 8, 61. [Google Scholar] [CrossRef]
- Nachtigall, P.G.; Loboda, T.S.; Pinhal, D. Signatures of positive selection in the mitochondrial genome of neotropical freshwater stingrays provide clues about the transition from saltwater to freshwater environment. Mol. Genet. Genom. 2023, 298, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Kousteni, V.; Mazzoleni, S.; Vasileiadou, K.; Rovatsos, M. Complete Mitochondrial DNA Genome of Nine Species of Sharks and Rays and Their Phylogenetic Placement among Modern Elasmobranchs. Genes 2021, 12, 324. [Google Scholar] [CrossRef]
- Song, H.M.; Mu, X.D.; Wei, M.X.; Wang, X.J.; Luo, J.R.; Hu, Y.C. Complete mitochondrial genome of the ocellate river stingray (Potamotrygon motoro). Mitochondrial DNA 2015, 26, 857–858. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.T.; Petit, R.A.; Read, T.D.; Dove, A.D.M. The complete mitochondrial genome sequence of the world’s largest fish, the whale shark (Rhincodon typus), and its comparison with those of related shark species. Gene 2014, 539, 44–49. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Diaz-Jaimes, P.; Bayona-Vasquez, N.J.; Adams, D.H.; Uribe-Alcocer, M. Complete mitochondrial DNA genome of bonnethead shark, Sphyrna tiburo, and phylogenetic relationships among main superorders of modern elasmobranchs. Meta Gene 2016, 7, 48–55. [Google Scholar] [CrossRef]
- Formenti, G.; Rhie, A.; Balacco, J.; Haase, B.; Mountcastle, J.; Fedrigo, O.; Brown, S.; Capodiferro, M.R.; Al-Ajli, F.O.; Ambrosini, R.; et al. Complete vertebrate mitogenomes reveal widespread repeats and gene duplications. Genome Biol. 2021, 22, 120. [Google Scholar] [CrossRef] [PubMed]
- Foote, A.D.; Morin, P.A.; Durban, J.W.; Pitman, R.L.; Wade, P.; Willerslev, E.; Gilbert, M.T.; da Fonseca, R.R. Positive selection on the killer whale mitogenome. Biol. Lett. 2011, 7, 116–118. [Google Scholar] [CrossRef]
- Sebastian, W.; Sukumaran, S.; Gopalakrishnan, A. Comparative mitogenomics of Clupeoid fish provides insights into the adaptive evolution of mitochondrial oxidative phosphorylation (OXPHOS) genes and codon usage in the heterogeneous habitats. Heredity 2022, 128, 236–249. [Google Scholar] [CrossRef]
- Xia, J.H.; Li, H.L.; Zhang, Y.; Meng, Z.N.; Lin, H.R. Identifying selectively important amino acid positions associated with alternative habitat environments in fish mitochondrial genomes. Mitochondrial DNA A 2018, 29, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Consuegra, S.; John, E.; Verspoor, E.; de Leaniz, C.G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. 2015, 47, 58. [Google Scholar] [CrossRef] [PubMed]
- de Souza, É.M.S.; Freitas, L.; da Silva Ramos, E.K.; Selleghin-Veiga, G.; Rachid-Ribeiro, M.C.; Silva, F.A.; Marmontel, M.; dos Santos, F.R.; Laudisoit, A.; Verheyen, E.; et al. The evolutionary history of manatees told by their mitogenomes. Sci. Rep. 2021, 11, 3564. [Google Scholar] [CrossRef] [PubMed]
- Noll, D.; Leon, F.; Brandt, D.; Pistorius, P.; Le Bohec, C.; Bonadonna, F.; Trathan, P.N.; Barbosa, A.; Rey, A.R.; Dantas, G.P.M.; et al. Positive selection over the mitochondrial genome and its role in the diversification of gentoo penguins in response to adaptation in isolation. Sci. Rep. 2022, 12, 3767. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Zhu, T.; Sato, Y.; Sado, T.; Miya, M.; Iwasaki, W. MitoFish, MitoAnnotator, and MiFish Pipeline: Updates in 10 Years. Mol. Biol. Evol. 2023, 40, 511–524. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Vella, N.; Vella, A. Characterization and comparison of the complete mitochondrial genomes of two stingrays, Dasyatis pastinaca and Dasyatis tortonesei (Myliobatiformes: Dasyatidae) from the Mediterranean Sea. Mol. Biol. Rep. 2021, 48, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-P.; Zhi, T.-T.; Zhang, S.; Yan, S.; Yang, T. Complete mitochondrial genome of the pelagic stingray Pteroplatytrygon violacea (Myliobatiformes: Dasyatidae). Mitochondrial DNA A 2016, 27, 935–936. [Google Scholar] [CrossRef]
- Wang, J.; Si, R.; Chen, H.; Chen, X. The phylogenomic position of the smalleye whip ray Himantura microphthalma (Myliobatiformes: Dasyatidae) inferred from the mitochondrial genome. Mitochondrial DNA Part B 2016, 1, 564–565. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ai, W.; Xiang, D.; Chen, S. Mitochondrial genome of blotched fantail ray Taeniura meyeni (Myliobatiformes: Dasyatidae). Mitochondrial DNA 2013, 24, 663–664. [Google Scholar] [CrossRef]
- Austin, C.M.; Tan, M.H.; Croft, L.J.; Meekan, M.G.; Gan, H.Y.; Gan, H.M. The complete mitogenome of the bluespotted ribbontail ray Taeniura lymma (Forsskål, 1775) (Elasmobranchii: Myliobatiformes: Dasyatidae). Mitochondrial DNA A 2016, 27, 3205–3207. [Google Scholar] [CrossRef] [PubMed]
- Feutry, P.; Kyne, P.M.; Peng, Z.; Pan, L.; Chen, X. Complete mitochondrial genome of the Freshwater Whipray Himantura dalyensis. Mitochondrial DNA A 2016, 27, 1589–1590. [Google Scholar] [CrossRef]
- Palacios-Barreto, P.; Mar-Silva, A.F.; Bayona-Vasquez, N.J.; Adams, D.H.; Díaz-Jaimes, P. Characterization of the complete mitochondrial genome of the brazilian cownose ray Rhinoptera brasiliensis (Myliobatiformes, Rhinopteridae) in the western Atlantic and its phylogenetic implications. Mol. Biol. Rep. 2023, 50, 4083–4095. [Google Scholar] [CrossRef] [PubMed]
- Santillán-Lugo, B.; Llera-Herrera, R.; Corro-Espinosa, D.; Oñate-González, E.C.; Rodríguez-Domínguez, G.; Saavedra-Sotelo, N.C. Complete mitochondrial genome of the Devil Ray, Mobula thurstoni (Lloyd, 1908) (Myliobatiformes: Myliobatidae). Mitochondrial DNA B Resour. 2017, 2, 868–870. [Google Scholar] [CrossRef]
- Hsiao, S.-T.; Hsu, C.-H.; Liu, D.-C.; Chen, I.-S. The Complete Mitochondrial DNA Sequence of the Deepwater Stingray Plesiobatis daviesi (Wallace, 1967): Unique Features in the Mitochondrial D-loop Region. J. Taiwan Fish. Res. 2012, 20, 1–16. [Google Scholar]
- Chen, X.; Ai, W.; Xiang, D.; Shi, X. Mitochondrial genome of the longtail butterfly ray Gymnura poecilura (Myliobatiformes: Gymnuridae). Mitochondrial DNA A 2016, 27, 696–697. [Google Scholar] [CrossRef]
- Cady, T.; Bemis, K.E.; Baeza, J.A. The mitochondrial genome of the endangered Spiny Butterfly Ray Gymnura altavela (Linnaeus 1758) (Myliobatiformes: Gymnuridae) provides insights into cryptic lineages. Mitochondrial DNA A 2021, 32, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, B.; Yamaguchi, A.; Furumitsu, K.; Zhang, B. Mitochondrial genome of longheaded eagle ray Aetobatus flagellum (Chondrichthyes: Myliobatidae). Mitochondrial DNA 2015, 26, 763–764. [Google Scholar] [CrossRef] [PubMed]
- Si, R.; Chen, X.; Chen, H.; Ai, W. Complete mitochondrial genome and the phylogenetic position of the Leadhued skate Notoraja tobitukai (Rajiformes: Arhynchobatidae). Mitochondrial DNA B Resour. 2016, 1, 502–503. [Google Scholar] [CrossRef]
- Yang, L.; Naylor, G.J. Long-PCR based next generation sequencing of the whole mitochondrial genome of the peacock skate Pavoraja nitida (Elasmobranchii: Arhynchobatidae). Mitochondrial DNA A 2016, 27, 943–944. [Google Scholar] [CrossRef] [PubMed]
- Duckett, D.J.; Naylor, G.J. The complete mitochondrial genome of the endangered spotback skate, Atlantoraja castelnaui. Mitochondrial DNA A 2016, 27, 2081–2082. [Google Scholar] [CrossRef]
- Jeong, D.; Lee, Y.H. Complete mitochondrial genome of the Yellownose skate: Zearaja chilensis (Rajiformes, Rajidae). Mitochondrial DNA A 2016, 27, 293–294. [Google Scholar] [CrossRef]
- Jeong, D.; Kim, S.; Kim, C.G.; Lee, Y.H. Complete mitochondrial genome of the Kwangtung skate: Dipturus kwangtungensis (Rajiformes, Rajidae). Mitochondrial DNA A 2015, 26, 873–874. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.; Kim, S.; Kim, C.G.; Lee, Y.H. The complete mitochondrial genome of the Korean skate: Hongeo koreana (Rajiformes, Rajidae). Mitochondrial DNA A 2014, 25, 437–438. [Google Scholar] [CrossRef]
- Choi, S.; Kim, S.J.; Kim, E.B. Complete mitochondrial DNA sequence of Raja pulchra from Yellow Sea and Alaska. Mitochondrial DNA Part B Resour. 2019, 4, 384–385. [Google Scholar] [CrossRef]
- Si, R.; Ding, W.; Ai, W.; Chen, X. Complete mitochondrial genome and the phylogenetic position of the Borneo leg skate Sinobatis borneensis (Rajiformes: Anacanthobatidae). Mitochondrial DNA B Resour. 2016, 1, 443–444. [Google Scholar] [CrossRef]
- Feutry, P.; Kyne, P.M.; Grewe, P.M.; Chen, X.; Liu, M. Whole mitogenome of the Endangered dwarf sawfish Pristis clavata (Rajiformes: Pristidae). Mitochondrial DNA 2015, 26, 329–330. [Google Scholar] [CrossRef] [PubMed]
- Kyne, P.M.; Wang, J.-J.; Xiang, D.; Chen, X.; Feutry, P. The phylogenomic position of the Critically Endangered Largetooth Sawfish Pristis pristis (Rhinopristiformes, Pristidae), inferred from the complete mitochondrial genome. Mitochondrial DNA Part B 2018, 3, 970–971. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kyne, P.M.; Pillans, R.D.; Feutry, P. Complete mitochondrial genome of the Endangered Narrow Sawfish Anoxypristis cuspidata (Rajiformes: Pristidae). Mitochondrial DNA A 2016, 27, 4172–4173. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, M.J.; Klein, J.D.; Bennett, R.H.; Bester-van der Merwe, A.E. Characterization of the complete mitochondrial genomes of two Critically Endangered wedgefishes: Rhynchobatus djiddensis and Rhynchobatus australiae. Mitochondrial DNA B Resour. 2023, 8, 352–358. [Google Scholar] [CrossRef]
- Johri, S.; Tiwari, A.; Kerr, E.N.; Dinsdale, E.A. Mitochondrial genome of the Smoothnose wedgefish Rhynchobatus laevis from the Western Indian Ocean. Mitochondrial DNA B Resour. 2020, 5, 2083–2084. [Google Scholar] [CrossRef]
- Chen, X.; Ai, W.; Xiang, D.; Pan, L.; Shi, X. Complete mitogenome of the brown guitarfish Rhinobatos schlegelii (Rajiformes, Rhinobatidae). Mitochondrial DNA A 2016, 27, 310–311. [Google Scholar] [CrossRef]
- Castillo-Páez, A.; Del Río-Portilla, M.A.; Oñate-González, E.; Rocha-Olivares, A. The mitochondrial genome of the banded guitarfish, Zapteryx exasperata (Jordan and Gilbert, 1880), possesses a non-coding duplication remnant region. Mitochondrial DNA A 2016, 27, 1668–1670. [Google Scholar] [CrossRef]
- Blower, D.C.; Hereward, J.P.; Ovenden, J.R. The complete mitochondrial genome of the dusky shark Carcharhinus obscurus. Mitochondrial DNA 2013, 24, 619–621. [Google Scholar] [CrossRef]
- Chen, X.; Peng, Z.; Cai, L.; Xu, Y. Mitochondrial genome of the spot-tail shark Carcharhinus sorrah (Carcharhiniformes: Carcharhinidae). Mitochondrial DNA 2015, 26, 734–735. [Google Scholar] [CrossRef]
- Blower, D.C.; Ovenden, J.R. The complete mitochondrial genome of the sandbar shark Carcharhinus plumbeus. Mitochondrial DNA A 2016, 27, 923–924. [Google Scholar] [CrossRef]
- Chen, X.; Liu, M.; Peng, Z.; Shi, X. Mitochondrial genome of the bull shark Carcharhinus leucas (Carcharhiniformes: Carcharhinidae). Mitochondrial DNA 2015, 26, 813–814. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shen, X.J.; Arunrugstichai, S.; Ai, W.; Xiang, D. Complete mitochondrial genome of the blacktip reef shark Carcharhinus melanopterus (Carcharhiniformes: Carcharhinidae). Mitochondrial DNA A 2016, 27, 873–874. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1. Institue of Evolutionary Biology, University of Endinburgh, 2010. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 2 December 2024).
- Li, W.L.S.; Drummond, A.J. Model Averaging and Bayes Factor Calculation of Relaxed Molecular Clocks in Bayesian Phylogenetics. Mol. Biol. Evol. 2011, 29, 751–761. [Google Scholar] [CrossRef]
- Aschliman, N.C.; Nishida, M.; Miya, M.; Inoue, J.G.; Rosana, K.M.; Naylor, G.J.P. Body plan convergence in the evolution of skates and rays (Chondrichthyes: Batoidea). Mol. Phylogen. Evol. 2012, 63, 28–42. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Xia, X. DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef]
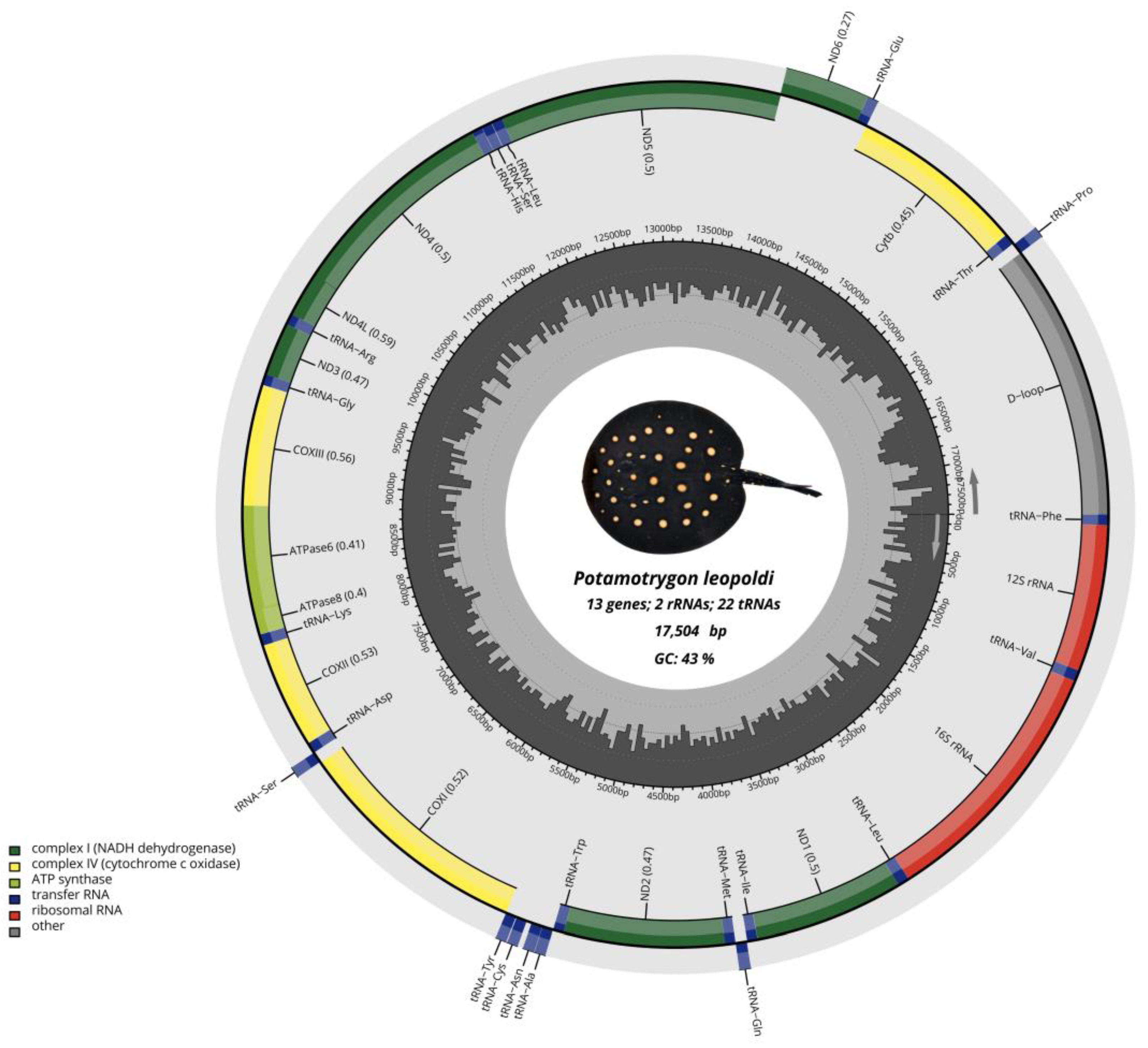
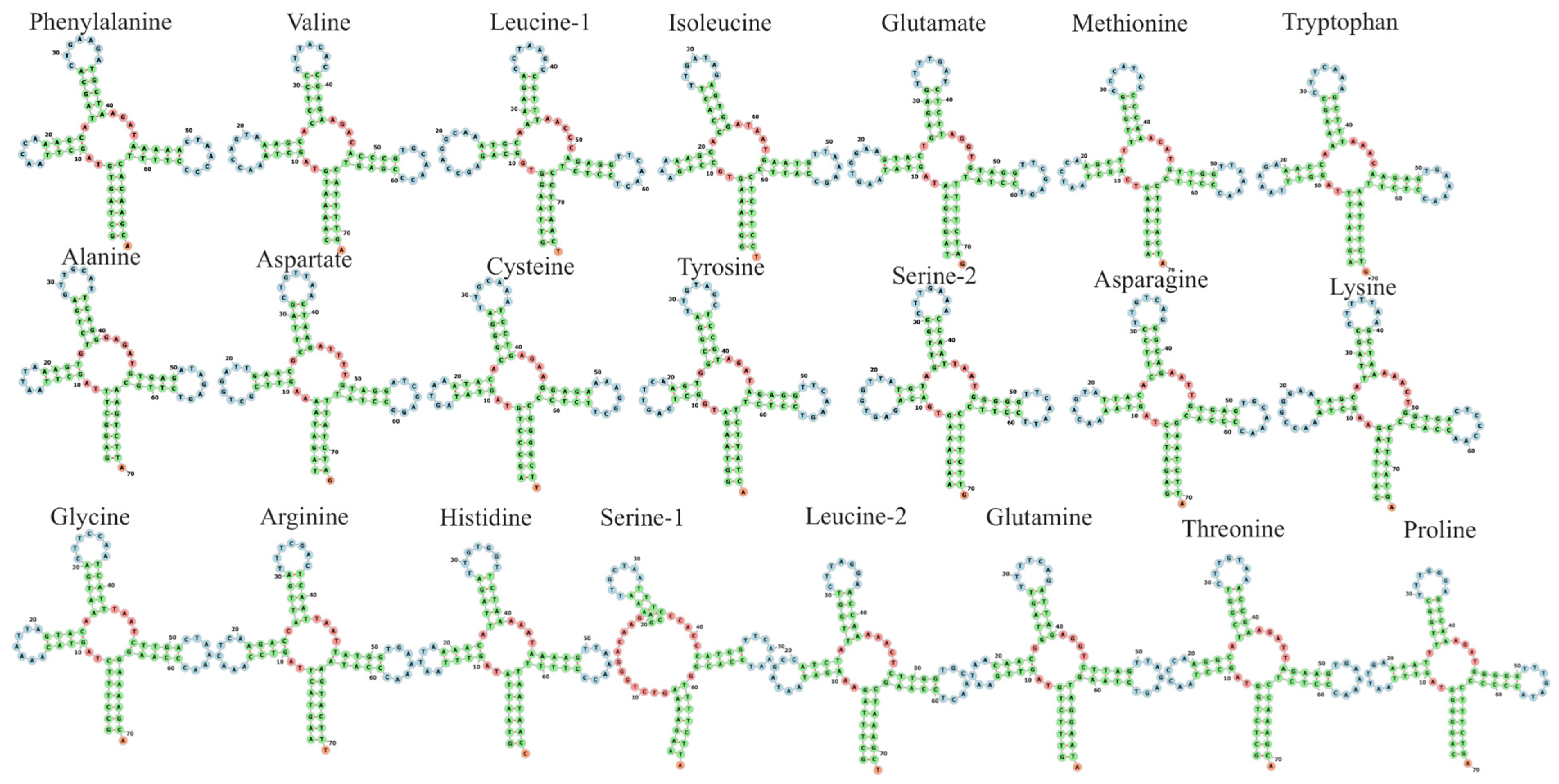
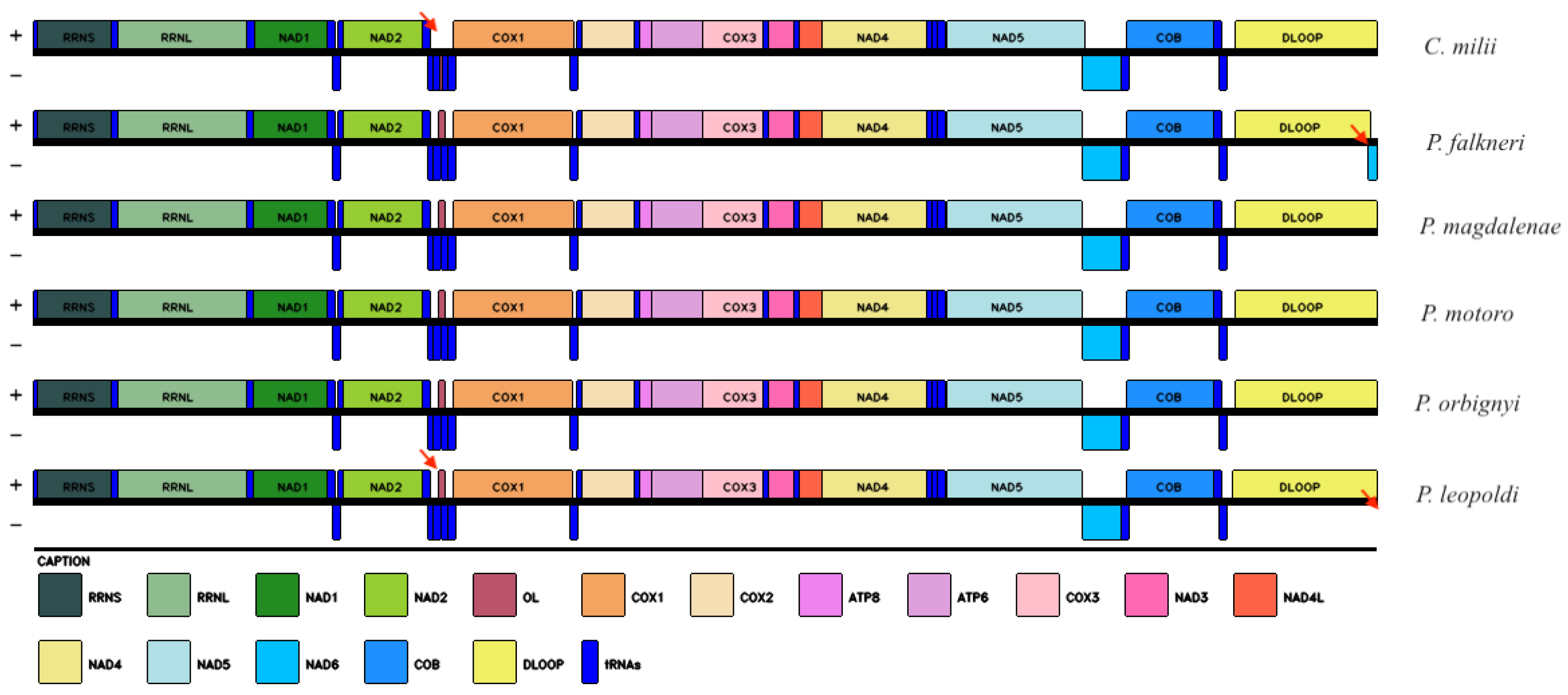
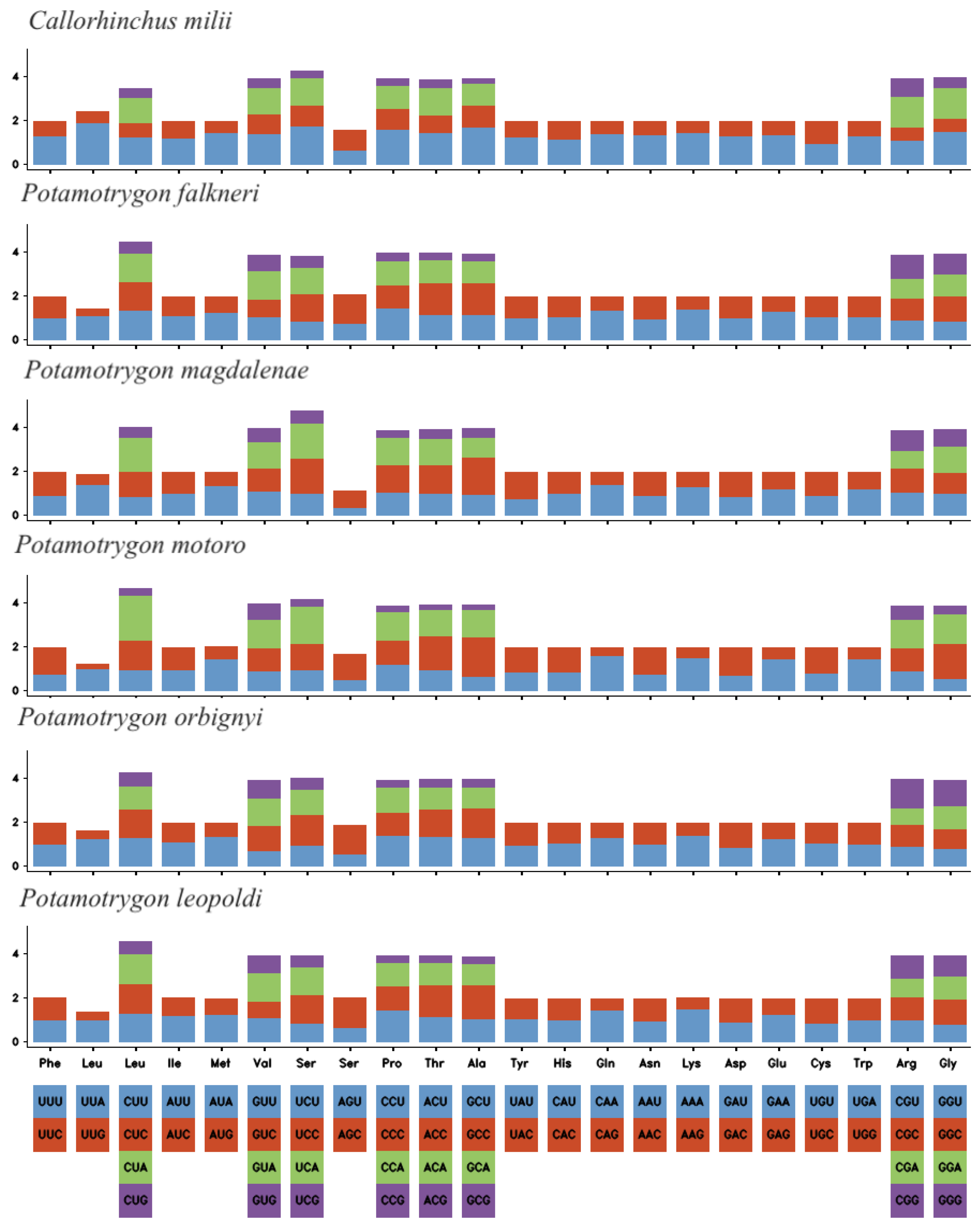
| Name | Type | Strand | Start | Stop | Intergenic Nucleotide | Length | Anticodon and Start Codon/Stop Codon |
|---|---|---|---|---|---|---|---|
| tRNA-Phe | tRNA | + | 1 | 69 | 0 | 69 | - |
| 12S rRNA | rRNA | + | 70 | 1030 | 0 | 961 | - |
| tRNA-Val | tRNA | + | 1031 | 1102 | 19 | 72 | - |
| 16S rRNA | rRNA | + | 1122 | 2788 | 3 | 1667 | - |
| tRNA-Leu | tRNA | + | 2792 | 2866 | 0 | 75 | - |
| ND1 | PCG | + | 2867 | 3841 | 1 | 975 | ATG/TAA |
| tRNA-Ile | tRNA | + | 3843 | 3910 | 4 | 68 | - |
| tRNA-Gln | tRNA | − | 3915 | 3986 | 0 | 72 | - |
| tRNA-Met | tRNA | + | 3987 | 4056 | 0 | 70 | - |
| ND2 | PCG | + | 4057 | 5103 | −1 | 1047 | ATG/TAA |
| tRNA-Trp | tRNA | + | 5103 | 5172 | 1 | 70 | - |
| tRNA-Ala | tRNA | − | 5174 | 5243 | 1 | 70 | - |
| tRNA-Asn | tRNA | − | 5245 | 5317 | 4 | 73 | - |
| tRNA-Cys | tRNA | − | 5351 | 5419 | 5 | 69 | - |
| tRNA-Tyr | tRNA | − | 5425 | 5493 | 1 | 69 | - |
| COX1 | PCG | + | 5495 | 7048 | 6 | 1554 | GTG/TAG |
| tRNA-Ser 2 | tRNA | − | 7055 | 7125 | 0 | 71 | - |
| tRNA-Asp | tRNA | + | 7126 | 7196 | 3 | 71 | - |
| COX2 | PCG | + | 7200 | 7890 | 0 | 691 | ATG/T(AA) |
| tRNA-Lys | tRNA | + | 7891 | 7964 | 1 | 74 | - |
| ATP8 | PCG | + | 7966 | 8133 | −10 | 168 | ATG/TAA |
| ATP6 | PCG | + | 8124 | 8807 | −1 | 683 | ATG/TAA |
| COX3 | PCG | + | 8807 | 9592 | 5 | 786 | ATG/TAA |
| tRNA-Gly | tRNA | + | 9598 | 9668 | 0 | 71 | - |
| ND3 | PCG | + | 9669 | 10,019 | −1 | 351 | ATG/TAA |
| tRNA-Arg | tRNA | + | 10,019 | 10,089 | 0 | 71 | - |
| ND4L | PCG | + | 10,090 | 10,386 | −7 | 297 | ATG/TAA |
| ND4 | PCG | + | 10,380 | 11,760 | 3 | 1381 | ATG/T(AA) |
| tRNA-His | tRNA | + | 11,764 | 11,832 | 1 | 69 | - |
| tRNA-Ser | tRNA | + | 11,834 | 11,900 | 0 | 72 | - |
| tRNA-Leu | tRNA | + | 11,903 | 11,974 | 0 | 72 | - |
| ND5 | PCG | + | 11,975 | 13,810 | −4 | 1836 | ATG/TAA |
| ND6 | PCG | − | 13,807 | 14,328 | 0 | 522 | ATG/TAG |
| tRNA-Glu | tRNA | − | 14,319 | 14,397 | 1 | 69 | - |
| CYTB | PCG | + | 14,399 | 15,541 | 4 | 1143 | ATG/TAA |
| tRNA-Thr | tRNA | + | 15,546 | 15,616 | 8 | 71 | - |
| tRNA-Pro | tRNA | − | 15,652 | 15,694 | 0 | 70 | - |
| D-loop | CR | 15,694 | 17,504 | 1810 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerreiro, S.L.M.; Vidal, A.F.; Silva, C.S.; Cavalcante, G.C.; Magalhães, L.; Gomes, D.H.F.; Filho, J.C.d.S.; Souza, J.E.S.d.; Pires, É.; Oliveira, G.; et al. Analysis of the Entire Mitogenome of the Threatened Freshwater Stingray Potamotrygon leopoldi (Myliobatiformes: Potamotrygonidae) and Comprehensive Phylogenetic Assessment in the Xingu River, Brazilian Amazon. Int. J. Mol. Sci. 2025, 26, 8252. https://doi.org/10.3390/ijms26178252
Guerreiro SLM, Vidal AF, Silva CS, Cavalcante GC, Magalhães L, Gomes DHF, Filho JCdS, Souza JESd, Pires É, Oliveira G, et al. Analysis of the Entire Mitogenome of the Threatened Freshwater Stingray Potamotrygon leopoldi (Myliobatiformes: Potamotrygonidae) and Comprehensive Phylogenetic Assessment in the Xingu River, Brazilian Amazon. International Journal of Molecular Sciences. 2025; 26(17):8252. https://doi.org/10.3390/ijms26178252
Chicago/Turabian StyleGuerreiro, Sávio L. M., Amanda F. Vidal, Caio S. Silva, Giovanna C. Cavalcante, Leandro Magalhães, Daniel H. F. Gomes, Júlio César da Silva Filho, Jorge E. S. de Souza, Éder Pires, Guilherme Oliveira, and et al. 2025. "Analysis of the Entire Mitogenome of the Threatened Freshwater Stingray Potamotrygon leopoldi (Myliobatiformes: Potamotrygonidae) and Comprehensive Phylogenetic Assessment in the Xingu River, Brazilian Amazon" International Journal of Molecular Sciences 26, no. 17: 8252. https://doi.org/10.3390/ijms26178252
APA StyleGuerreiro, S. L. M., Vidal, A. F., Silva, C. S., Cavalcante, G. C., Magalhães, L., Gomes, D. H. F., Filho, J. C. d. S., Souza, J. E. S. d., Pires, É., Oliveira, G., Melo, D. S. D., Sá, A. L. A. d., Hamoy, I., Ribeiro-dos-Santos, Â., & Santos, S. E. B. (2025). Analysis of the Entire Mitogenome of the Threatened Freshwater Stingray Potamotrygon leopoldi (Myliobatiformes: Potamotrygonidae) and Comprehensive Phylogenetic Assessment in the Xingu River, Brazilian Amazon. International Journal of Molecular Sciences, 26(17), 8252. https://doi.org/10.3390/ijms26178252






