Genome-Wide Identification of the TIFY Family in Cannabis sativa L. and Its Potential Functional Analysis in Response to Alkaline Stress and in Cannabinoid Metabolism
Abstract
1. Introduction
2. Results
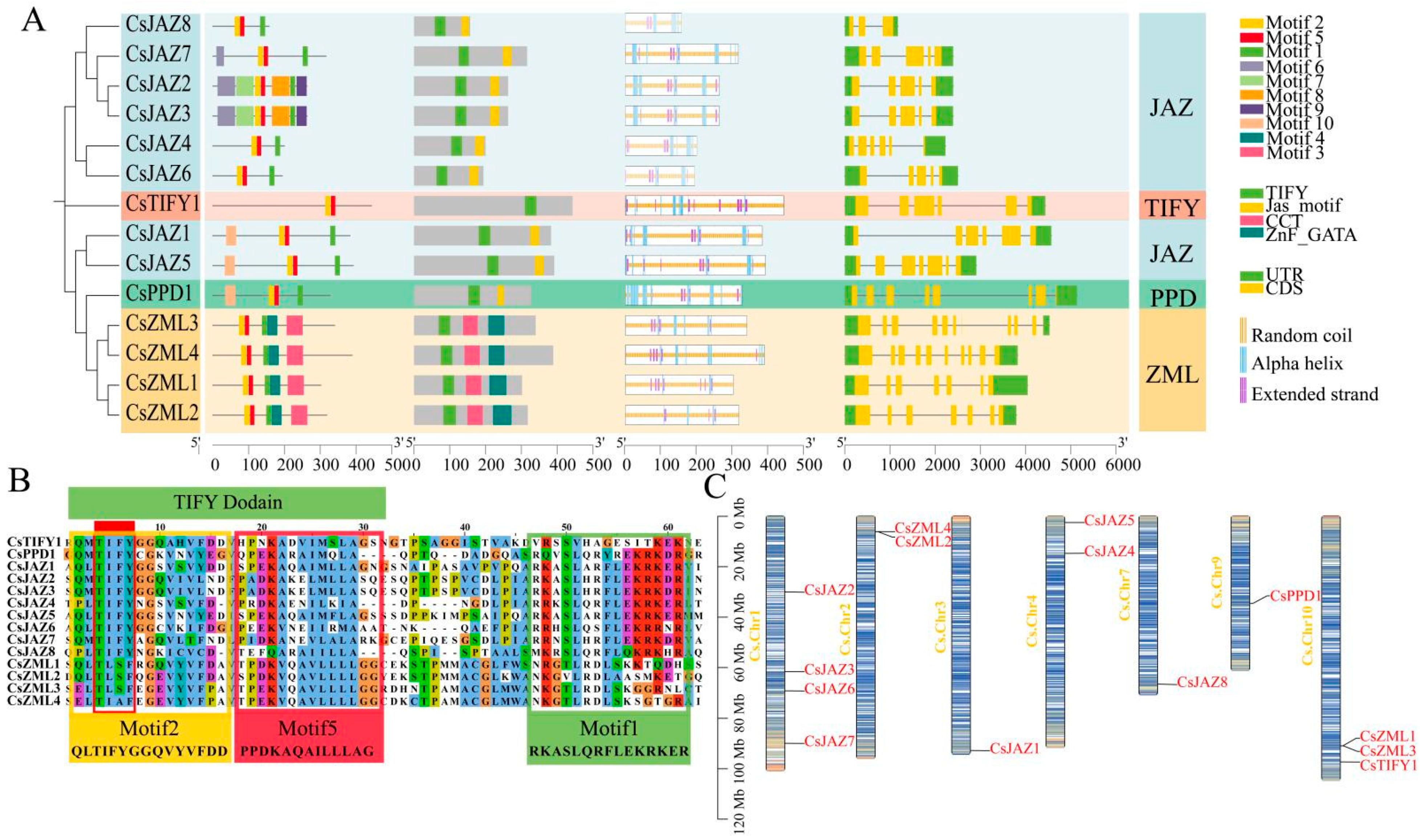
2.1. Physicochemical Properties and Evolutionary Relationship Analysis of TIFY Transcription Factors
2.2. Gene Structure and Secondary Protein Structure of TIFY Transcription Factors
2.3. Chromosomal Localization of TIFY Transcription Factors
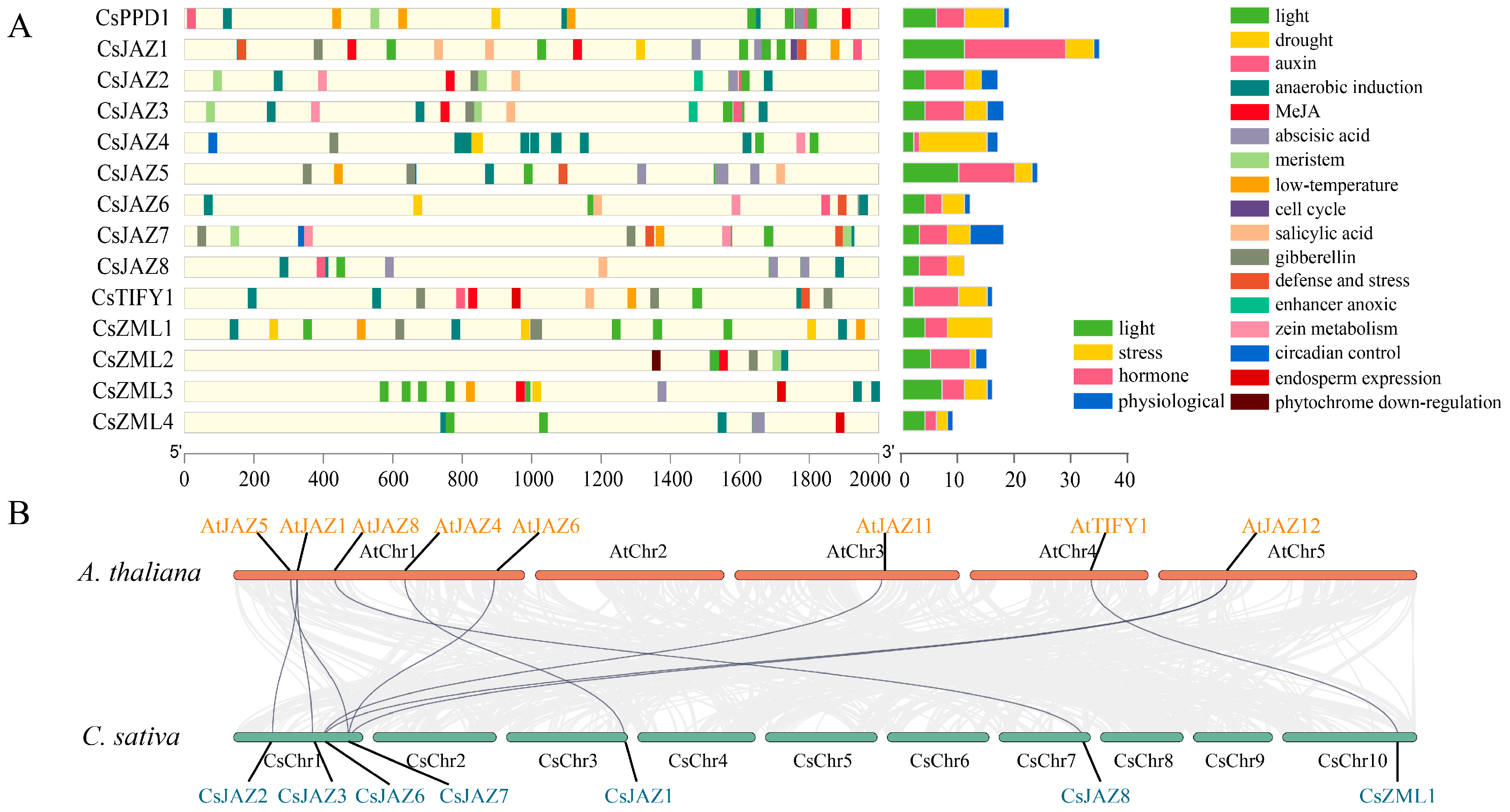
2.4. Cis-Acting Elements of TIFY in C. sativa
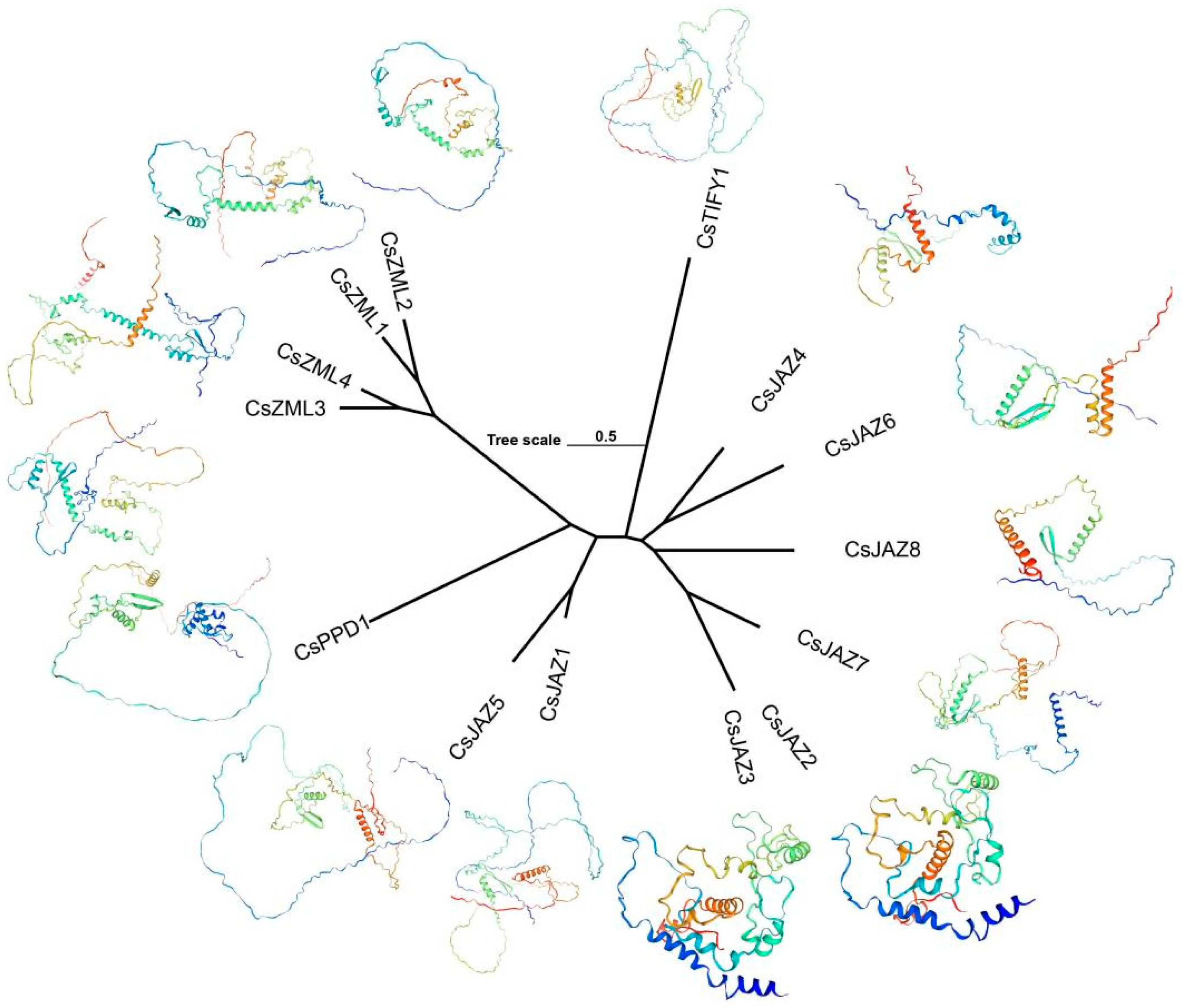
2.5. Collinearity and Protein Tertiary Structure of TIFY
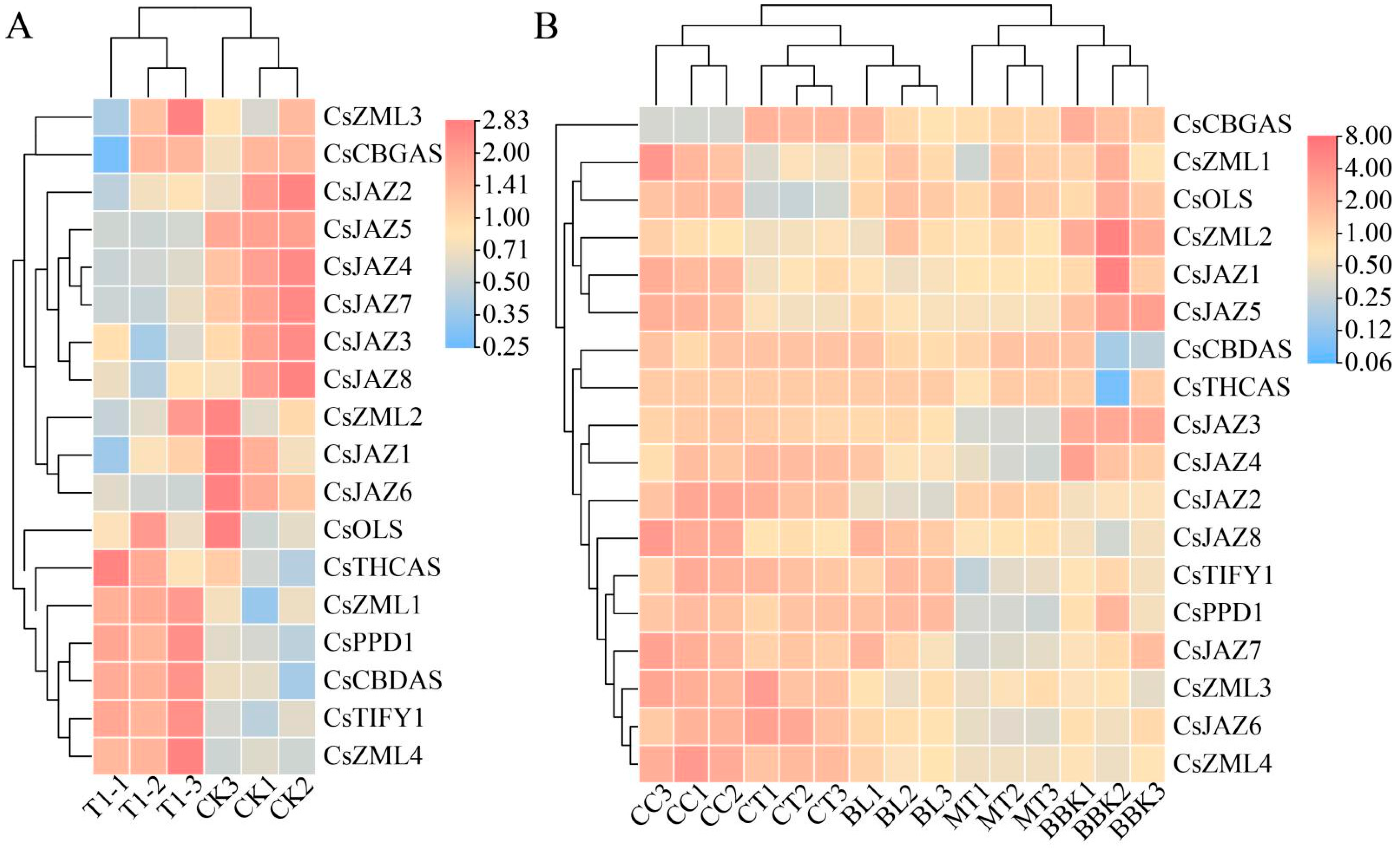
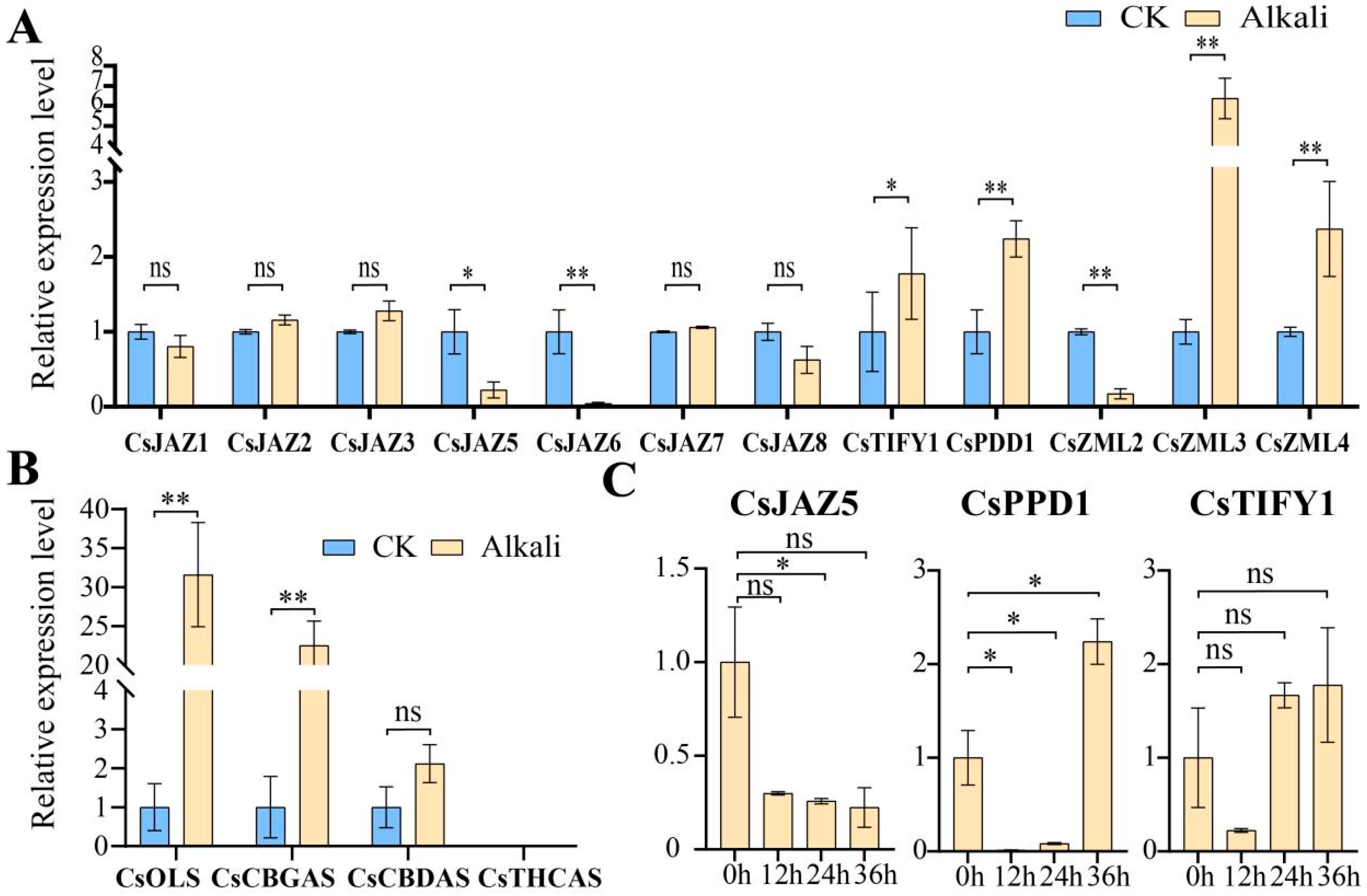
2.6. Expression Patterns of TIFY Transcription Factors in Alkali Stress and Inflorescence Tissue
2.7. Relative Expression Levels of TIFY Under Adverse Stress and MeJA Treatment
3. Discussion
3.1. Classification of the TIFY Family in C. sativa
3.2. Genetic Structure and Classification of TIFY in C. sativa
3.3. Cis-Acting Elements of the TIFY Family
3.4. Homology of the TIFY Family in C. sativa and A. thaliana
3.5. Function Prediction of TIFY Transcription Factors
3.6. RT-qPCR Analysis of Cannabinoid Metabolism Genes
3.7. RT-qPCR Analysis of TIFY Transcription Factors
4. Materials and Methods
4.1. Genome-Wide Identification of TIFY Transcription Factors in C. sativa
4.2. Physicochemical Property Prediction, Chromosome Localization, and Evolutionary Analysis of TIFY Transcription Factor
4.3. Gene Structure and Collinearity Analysis of TIFY Transcription Factors
4.4. Cis-Acting Element Prediction of TIFY Transcription Factors
4.5. Expression Patterns of CsTIFYs in Gender Differences and Adverse Stress
4.6. Stress Treatment and RNA Extraction
4.7. Primer Design and RT-qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shikata, M.; Matsuda, Y.; Ando, K.; Nishii, A.; Takemura, M.; Yokota, A.; Kohchi, T. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 2004, 55, 631–639. [Google Scholar] [CrossRef]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef]
- Bai, Y.; Meng, Y.; Huang, D.; Qi, Y.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [CrossRef]
- Zhao, G.; Song, Y.; Wang, C.; Butt, H.I.; Wang, Q.; Zhang, C.; Yang, Z.; Liu, Z.; Chen, E.; Zhang, X.; et al. Genome-wide identification and functional analysis of the TIFY gene family in response to drought in cotton. Mol. Genet. Genom. 2016, 291, 2173–2187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, M.; Singer, S.D.; Fei, Z.; Wang, H.; Wang, X. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 2012, 7, e44465. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wen, K.; Zhu, L.; Chen, C.; Yin, T.; Yang, X.; Zhao, K.; Zi, Y.; Zhang, H.; Luo, X.; et al. Genome-wide identification and expression analysis of the Eriobotrya japonica TIFY gene family reveals its functional diversity under abiotic stress conditions. BMC Genom. 2024, 25, 468. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wan, Q.; Wang, H.; Guo, C.; Niu, X.; Zhang, X.; Zhang, R.; Chen, Y.; Luo, K. Genome-wide identification and expression of TIFY family in cassava (Manihot esculenta Crantz). Front. Plant Sci. 2022, 13, 1017840. [Google Scholar] [CrossRef]
- Lian, C.; Zhang, B.; Li, J.; Yang, H.; Liu, X.; Ma, R.; Zhang, F.; Liu, J.; Yang, J.; Lan, J.; et al. Genome-wide identification, characterization and expression pattern analysis of TIFY family members in Artemisia argyi. BMC Genom. 2024, 25, 925. [Google Scholar] [CrossRef]
- Guan, Y.; Zhang, Q.; Li, M.; Zhai, J.; Wu, S.; Ahmad, S.; Lan, S.; Peng, D.; Liu, Z.J. Genome-Wide Identification and Expression Pattern Analysis of TIFY Family Genes Reveal Their Potential Roles in Phalaenopsis aphrodite Flower Opening. Int. J. Mol. Sci. 2024, 25, 5422. [Google Scholar] [CrossRef]
- Ebel, C.; BenFeki, A.; Hanin, M.; Solano, R.; Chini, A. Characterization of wheat (Triticum aestivum) TIFY family and role of Triticum Durum TdTIFY11a in salt stress tolerance. PLoS ONE 2018, 13, e0200566. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, F.; Chen, D.; Chu, W.; Liu, H.; Xiang, Y. Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiol. Biochem. 2017, 115, 360–371. [Google Scholar] [CrossRef]
- He, X.; Kang, Y.; Li, W.; Liu, W.; Xie, P.; Liao, L.; Huang, L.; Yao, M.; Qian, L.; Liu, Z.; et al. Genome-wide identification and functional analysis of the TIFY gene family in the response to multiple stresses in Brassica napus L. BMC Genom. 2020, 21, 736. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Ban, Q.; Zhou, L.; Cui, Q. Cloning and tissue expression analysis of TIFY genes in Luffa cylindrica. Acta Bot. Boreali-Occident. Sin. 2024, 44, 1752–1759. [Google Scholar]
- Zhang, H.; Liu, Z.; Geng, R.; Ren, M.; Cheng, L.; Liu, D.; Jiang, C.; Wen, L.; Xiao, Z.; Yang, A. Genome-wide identification of the TIFY gene family in tobacco and expression analysis in response to Ralstonia solanacearum infection. Genomics 2024, 116, 110823. [Google Scholar] [CrossRef] [PubMed]
- White, D.W. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 13238–13243. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Luo, X.; Liu, X.; Wu, W.; Cui, X.; He, Y.; Huang, J. Arabidopsis PEAPODs function with LIKE HETEROCHROMATIN PROTEIN1 to regulate lateral organ growth. J. Integr. Plant Biol. 2020, 62, 812–831. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wen, T.; Ren, A.; Li, D.; Dawood, M.; Wu, J.; Zhao, G. Gossypium arboreum PPD2 facilitates root architecture development to increase plant resilience to salt stress. Physiol. Plant. 2024, 176, e14473. [Google Scholar] [CrossRef]
- White, D.W.R. PEAPOD repressors modulate and coordinate developmental responses to light intensity in Arabidopsis. New Phytol. 2022, 235, 1470–1485. [Google Scholar] [CrossRef]
- Naito, K.; Takahashi, Y.; Chaitieng, B.; Hirano, K.; Kaga, A.; Takagi, K.; Ogiso-Tanaka, E.; Thavarasook, C.; Ishimoto, M.; Tomooka, N. Multiple organ gigantism caused by mutation in VmPPD gene in blackgram (Vigna mungo). Breed. Sci. 2017, 67, 151–158. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Seong, K.; Pang, S.; Song, J.; Gao, H.; Wang, C.; Zhai, J.; Zhang, Y.; Gao, S.; Li, X.; et al. Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in jasmonate and COI1-regulated growth, development, and defense. New Phytol. 2021, 231, 1525–1545. [Google Scholar] [CrossRef]
- Zhang, T.; Meng, L.; Kong, W.; Yin, Z.; Wang, Y.; Schneider, J.D.; Chen, S. Quantitative proteomics reveals a role of JAZ7 in plant defense response to Pseudomonas syringae DC3000. J. Proteom. 2018, 175, 114–126. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Cevik, V.; Grant, M.; Zhai, B.; Jones, J.D.; Manners, J.M.; Kazan, K. Characterization of a JAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. J. Exp. Bot. 2016, 67, 2367–2386. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zheng, D.; Gao, Y.; She, M.; Wu, Z.; Lu, Y.; Zhang, Z. The TIFY transcription factor ZmJAZ13 enhances plant tolerance to drought and salt stress by interacting with ZmbHLH161 and ZmA0A1D6GLB9. Plant Sci. 2025, 352, 112388. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Wang, J.; Xu, K.; Xu, Q.; Liu, X.; Tian, T.; Du, Z.; Wang, J.; Liao, Z.; Wang, B.; et al. Genome-wide insights into the nomenclature, evolution and expression of tobacco TIFY/JAZ genes. Planta 2025, 261, 103. [Google Scholar] [CrossRef]
- Qing, H.; Wu, Z.; Mo, X.; Wei, J.; Shi, Y.; Guo, H.; Xu, J.; Ding, F.; Zhang, S. Genome-Wide Identification of the TIFY Family in Longan and Their Potential Functional Analysis in Anthocyanin Synthesis. Biology 2025, 14, 364. [Google Scholar] [CrossRef]
- Yang, F.; Wang, Z.; Ma, H.; Yang, F.; Tian, C.; An, F. Research and integrated demonstration of ecological amelioration techniques of saline-sodic land in northeast China. Acta Ecol. Sin. 2016, 36, 7054–7058. [Google Scholar] [CrossRef]
- Lin, J.; Song, G.; Zhang, Y. Regionalization planning for prevention and control of saline-alkali land based on a landscape ecology risk pattern theory: A case study in Lindian County, Heilongjiang Province. Acta Ecol. Sin. 2018, 38, 5509–5518. [Google Scholar] [CrossRef]
- Xia, N.; Wang, Y.; Xu, H.; Sun, Y.; Yuan, Y.; Cheng, L.; Jiang, P.; Li, M. Demarcation of Prime Farmland Protection Areas around a Metropolis Based on High-Resolution Satellite Imagery. Sci. Rep. 2016, 6, 37634. [Google Scholar] [CrossRef]
- Cao, K.; Sun, Y.F.; Zhang, X.; Zhao, Y.; Bian, J.; Zhu, H.; Wang, P.; Gao, B.; Sun, X.; Hu, M.; et al. The miRNA-mRNA regulatory networks of the response to NaHCO3 stress in industrial hemp (Cannabis sativa L.). BMC Plant Biol. 2023, 23, 509. [Google Scholar] [CrossRef]
- Cao, K.; Sun, Y.F.; Han, C.W.; Zhang, X.O.; Zhao, Y.; Jiang, Y.; Jiang, Y.Z.; Sun, X.L.; Guo, Y.X.; Wang, X.N. The transcriptome of saline-alkaline resistant industrial hemp (Cannabis sativa L.) exposed to NaHCO3 stress. Ind. Crops Products 2021, 170, 113766. [Google Scholar] [CrossRef]
- Lv, Z.; Niu, L.; Hao, J. A Review of Research on China′s Prime Farmland. Jiangsu Agric. Sci. 2017, 45, 24–27. [Google Scholar]
- Zhang, P.; Duo, T.; Wang, F.; Zhang, X.; Yang, Z.; Hu, G. De novo transcriptome in roots of switchgrass (Panicum virgatum L.) reveals gene expression dynamic and act network under alkaline salt stress. BMC Genom. 2021, 22, 82. [Google Scholar] [CrossRef]
- Jiao, Y.; Xie, R.J.; Jia, H.M. Identification of Potential Pathways of Morella cerifera Seedlings in Response to Alkali Stress via Transcriptomic Analysis. Plants 2022, 11, 1053. [Google Scholar] [CrossRef]
- Zhu, D.; Bai, X.; Chen, C.; Chen, Q.; Cai, H.; Li, Y.; Ji, W.; Zhai, H.; Lv, D.; Luo, X.; et al. GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol. Biol. 2011, 77, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Van Bakel, H.; Stout, J.M.; Cote, A.G.; Tallon, C.M.; Sharpe, A.G.; Hughes, T.R.; Page, J.E. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011, 12, R102. [Google Scholar] [CrossRef] [PubMed]
- Haiden, S.R.; Apicella, P.V.; Ma, Y.; Berkowitz, G.A. Overexpression of CsMIXTA, a Transcription Factor from Cannabis sativa, Increases Glandular Trichome Density in Tobacco Leaves. Plants 2022, 11, 1519. [Google Scholar] [CrossRef]
- Elsohly, M.A.; Slade, D. Chemical constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef]
- Apicella, P.V.; Sands, L.B.; Ma, Y.; Berkowitz, G.A. Delineating genetic regulation of cannabinoid biosynthesis during female flower development in Cannabis sativa. Plant Direct 2022, 6, e412. [Google Scholar] [CrossRef] [PubMed]
- Wieckiewicz, G.; Stokłosa, I.; Stokłosa, M.; Gorczyca, P.; Pudlo, R. Cannabidiol (CBD) in the Self-Treatment of Depression-Exploratory Study and a New Phenomenon of Concern for Psychiatrists. Front. Psychiatry 2022, 13, 837946. [Google Scholar] [CrossRef] [PubMed]
- Dahlgren, M.K.; Lambros, A.M.; Smith, R.T.; Sagar, K.A.; El-Abboud, C.; Gruber, S.A. Clinical and cognitive improvement following full-spectrum, high-cannabidiol treatment for anxiety: Open-label data from a two-stage, phase 2 clinical trial. Commun. Med. 2022, 2, 139. [Google Scholar] [CrossRef] [PubMed]
- Roebuck, A.J.; Greba, Q.; Onofrychuk, T.J.; McElroy, D.L.; Sandini, T.M.; Zagzoog, A.; Simone, J.; Cain, S.M.; Snutch, T.P.; Laprairie, R.B.; et al. Dissociable changes in spike and wave discharges following exposure to injected cannabinoids and smoked cannabis in Genetic Absence Epilepsy Rats from Strasbourg. Eur. J. Neurosci. 2022, 55, 1063–1078. [Google Scholar] [CrossRef]
- Bonn-Miller, M.O.; Sisley, S.; Riggs, P.; Yazar-Klosinski, B.; Wang, J.B.; Loflin, M.J.E.; Shechet, B.; Hennigan, C.; Matthews, R.; Emerson, A.; et al. The short-term impact of 3 smoked cannabis preparations versus placebo on PTSD symptoms: A randomized cross-over clinical trial. PLoS ONE 2021, 16, e0246990. [Google Scholar] [CrossRef]
- Wang, B.; Li, D.; Fiselier, A.; Kovalchuk, I.; Kovalchuk, O. New AKT-dependent mechanisms of anti-COVID-19 action of high-CBD Cannabis sativa extracts. Cell Death Discov. 2022, 8, 110. [Google Scholar] [CrossRef]
- Borroto Fernandez, E.; Peterseil, V.; Hackl, G.; Menges, S.; de Meijer, E.; Staginnus, C. Distribution of Chemical Phenotypes (Chemotypes) in European Agricultural Hemp. (Cannabis sativa L.) Cultivars. J. Forensic Sci. 2020, 65, 715–721. [Google Scholar] [CrossRef]
- Zubcevic, K.; Petersen, M.; Bach, F.W.; Heinesen, A.; Enggaard, T.P.; Almdal, T.P.; Holbech, J.V.; Vase, L.; Jensen, T.S.; Hansen, C.S.; et al. Oral capsules of tetra-hydro-cannabinol (THC), cannabidiol (CBD) and their combination in peripheral neuropathic pain treatment. Eur. J. Pain 2023, 27, 492–506. [Google Scholar] [CrossRef]
- Saloner, A.; Bernstein, N. Nitrogen supply affects cannabinoid and terpenoid profile in medical cannabis (Cannabis sativa L.). Ind. Crops Prod. 2021, 167, 113516. [Google Scholar] [CrossRef]
- Saloner, A.; Bernstein, N. Nitrogen Source Matters: High NH4/NO3 Ratio Reduces Cannabinoids, Terpenoids, and Yield in Medical Cannabis. Front. Plant Sci. 2022, 13, 830224. [Google Scholar] [CrossRef]
- Shiponi, S.; Bernstein, N. The Highs and Lows of P Supply in Medical Cannabis: Effects on Cannabinoids, the Ionome, and Morpho-Physiology. Front. Plant Sci. 2021, 12, 657323. [Google Scholar] [CrossRef] [PubMed]
- Baas, R.; Wijnen, D. Salinity effects on yield and nutrient uptake in Cannabis sativa L. In XXXI International Horticultural Congress (IHC2022): International Symposium on Innovative Technologies and Production; ISHS Acta Horticulturae: Leuven, Belgium, 2023; Volume 1377, pp. 785–792. [Google Scholar]
- Saloner, A.; Sade, Y.; Bernstein, N. To flush or not to flush: Does flushing the growing media affect cannabinoid and terpenoid production in cannabis? Ind. Crops Prod. 2024, 220, 119157. [Google Scholar] [CrossRef]
- Rodriguez-Morrison, V.; Llewellyn, D.; Zheng, Y. Cannabis Yield, Potency, and Leaf Photosynthesis Respond Differently to Increasing Light Levels in an Indoor Environment. Front Plant Sci. 2021, 12, 646020. [Google Scholar] [CrossRef] [PubMed]
- Booth, J.K.; Page, J.E.; Bohlmann, J. Terpene synthases from Cannabis sativa. PLoS ONE 2017, 12, e0173911. [Google Scholar] [CrossRef]
- Booth, J.K.; Yuen, M.M.S.; Jancsik, S.; Madilao, L.L.; Page, J.E.; Bohlmann, J. Terpene Synthases and Terpene Variation in Cannabis sativa. Plant Physiol. 2020, 184, 130–147. [Google Scholar] [CrossRef]
- Zager, J.J.; Lange, I.; Srividya, N.; Smith, A.; Lange, B.M. Gene Networks Underlying Cannabinoid and Terpenoid Accumulation in Cannabis. Plant Physiol. 2019, 180, 1877–1897. [Google Scholar] [CrossRef]
- Lynch, R.C.; Padgitt-Cobb, L.K.; Garfinkel, A.R.; Knaus, B.J.; Hartwick, N.T.; Allsing, N.; Aylward, A.; Bentz, P.C.; Carey, S.B.; Mamerto, A.; et al. Domesticated cannabinoid synthases amid a wild mosaic cannabis pangenome. Nature 2025, 643, 1001–1010. [Google Scholar] [CrossRef]
- Li, J.; Terzaghi, W.; Deng, X.W. Genomic basis for light control of plant development. Protein Cell 2012, 3, 106–116. [Google Scholar] [CrossRef]
- Zhao, Z.; Meng, G.; Zamin, I.; Wei, T.; Ma, D.; An, L.; Yue, X. Genome-Wide Identification and Functional Analysis of the TIFY Family Genes in Response to Abiotic Stresses and Hormone Treatments in Tartary Buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2023, 24, 10916. [Google Scholar] [CrossRef]
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.; Zhang, W.; Su, Y.; Xiao, L.; Deng, H.; Xie, D. Injury Activates Ca2+/Calmodulin-Dependent Phosphorylation of JAV1-JAZ8-WRKY51 Complex for Jasmonate Biosynthesis. Mol. Cell 2018, 70, 136–149. [Google Scholar] [CrossRef]
- Thireault, C.; Shyu, C.; Yoshida, Y.; St Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Sng, B.J.R.; Jeong, Y.J.; Leong, S.H.; Jeong, J.C.; Lee, J.; Rajani, S.; Kim, C.Y.; Jang, I.C. Genome-wide identification of cannabinoid biosynthesis genes in non-drug type Cannabis (Cannabis sativa L.) cultivar. J. Cannabis Res. 2024, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Sun, X. Research Progress on Cannabinoids in Cannabis (Cannabis sativa L.) in China. Molecules 2023, 28, 3806. [Google Scholar] [CrossRef] [PubMed]
- Szalata, M.; Dreger, M.; Zielińska, A.; Banach, J.; Szalata, M.; Wielgus, K. Simple Extraction of Cannabinoids from Female Inflorescences of Hemp (Cannabis sativa L.). Molecules 2022, 27, 5868. [Google Scholar] [CrossRef]
- Fulvio, F.; Paris, R.; Montanari, M.; Citti, C.; Cilento, V.; Bassolino, L.; Moschella, A.; Alberti, I.; Pecchioni, N.; Cannazza, G.; et al. Analysis of Sequence Variability and Transcriptional Profile of Cannabinoid synthase Genes in Cannabis sativa L. Chemotypes with a Focus on Cannabichromenic acid synthase. Plants 2021, 10, 1857. [Google Scholar] [CrossRef]
- Welling, M.T.; Liu, L.; Kretzschmar, T.; Mauleon, R.; Ansari, O.; King, G.J. An extreme-phenotype genome-wide association study identifies candidate cannabinoid pathway genes in Cannabis. Sci. Rep. 2020, 10, 18643. [Google Scholar] [CrossRef]
- Stout, J.M.; Boubakir, Z.; Ambrose, S.J.; Purves, R.W.; Page, J.E. The hexanoyl-CoA precursor for cannabinoid biosynthesis is formed by an acyl-activating enzyme in Cannabis sativa trichomes. Plant J. 2012, 71, 353–365. [Google Scholar] [CrossRef]
- Park, S.H.; Pauli, C.S.; Gostin, E.L.; Staples, S.K.; Seifried, D.; Kinney, C.; Vanden Heuvel, B.D. Effects of short-term environmental stresses on the onset of cannabinoid production in young immature flowers of industrial hemp (Cannabis sativa L.). J. Cannabis Res. 2022, 4, 1. [Google Scholar] [CrossRef]
- Laverty, K.U.; Stout, J.M.; Sullivan, M.J.; Shah, H.; Gill, N.; Holbrook, L.; Deikus, G.; Sebra, R.; Hughes, T.R.; Page, J.E.; et al. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Res. 2019, 29, 146–156. [Google Scholar] [CrossRef]
- Taura, F.; Morimoto, S.; Shoyama, Y. Purification and characterization of cannabidiolic-acid synthase from Cannabis sativa L. Biochemical analysis of a novel enzyme that catalyzes the oxidocyclization of cannabigerolic acid to cannabidiolic acid. J. Biol. Chem. 1996, 271, 17411–17416. [Google Scholar] [CrossRef]
- Zirpel, B.; Kayser, O.; Stehle, F. Elucidation of structure-function relationship of THCA and CBDA synthase from Cannabis sativa L. J. Biotechnol. 2018, 284, 17–26. [Google Scholar] [CrossRef]
- Rodziewicz, P.; Loroch, S.; Marczak, Ł.; Sickmann, A.; Kayser, O. Cannabinoid synthases and osmoprotective metabolites accumulate in the exudates of Cannabis sativa L. glandular trichomes. Plant Sci. 2019, 284, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Mahlberg, P.G.; Kim, E.S. Accumulation of Cannabinoids in Glandular Trichomes of Cannabis (Cannabaceae). J. Ind. Hemp 2004, 9, 15–36. [Google Scholar] [CrossRef]
- Wilson, W.B.; Yarberry, A.J.; Goldman, S. Chromatographic interferences potentially inflating the levels of Δ9-THC in Cannabis Sativa plant samples and possible solutions. J. Chromatogr. A 2025, 1748, 465871. [Google Scholar] [CrossRef] [PubMed]
- Timoteo Junior, A.A.; Oswald, I.W. Optimized guidelines for feminized seed production in high-THC Cannabis cultivars. Front. Plant Sci. 2024, 15, 1384286. [Google Scholar] [CrossRef]
- Wu, S.; Hu, C.; Zhu, C.; Fan, Y.; Zhou, J.; Xia, X.; Shi, K.; Zhou, Y.; Foyer, C.H.; Yu, J. The MYC2-PUB22-JAZ4 module plays a crucial role in jasmonate signaling in tomato. Mol. Plant 2024, 17, 598–613. [Google Scholar] [CrossRef]
- Gupta, A.; Bhardwaj, M.; Tran, L.P. JASMONATE ZIM-DOMAIN Family Proteins: Important Nodes in Jasmonic Acid-Abscisic Acid Crosstalk for Regulating Plant Response to Drought. Curr. Protein Pept. Sci. 2021, 22, 759–766. [Google Scholar] [CrossRef]
- Li, X.; Bashir, A.; Yang, H.; Abbas, A.; Li, Y.; Zeng, X.; Zhu, L.; Shi, Q.; Tursunniyaz, M.; Zhang, L. Genome-Wide Analysis and Expression Profiling of the JAZ Gene Family in Response to Abiotic Stress in Alfalfa. Int. J. Mol. Sci. 2025, 26, 4684. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, X.; Li, Y.; Zhang, L.; Zhang, Y.; Wang, X.; He, F.; Li, M.; Zhang, T.; Kang, J. Genome-Wide Identification, Phylogenetic, and Expression Analysis of Jasmonate ZIM-Domain Gene Family in Medicago sativa L. Int. J. Mol. Sci. 2024, 25, 10589. [Google Scholar] [CrossRef]
- Liu, Y.L.; Zheng, L.; Jin, L.G.; Liu, Y.X.; Kong, Y.N.; Wang, Y.X.; Yu, T.F.; Chen, J.; Zhou, Y.B.; Chen, M.; et al. Genome-Wide Analysis of the Soybean TIFY Family and Identification of GmTIFY10e and GmTIFY10g Response to Salt Stress. Front Plant Sci. 2022, 13, 845314. [Google Scholar] [CrossRef]
- Li, G.; Manzoor, M.A.; Chen, R.; Zhang, Y.; Song, C. Genome-wide identification and expression analysis of TIFY genes under MeJA, cold and PEG-induced drought stress treatment in Dendrobium huoshanense. Physiol. Mol. Biol. Plants 2024, 30, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liang, S.; Yang, W.; Yu, K.; Liang, F.; Zhao, B.; Zhu, X.; Zhou, C.; Mur, L.A.J.; Roberts, J.A.; et al. MetMiner: A user-friendly pipeline for large-scale plant metabolomics data analysis. J. Integr. Plant Biol. 2024, 66, 2329–2345. [Google Scholar] [CrossRef]
- Yuan, H.; Si, H.; Ye, Y.; Ji, Q.; Wang, H.; Zhang, Y. Arbuscular Mycorrhizal Fungi-Mediated Modulation of Physiological, Biochemical, and Secondary Metabolite Responses in Hemp (Cannabis sativa L.) under Salt and Drought Stress. J. Fungi 2024, 10, 283. [Google Scholar] [CrossRef]
- Guo, R.; Guo, H.; Zhang, Q.; Guo, M.; Xu, Y.; Zeng, M.; Lv, P.; Chen, X.; Yang, M. Evaluation of reference genes for RT-qPCR analysis in wild and cultivated Cannabis. Biosci. Biotechnol. Biochem. 2018, 82, 1902–1910. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]


| Name | Gene ID | Number of Amino Acid/Amino Acid | Molecular Weight/kDa | pI | Instability Index | GRAVY | Alpha Helix | Extended Strand | Random Coil |
|---|---|---|---|---|---|---|---|---|---|
| CsPPD1 | LOC115723116 | 326 | 35,518.60 | 8.06 | 53.57 | −0.728 | 21.78% | 5.21% | 73.01% |
| CsJAZ1 | LOC115711241 | 382 | 40,204.84 | 9.35 | 53.22 | −0.387 | 10.73% | 4.71% | 84.55% |
| CsJAZ2 | LOC115706817 | 262 | 28,721.44 | 9.16 | 51.54 | −0.528 | 14.89% | 5.73% | 79.39% |
| CsJAZ3 | LOC115706226 | 262 | 28,672.37 | 9.18 | 50.96 | −0.537 | 13.36% | 6.11% | 80.53% |
| CsJAZ4 | LOC115712145 | 199 | 22,310.31 | 9.04 | 58.72 | −0.459 | 14.57% | 8.04% | 77.39% |
| CsJAZ5 | LOC115714189 | 391 | 42,034.53 | 8.74 | 49.53 | −0.295 | 7.67% | 5.63% | 86.70% |
| CsJAZ6 | LOC115705512 | 193 | 21,190.96 | 5.83 | 75.37 | −0.577 | 9.84% | 7.77% | 82.38% |
| CsJAZ7 | LOC115707122 | 315 | 34,251.27 | 8.93 | 49.35 | −0.688 | 12.38% | 4.13% | 83.49% |
| CsJAZ8 | LOC115696976 | 156 | 17,524.89 | 9.16 | 63.53 | −0.552 | 17.31% | 6.41% | 76.28% |
| CsTIFY1 | LOC115699142 | 442 | 46,479.54 | 9.13 | 51.16 | −0.587 | 6.56% | 7.01% | 86.43% |
| CsZML1 | LOC115697696 | 301 | 32,461.79 | 6.21 | 38.01 | −0.654 | 4.32% | 6.98% | 88.70% |
| CsZML2 | LOC115719706 | 317 | 33,897.48 | 5.57 | 34.85 | −0.656 | 3.79% | 3.15% | 93.06% |
| CsZML3 | LOC115697707 | 339 | 37,386.45 | 5.01 | 43.30 | −0.703 | 7.67% | 4.13% | 88.20% |
| CsZML4 | LOC115718519 | 388 | 42,163.76 | 4.75 | 49.51 | −0.542 | 7.47% | 4.12% | 88.40% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhang, M.; Fang, Y.; Zheng, N.; Yan, B.; Sui, Y.; Zhang, L. Genome-Wide Identification of the TIFY Family in Cannabis sativa L. and Its Potential Functional Analysis in Response to Alkaline Stress and in Cannabinoid Metabolism. Int. J. Mol. Sci. 2025, 26, 8171. https://doi.org/10.3390/ijms26178171
Zhang Y, Zhang M, Fang Y, Zheng N, Yan B, Sui Y, Zhang L. Genome-Wide Identification of the TIFY Family in Cannabis sativa L. and Its Potential Functional Analysis in Response to Alkaline Stress and in Cannabinoid Metabolism. International Journal of Molecular Sciences. 2025; 26(17):8171. https://doi.org/10.3390/ijms26178171
Chicago/Turabian StyleZhang, Yuanye, Ming Zhang, Yuyan Fang, Nan Zheng, Bowei Yan, Yue Sui, and Liguo Zhang. 2025. "Genome-Wide Identification of the TIFY Family in Cannabis sativa L. and Its Potential Functional Analysis in Response to Alkaline Stress and in Cannabinoid Metabolism" International Journal of Molecular Sciences 26, no. 17: 8171. https://doi.org/10.3390/ijms26178171
APA StyleZhang, Y., Zhang, M., Fang, Y., Zheng, N., Yan, B., Sui, Y., & Zhang, L. (2025). Genome-Wide Identification of the TIFY Family in Cannabis sativa L. and Its Potential Functional Analysis in Response to Alkaline Stress and in Cannabinoid Metabolism. International Journal of Molecular Sciences, 26(17), 8171. https://doi.org/10.3390/ijms26178171






