Dynamic Genetic Changes Reveal: Intra-Lineage Diversity, Not Admixture, Explains Amaranthus palmeri’s Success in China
Abstract
1. Introduction
2. Results
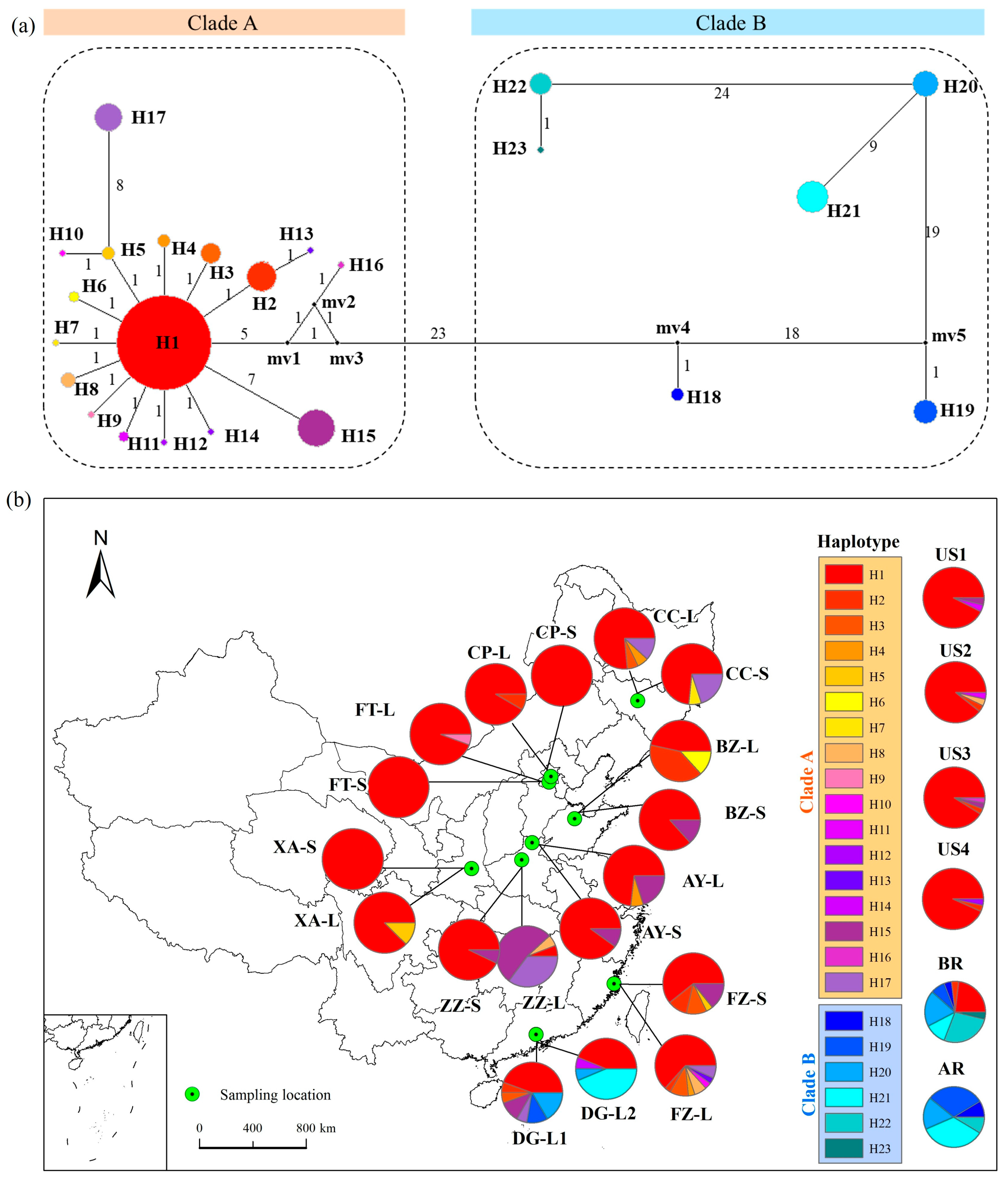
2.1. Genetic Lineages and Pre-Introduction Source of cpDNA Haplotypes
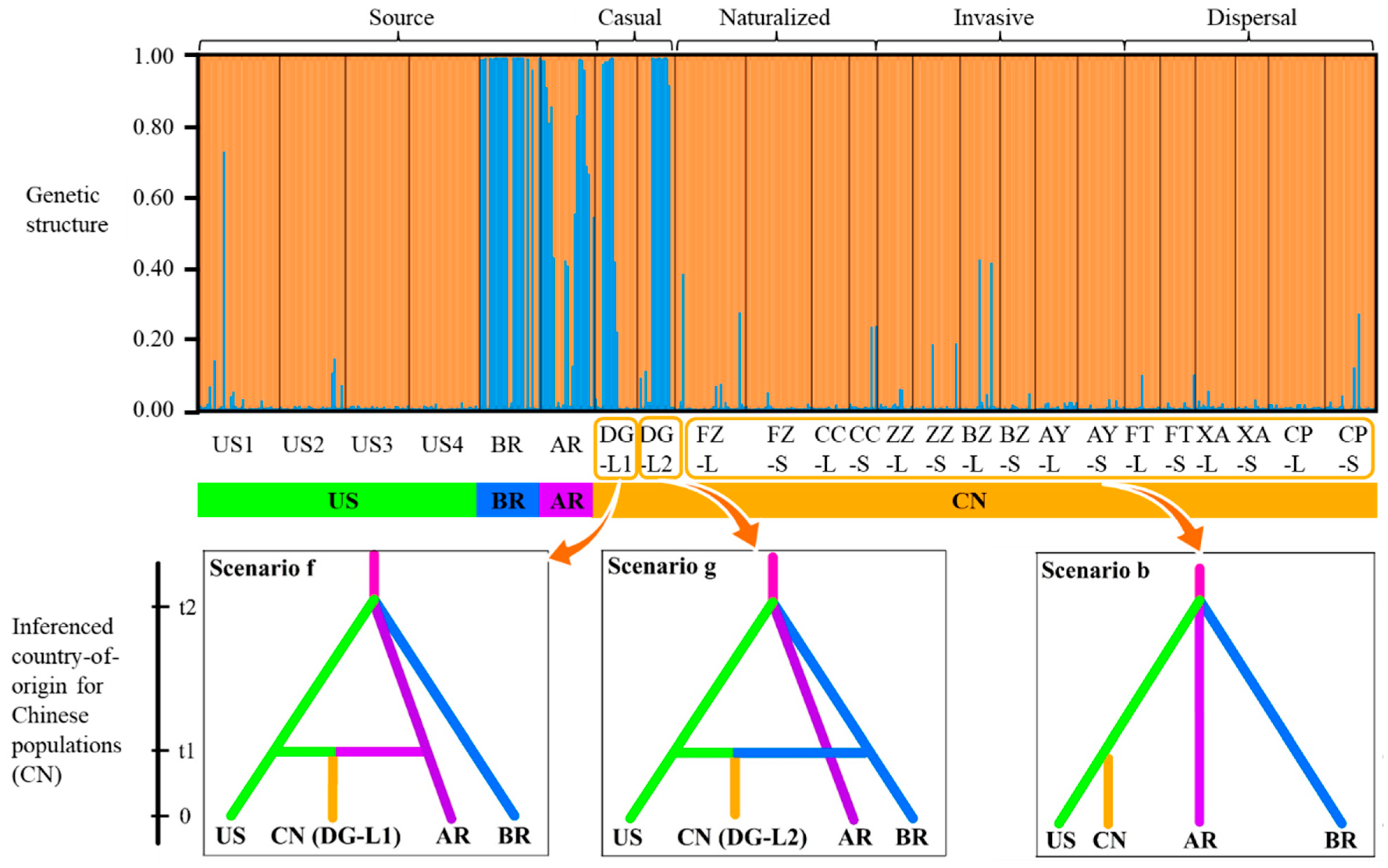
2.2. Post-Introduction Population Structure and Genetic Differentiation
2.3. Inferenced Country-of-Origin and Introduction Histories
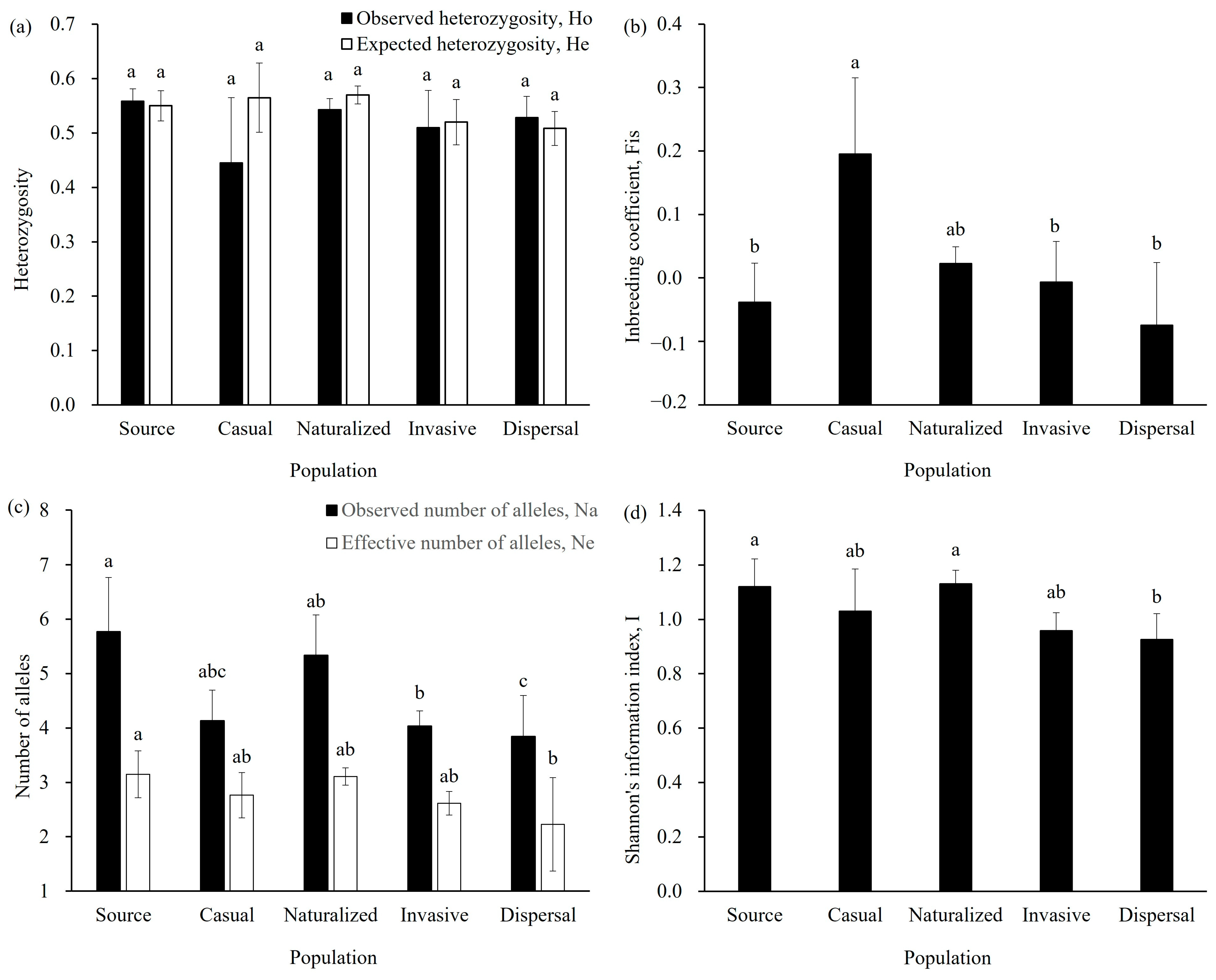
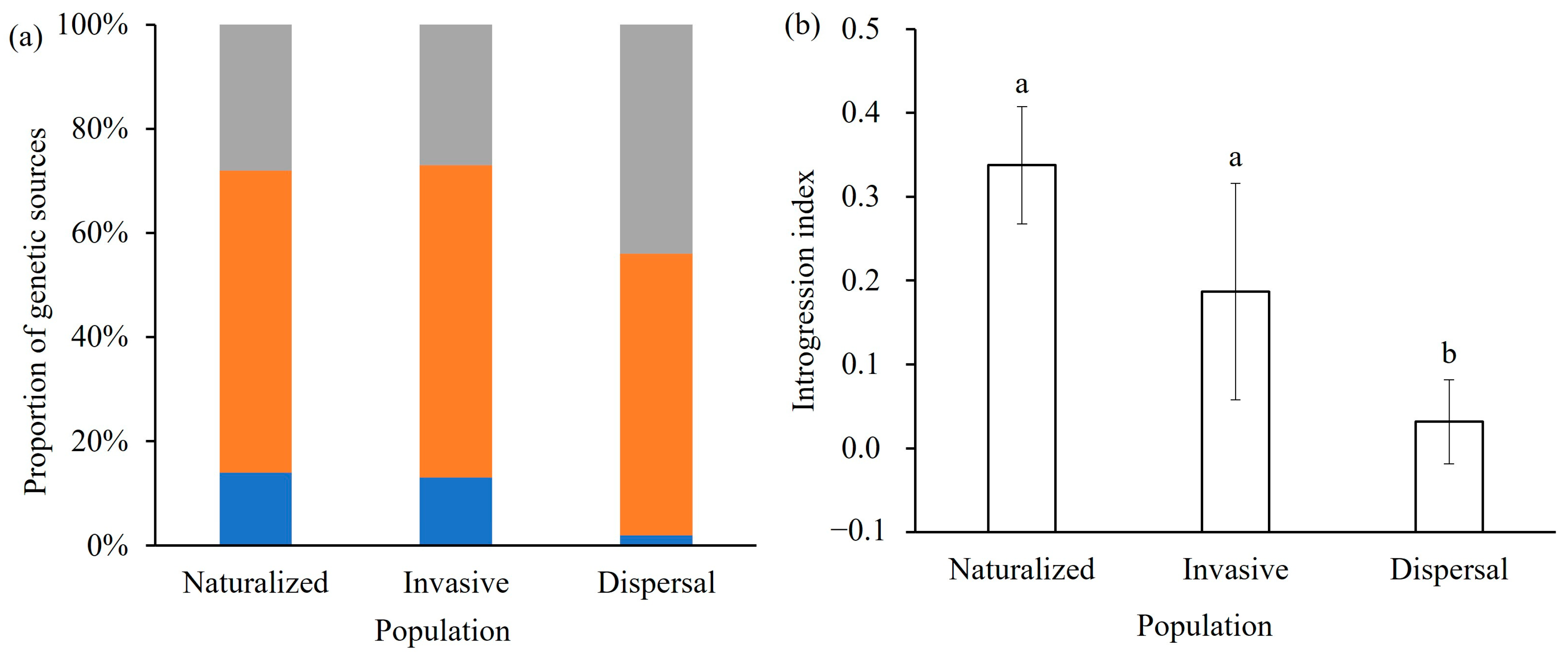
2.4. Genetic Diversity and Hybridization Proportion
2.5. Spatial-Temporal Dynamics with Bottlenecks and Expansions
3. Discussion
3.1. North American Lineage Solely Drives Invasion Success
3.2. Declining South American Genetic Introgression During Invasion
3.3. Inter-Population Crosses Within North American Lineage Facilitates Invasion
4. Materials and Methods
4.1. Study Species and Sample Collection
4.2. DNA Extraction and Molecular Identification
4.3. Sequencing and Data Analysis
4.3.1. Genetic Lineages and Pre-Introduction Source Analysis
4.3.2. Post-Introduction Population Structure and Genetic Differentiation Analysis
4.3.3. Inferring Country-of-Origin and History of Introduced Populations
4.3.4. Genetic Diversity and Hybridization Detection
4.3.5. Spatial-Temporal Dynamics of Genetic Bottleneck and Expansion Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diagne, C.; Leroy, B.; Vaissière, A.C.; Gozlan, R.E.; Roiz, D.; Jarić, I.; Salles, J.M.; Bradshaw, C.J.A.; Courchamp, F. High and rising economic costs of biological invasions worldwide. Nature 2021, 592, 571–576. [Google Scholar] [CrossRef]
- Pyšek, P.; Hulme, P.E.; Simberloff, D.; Bacher, S.; Blackburn, T.M.; Carlton, J.T.; Dawson, W.; Essl, F.; Foxcroft, L.C.; Genovesi, P.; et al. Scientists’ warning on invasive alien species. Biol. Rev. Camb. Philos. Soc. 2021, 95, 1511–1534. [Google Scholar] [CrossRef] [PubMed]
- van Boheemen, L.A.; Lombaert, E.; Nurkowski, K.A.; Gauffre, B.; Rieseberg, L.H.; Hodgins, K.A. Multiple introductions, admixture and bridgehead invasion characterize the introduction history of Ambrosia artemisiifolia in Europe and Australia. Mol. Ecol. 2017, 26, 5421–5434. [Google Scholar] [CrossRef] [PubMed]
- Rius, M.; Darling, J.A. How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol. Evol. 2014, 29, 233–242. [Google Scholar] [CrossRef]
- Verhoeven, K.J.; Macel, M.; Wolfe, L.M.; Biere, A. Population admixture, biological invasions and the balance between local adaptation and inbreeding depression. Proc. R. Soc. Biol. Sci. 2011, 278, 2–8. [Google Scholar] [CrossRef]
- Shi, J.; Joshi, J.; Tielbörger, K.; Verhoeven, K.J.F.; Macel, M. Costs and benefits of admixture between foreign genotypes and local populations in the field. Ecol. Evol. 2018, 8, 3675–3684. [Google Scholar] [CrossRef]
- VanWallendael, A.; Alvarez, M.; Franks, S.J. Patterns of population genomic diversity in the invasive Japanese knotweed species complex. Am. J. Bot. 2021, 108, 857–868. [Google Scholar] [CrossRef]
- Lenormand, T. Gene flow and the limits to natural selection. Trends Ecol. Evol. 2002, 17, 183–189. [Google Scholar] [CrossRef]
- Hahn, M.A.; Rieseberg, L.H. Genetic admixture and heterosis may enhance the invasiveness of common ragweed. Evol. Appl. 2016, 10, 241–250. [Google Scholar] [CrossRef]
- Brandenburg, J.T.; Mary-Huard, T.; Rigaill, G.; Hearne, S.J.; Corti, H.; Joets, J.; Vitte, C.; Charcosset, A.; Nicolas, S.D.; Tenaillon, M.I. Independent introductions and admixtures have contributed to adaptation of European maize and its American counterparts. PLoS Genet. 2017, 13, e1006666. [Google Scholar] [CrossRef] [PubMed]
- Johansen-Morris, A.D.; Latta, R.G. Fitness consequences of hybridization between ecotypes of Avena barbata: Hybrid breakdown, hybrid vigor, and transgressive segregation. Evolution 2006, 60, 1585–1595. [Google Scholar] [CrossRef]
- Barker, B.S.; Cocio, J.E.; Anderson, S.R.; Braasch, J.E.; Cang, F.A.; Gillette, H.D.; Dlugosch, K.M. Potential limits to the benefits of admixture during biological invasion. Mol. Ecol. 2019, 28, 100–113. [Google Scholar] [CrossRef]
- Schneemann, H.; Munzur, A.D.; Thompson, K.A.; Welch, J.J. The diverse effects of phenotypic dominance on hybrid fitness. Evolution 2022, 76, 2846–2863. [Google Scholar] [CrossRef]
- Shi, J.; Macel, M.; Tielbörger, K.; Verhoeven, K.J.F. Effects of admixture in native and invasive populations of Lythrum salicaria. Biol. Invasions 2018, 20, 2381–2393. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, J.J. (Ed.) Invasion genetics: Molecular genetic insights into the spatial and temporal dynamics of biological invasions. In The Evolutionary Ecology of Invasive Species; Elsevier: Amsterdam, The Netherlands, 2022; pp. 159–188. [Google Scholar] [CrossRef]
- Mabin, C.A.; Robinson, T.B.; Wilson, J.R.; Hirsch, H.; Castillo, M.L.; Jooste, M.; Le Roux, J.J. Molecular insights into the invasion dynamics of Carcinus crabs in South Africa. Biol. Invasions 2022, 24, 3597–3613. [Google Scholar] [CrossRef]
- Sherpa, S.; Després, L. The evolutionary dynamics of biological invasions: A multi-approach perspective. Evol. Appl. 2021, 14, 1463–1484. [Google Scholar] [CrossRef]
- Blackburn, T.M.; Pyšek, P.; Bacher, S.; Carlton, J.T.; Duncan, R.P.; Jarošík, V.; Wilson, J.R.; Richardson, D.M. A proposed unified framework for biological invasions. Trends Ecol. Evol. 2011, 26, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.M.; Pyšek, P.; Rejmánek, M.; Barbour, M.G.; Panetta, F.D.; West, C.J. Naturalization and invasion of alien plants: Concepts and definitions. Divers. Distrib. 2000, 6, 93–107. [Google Scholar] [CrossRef]
- Bock, D.G.; Caseys, C.; Cousens, R.D.; Hahn, M.A.; Heredia, S.M.; Hübner, S.; Turner, K.G.; Whitney, K.D.; Rieseberg, L.H. What we still don’t know about invasion genetics. Mol. Ecol. 2015, 24, 2277–2297. [Google Scholar] [CrossRef]
- van Kleunen, M.; Dawson, W.; Essl, F.; Pergl, J.; Winter, M.; Weber, E.; Kreft, H.; Weigelt, P.; Kartesz, J.; Nishino, M.; et al. Global exchange and accumulation of non-native plants. Nature 2015, 525, 100–103. [Google Scholar] [CrossRef]
- Jackowiak, B.; Celka, Z.; Chmiel, J.; Latowski, K.; Żukowski, W. Checklist of the vascular flora of Wielkopolska (Poland): Casual alien species. Biodivers. Res. Conserv. 2017, 46, 35–55. [Google Scholar] [CrossRef]
- Brandes, U.; Furevik, B.B.; Nielsen, L.R.; Kjær, E.D.; Rosef, L.; Fjellheim, S. Introduction history and population genetics of intracontinental scotch broom (Cytisus scoparius) invasion. Divers. Distrib. 2019, 25, 1773–1786. [Google Scholar] [CrossRef]
- Ji, M.; Ze, S.; Tang, S.; Hu, L.; Li, J. Independent origins of populations from Dehong State, Yunnan Province, and the multiple introductions and post-introduction admixture sources of mile-a-minute (Mikania micrantha) in China. Weed Sci. 2021, 69, 439–445. [Google Scholar] [CrossRef]
- Marchi, N.; Schlichta, F.; Excoffier, L. Demographic inference. Curr. Biol. 2021, 31, R276–R279. [Google Scholar] [CrossRef]
- Curtis, S.E.; Clegg, M.T. Molecular evolution of chloroplast DNA sequences. Mol. Biol. Evol. 1984, 1, 291–301. [Google Scholar] [CrossRef][Green Version]
- Hufbauer, R.A.; Sforza, R. Multiple introductions of two invasive Centaurea taxa inferred from cpDNA haplotypes. Divers. Distrib. 2008, 14, 252–261. [Google Scholar] [CrossRef]
- Bairu, M.W.; Amelework, A.B.; Coetzer, W.G. Genetic diversity and population structure of six South African Acacia mearnsii breeding populations based on SSR markers. J. Plant Res. 2021, 134, 1243–1252. [Google Scholar] [CrossRef]
- Kropf, M.; Huppenberger, A.; Karrer, G. Genetic structuring and diversity patterns along rivers—Local invasion history of Ambrosia artemisiifolia (Asteraceae) along the Danube River in Vienna (Austria) shows non-linear pattern. Weed Res. 2018, 58, 131–140. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.L.; Pei, S.Y.; Ning, M.M.; Tang, S.Q. Genetic diversity and population structure of Camellia huana (Theaceae), a limestone species with narrow geographic range, based on chloroplast DNA sequence and microsatellite markers. Plant Divers. 2020, 42, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.Q.; Wei, F.; Zeng, L.Y.; Li, X.K.; Tang, S.C.; Zhong, Y.; Geng, Y.P. Multiple introductions are responsible for the disjunct distributions of invasive Parthenium hysterophorus in China: Evidence from nuclear and chloroplast DNA. Weed Res. 2009, 49, 373–380. [Google Scholar] [CrossRef]
- Keller, S.R.; Gilbert, K.J.; Fields, P.D.; Taylor, D.R. Bayesian inference of a complex invasion history revealed by nuclear and chloroplast genetic diversity in the colonizing plant, Silene latifolia. Mol. Ecol. 2012, 21, 4721–4734. [Google Scholar] [CrossRef] [PubMed]
- Wahocho, S.A.; Cao, Y.; Xu, J.; Qi, D.; Wahocho, N.A.; Gul, H.; Dong, X.; Tian, L.; Huo, H.; Liu, C.; et al. Origin and dissemination route of pear accessions from Western China to abroad based on combined analysis of SSR and cpDNA markers. Genet. Resour. Crop Evol. 2019, 67, 107–128. [Google Scholar] [CrossRef]
- Li, Z.Y. Amaranthus palmeri S. Watson, a newly naturalized species in China. Chin. Bull. Bot. 2003, 20, 734–735, (In Chinese with English abstract). [Google Scholar]
- Cao, J.; Wu, Q.; Wan, F.; Guo, J.; Wang, R. Reliable and rapid identification of glyphosate-resistance in the invasive weed Amaranthus palmeri in China. Pest Manag. Sci. 2022, 78, 2173–2182. [Google Scholar] [CrossRef]
- A growing problem. Nature 2014, 510, 187. [CrossRef]
- Gaines, T.A.; Slavov, G.T.; Hughes, D.; Küpper, A.; Sparks, C.D.; Oliva, J.; Vila-Aiub, M.M.; Garcia, M.A.; Merotto, A., Jr.; Neve, P. Investigating the origins and evolution of a glyphosate-resistant weed invasion in South America. Mol. Ecol. 2021, 30, 5360–5372. [Google Scholar] [CrossRef]
- Shimono, A.; Kanbe, H.; Nakamura, S.; Ueno, S.; Yamashita, J.; Asai, M. Initial invasion of glyphosate-resistant Amaranthus palmeri around grain-import ports in Japan. Plants People Planet 2020, 2, 640–648. [Google Scholar] [CrossRef]
- Vallejo-Marín, M.; Friedman, J.; Twyford, A.D.; Lepais, O.; Ickert-Bond, S.M.; Streisfeld, M.A.; Yant, L.; van Kleunen, M.; Rotter, M.C.; Puzey, J.R. Population genomic and historical analysis suggests a global invasion by bridgehead processes in Mimulus guttatus. Commun. Biol. 2021, 4, 327. [Google Scholar] [CrossRef]
- Cao, J.; Wang, R.; Li, Y.; Zhang, G.; Guo, J.; Wan, F. The phenotypic variation and environmental adaptability among different geographical populations of Amaranthus palmeri in China. Plant Quar. 2020, 34, 25–31, (In Chinese with English abstract). [Google Scholar] [CrossRef]
- Chapple, D.G.; Miller, K.A.; Kraus, F.; Thompson, M.B. Divergent introduction histories among invasive populations of the delicate skink (Lampropholis delicata): Has the importance of genetic admixture in the success of biological invasions been overemphasized? Divers. Distrib. 2013, 19, 134–146. [Google Scholar] [CrossRef]
- Genton, B.J.; Shykoff, J.A.; Giraud, T. High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Mol. Ecol. 2005, 14, 4275–4285. [Google Scholar] [CrossRef]
- Roberts, J.D.; Florentine, S.K. A review of the biology, distribution patterns and management of the invasive species Amaranthus palmeri S. Watson (Palmer amaranth): Current and future management challenges. Weed Res. 2021, 62, 113–122. [Google Scholar] [CrossRef]
- Keller, S.R.; Taylor, D.R. Genomic admixture increases fitness during a biological invasion. J. Evol. Biol. 2010, 23, 1720–1731. [Google Scholar] [CrossRef]
- McFarlane, S.E.; Pemberton, J.M. Detecting the true extent of introgression during anthropogenic hybridization. Trends Ecol. Evol. 2019, 34, 315–326. [Google Scholar] [CrossRef]
- Gaines, T.A.; Ward, S.M.; Bukun, B.; Preston, C.; Leach, J.E.; Westra, P. Interspecific hybridization transfers a previously unknown glyphosate resistance mechanism in Amaranthus species. Evol. Appl. 2012, 5, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez, V.L.; Pascual, M.; Rius, M.; Turon, X. Mixed but not admixed: A spatial analysis of genetic variation of an invasive ascidian on natural and artificial substrates. Mar. Biol. 2013, 160, 1645–1660. [Google Scholar] [CrossRef]
- Qiao, H.; Liu, W.; Zhang, Y.; Zhang, Y.Y.; Li, Q.Q. Genetic admixture accelerates invasion via provisioning rapid adaptive evolution. Mol. Ecol. 2019, 28, 4012–4027. [Google Scholar] [CrossRef] [PubMed]
- Oakley, C.G.; Ågren, J.; Schemske, D.W. Heterosis and outbreeding depression in crosses between natural populations of Arabidopsis thaliana. Heredity 2015, 115, 73–82. [Google Scholar] [CrossRef]
- Li, Y.; Stift, M.; van Kleunen, M. Admixture increases performance of an invasive plant beyond first-generation heterosis. J. Ecol. 2018, 106, 1595–1606. [Google Scholar] [CrossRef]
- Koski, M.H.; Galloway, L.F.; Busch, J.W. Hybrid breakdown is elevated near the historical cores of a species’ range. Proc. Biol. Sci. 2022, 289, 20220070. [Google Scholar] [CrossRef]
- Zenni, R.D.; Nuñez, M.A. The elephant in the room: The role of failed invasions in understanding invasion biology. Oikos 2013, 122, 801–815. [Google Scholar] [CrossRef]
- Burrell, A.M.; Pepper, A.E.; Hodnett, G.; Goolsby, J.A.; Overholt, W.A.; Racelis, A.E.; Diaz, R.; Klein, P.E. Exploring origins, invasion history and genetic diversity of Imperata cylindrica (L.) P. Beauv. (Cogongrass) in the United States using genotyping by sequencing. Mol. Ecol. 2015, 24, 2177–2193. [Google Scholar] [CrossRef] [PubMed]
- Dlugosch, K.M.; Parker, I.M. Founding events in species invasions: Genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 2008, 17, 431–449. [Google Scholar] [CrossRef]
- Aguillon, S.M.; Dodge, T.O.; Preising, G.A.; Schumer, M. Introgression. Curr. Biol. 2022, 32, R865–R868. [Google Scholar] [CrossRef]
- Vicente, S.; Máguas, C.; Richardson, D.M.; Trindade, H.; Wilson, J.R.U.; Le Roux, J.J. Highly diverse and highly successful: Invasive Australian acacias have not experienced genetic bottlenecks globally. Ann. Bot. 2021, 128, 149–157. [Google Scholar] [CrossRef]
- Falahati-Anbaran, M.; Rijal, D.P.; Lundemo, S.; Alsos, I.G.; Stenoien, H.K. Disentangling the genetic origin of Heracleum persicum (Apiaceae) in Europe: Multiple introductions from multiple source populations. Biol. Invasions 2021, 23, 3871–3890. [Google Scholar] [CrossRef]
- Lippman, Z.B.; Zamir, D. Heterosis: Revisiting the magic. Trends Genet. 2007, 23, 60–66. [Google Scholar] [CrossRef]
- Rosenthal, D.M.; Ramakrishnan, A.P.; Cruzan, M.B. Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol. Ecol. 2008, 17, 4657–4669. [Google Scholar] [CrossRef]
- Wagner, N.K.; Ochocki, B.M.; Crawford, K.M.; Compagnoni, A.; Miller, T.E. Genetic mixture of multiple source populations accelerates invasive range expansion. J. Anim. Ecol. 2017, 86, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Dormontt, E.E.; Gardner, M.G.; Breed, M.F.; Rodger, J.G.; Prentis, P.J.; Lowe, A.J. Genetic bottlenecks in time and space: Reconstructing invasions from contemporary and historical collections. PLoS ONE 2014, 9, e106874. [Google Scholar] [CrossRef]
- Mairal, M.; Chown, S.L.; Shaw, J.; Chala, D.; Chau, J.H.; Hui, C.; Kalwij, J.M.; Münzbergová, Z.; Jansen van Vuuren, B.; Le Roux, J.J. Human activity strongly influences genetic dynamics of the most widespread invasive plant in the sub-Antarctic. Mol. Ecol. 2022, 31, 1649–1665. [Google Scholar] [CrossRef]
- Urquía, D.; Gutierrez, B.; Pozo, G.; Pozo, M.J.; Espín, A.; Torres, M.L. Psidium guajava in the Galapagos Islands: Population genetics and history of an invasive species. PLoS ONE 2019, 14, e0203737. [Google Scholar] [CrossRef]
- Morais, P.; Reichard, M. Cryptic invasions: A review. Sci. Total Environ. 2018, 613, 1438–1448. [Google Scholar] [CrossRef]
- Usui, T.; Lerner, D.; Eckert, I.; Angert, A.L.; Garroway, C.J.; Hargreaves, A.; Lancaster, L.T.; Lessard, J.P.; Riva, F.; Schmidt, C.; et al. The evolution of plasticity at geographic range edges. Trends Ecol. Evol. 2023, 38, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, N.; Campbell, L.G.; Ahern, J.R.; Paine, K.C.; Giroldo, A.B.; Whitney, K.D. Correlates of hybridization in plants. Evol. Lett. 2019, 3, 570–585. [Google Scholar] [CrossRef]
- Sakata, Y.; Itami, J.; Isagi, Y.; Ohgushi, T. Multiple and mass introductions from limited origins: Genetic diversity and structure of Solidago altissima in the native and invaded range. J. Plant Res. 2015, 128, 909–921. [Google Scholar] [CrossRef]
- Kolbe, J.J.; Glor, R.E.; Rodríguez Schettino, L.; Lara, A.C.; Larson, A.; Losos, J.B. Genetic variation increases during biological invasion by a Cuban lizard. Nature 2004, 431, 177–181. [Google Scholar] [CrossRef]
- Xu, H.; Pan, X.; Wang, C.; Chen, Y.; Chen, K.; Zhu, S.; van Klinken, R.D. Species identification, phylogenetic analysis and detection of herbicide-resistant biotypes of Amaranthus based on ALS and ITS. Sci. Rep. 2020, 10, 11735. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.A. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Montgomery, J.S.; Giacomini, D.; Waithaka, B.; Lanz, C.; Murphy, B.P.; Campe, R.; Lerchl, J.; Landes, A.; Gatzmann, F.; Janssen, A.; et al. Draft Genomes of Amaranthus tuberculatus, Amaranthus hybridus, and Amaranthus palmeri. Genome Biol. Evol. 2020, 12, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.R.; Hong, G.Y.; Dixit, A.; Chung, J.W.; Ma, K.H.; Cho, Y.H.; Gwag, J.G.; Park, Y.J. Characterization of microsatellite loci developed for Amaranthus hypochondriacus and their cross-amplifications in wild species. Conserv. Genet. 2008, 9, 243–246. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Tsykun, T.; Prospero, S.; Schoebel, C.N.; Rea, A.; Burgess, T.I. Global invasion history of the emerging plant pathogen Phytophthora multivora. BMC Genom. 2022, 23, 153. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Collin, F.D.; Durif, G.; Raynal, L.; Lombaert, E.; Gautier, M.; Vitalis, R.; Marin, J.M.; Estoup, A. DIYABC Random Forest v1.0: Extending approximate Bayesian computation with supervised machine learning to infer demographic history from genetic polymorphisms. Mol. Ecol. Resour. 2021, 21, 2598–2613. [Google Scholar] [CrossRef]
- Yue, X.; Zheng, X.; Zong, Y.; Jiang, S.; Hu, C.; Yu, P.; Liu, G.; Cao, Y.; Hu, H.; Teng, Y. Combined analyses of chloroplast DNA haplotypes and microsatellite markers reveal new insights into the origin and dissemination route of cultivated pears native to East Asia. Front. Plant Sci. 2018, 9, 591. [Google Scholar] [CrossRef]
- Maebara, Y.; Tamaoki, M.; Iguchi, Y.; Nakahama, N.; Hanai, T.; Nishino, A.; Hayasaka, D. Genetic diversity of invasive Spartina alterniflora Loisel. (Poaceae) introduced unintentionally into Japan and its invasion pathway. Front. Plant Sci. 2020, 11, 556039. [Google Scholar] [CrossRef] [PubMed]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Jones, A.G.; Small, C.M.; Paczolt, K.A.; Ratterman, N.L. A practical guide to methods of parentage analysis. Mol. Ecol. Resour. 2010, 10, 6–30. [Google Scholar] [CrossRef] [PubMed]
- Bennuah, S.Y.; Wang, T.; Aitken, S.N. Genetic analysis of the Picea stichensis×glauca introgression zone in British Columbia. For. Ecol. Manag. 2004, 197, 65–77. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Mesgaran, M.B.; Lewis, M.A.; Ades, P.K.; Donohue, K.; Ohadi, S.; Li, C.; Cousens, R.D. Hybridization can facilitate species invasions, even without enhancing local adaptation. Proc. Natl. Acad. Sci. USA 2016, 113, 10210–10214. [Google Scholar] [CrossRef]
- Glisson, W.J.; Larkin, D.J. Hybrid watermilfoil (Myriophyllum spicatum × Myriophyllum sibiricum) exhibits traits associated with greater invasiveness than its introduced and native parental taxa. Biol. Invasions 2021, 23, 2417–2433. [Google Scholar] [CrossRef]
| Population | SMM | TPM | Mode Shift | ||||
|---|---|---|---|---|---|---|---|
| Deficiency | Excess | Deficiency or Excess | Deficiency | Excess | Deficiency or Excess | ||
| FZ-L | 0.04 * | 0.97 | 0.08 | 0.21 | 0.80 | 0.43 | L |
| FZ-S | 0.27 | 0.75 | 0.54 | 0.85 | 0.16 | 0.33 | L |
| CC-L | 0.55 | 0.48 | 0.95 | 0.87 | 0.15 | 0.30 | L |
| CC-S | 0.09 | 0.92 | 0.17 | 0.27 | 0.75 | 0.54 | L |
| AY-L | 0.07 | 0.94 | 0.14 | 0.40 | 0.62 | 0.81 | L |
| AY-S | 0.92 | 0.09 | 0.17 | 0.99 | 0.02 * | 0.04 * | Y |
| ZZ-L | 0.27 | 0.74 | 0.54 | 0.77 | 0.25 | 0.50 | L |
| ZZ-S | 0.02 * | 0.99 | 0.04 * | 0.05 | 0.95 | 0.10 | L |
| BZ-L | 0.50 | 0.52 | 1.00 | 0.85 | 0.16 | 0.33 | L |
| BZ-S | 0.87 | 0.15 | 0.30 | 0.99 | 0.01 * | 0.03 * | Y |
| XA-L | 0.32 | 0.71 | 0.64 | 0.81 | 0.21 | 0.41 | L |
| XA-S | 0.96 | 0.05 | 0.09 | 1.00 | 0.00 * | 0.01 * | Y |
| FT-L | 0.52 | 0.52 | 1.00 | 0.92 | 0.09 | 0.18 | L |
| FT-S | 0.05 | 0.95 | 0.10 | 0.43 | 0.60 | 0.86 | L |
| CP-L | 0.98 | 0.03 * | 0.06 | 0.99 | 0.01 * | 0.03 * | L |
| CP-S | 0.82 | 0.20 | 0.39 | 1.00 | 0.00 * | 0.01 * | L |
| Country | No. | Coordinates (N, E) | Location | Year | Collected Material | Code | Sample Size | Category |
|---|---|---|---|---|---|---|---|---|
| United States | 1 | - | 2019 | Seeds | US1 | 34 | Source | |
| - | 2020 | Seeds | US2 | 28 | ||||
| - | 2021 | Seeds | US3 | 27 | ||||
| - | 2022 | Seeds | US4 | 30 | ||||
| Brazil | 2 | - | 2019–2022 | Seeds | BR | 26 | Source | |
| Argentina | 3 | - | 2019–2022 | Seeds | AR | 23 | Source | |
| China | 4 | (22°59′41″, 113°32′58″) | Dongguan | 2019 | Leaves | DG-L1 | 18 | Casual |
| (22°59′41″, 113°32′58″) | 2019 | Leaves | DG-L2 | 16 | ||||
| 5 | (25°57′36″, 119°31′12″) | Fuzhou | 2021 | Leaves | FZ-L | 30 | Naturalized | |
| (25°57′36″, 119°31′12″) | 2021 | Seeds | FZ-S | 28 | ||||
| 6 | (43°58′57″, 125°27′50″) | Changchun | 2019 | Leaves | CC-L | 17 | Naturalized | |
| (43°58′57″, 125°27′50″) | 2019 | Seeds | CC-S | 15 | ||||
| 7 | (35°53′12″, 114°23′13″) | Anyang | 2021 | Leaves | AY-L | 15 | Invasive | |
| (35°53′12″, 114°23′13″) | 2021 | Seeds | AY-S | 20 | ||||
| 8 | (34°48′02″, 113°24′19″) | Zhengzhou | 2021 | Leaves | ZZ-L | 17 | Invasive | |
| (34°48′02″, 113°24′19″) | 2021 | Seeds | ZZ-S | 14 | ||||
| 9 | (37°08′38″, 118°08′42″) | Binzhou | 2019 | Leaves | BZ-L | 15 | Invasive | |
| (37°08′38″, 118°08′42″) | 2019 | Seeds | BZ-S | 15 | ||||
| 10 | (34°25′49″, 109°17′05″) | Xi’an | 2021 | Leaves | XA-L | 16 | Dispersal | |
| (34°25′49″, 109°17′05″) | 2021 | Seeds | XA-S | 12 | ||||
| 11 | (39°48′38″, 116°22′10″) | Fengtai | 2021 | Leaves | FT-L | 18 | Dispersal | |
| (39°48′38″, 116°22′10″) | 2021 | Seeds | FT-S | 20 | ||||
| 12 | (39°41′33″, 116°15′32″) | Changping | 2019 | Leaves | CP-L | 24 | Dispersal | |
| (39°41′33″, 116°15′32″) | 2019 | Seeds | CP-S | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.-J.; Wang, H.-W.; Fu, J.-G.; Wan, F.-H.; Guo, J.-Y.; Wang, R. Dynamic Genetic Changes Reveal: Intra-Lineage Diversity, Not Admixture, Explains Amaranthus palmeri’s Success in China. Int. J. Mol. Sci. 2025, 26, 8128. https://doi.org/10.3390/ijms26178128
Cao J-J, Wang H-W, Fu J-G, Wan F-H, Guo J-Y, Wang R. Dynamic Genetic Changes Reveal: Intra-Lineage Diversity, Not Admixture, Explains Amaranthus palmeri’s Success in China. International Journal of Molecular Sciences. 2025; 26(17):8128. https://doi.org/10.3390/ijms26178128
Chicago/Turabian StyleCao, Jing-Jing, Hong-Wei Wang, Jian-Guo Fu, Fang-Hao Wan, Jian-Ying Guo, and Rui Wang. 2025. "Dynamic Genetic Changes Reveal: Intra-Lineage Diversity, Not Admixture, Explains Amaranthus palmeri’s Success in China" International Journal of Molecular Sciences 26, no. 17: 8128. https://doi.org/10.3390/ijms26178128
APA StyleCao, J.-J., Wang, H.-W., Fu, J.-G., Wan, F.-H., Guo, J.-Y., & Wang, R. (2025). Dynamic Genetic Changes Reveal: Intra-Lineage Diversity, Not Admixture, Explains Amaranthus palmeri’s Success in China. International Journal of Molecular Sciences, 26(17), 8128. https://doi.org/10.3390/ijms26178128





