Cascading 58mer Alpha Satellite superHOR in Complete Orangutan Y Chromosome
Abstract
1. Introduction
2. Results
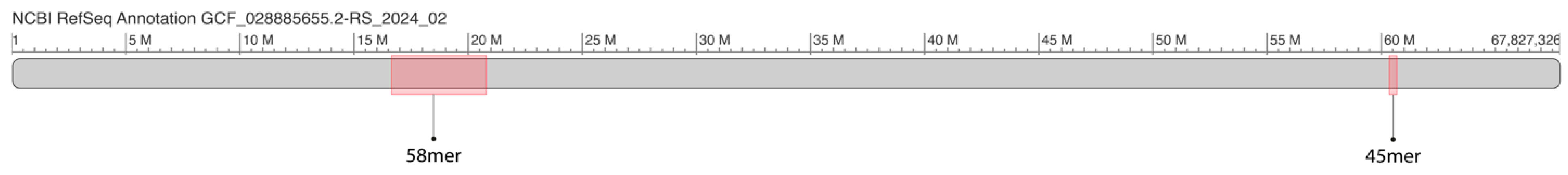
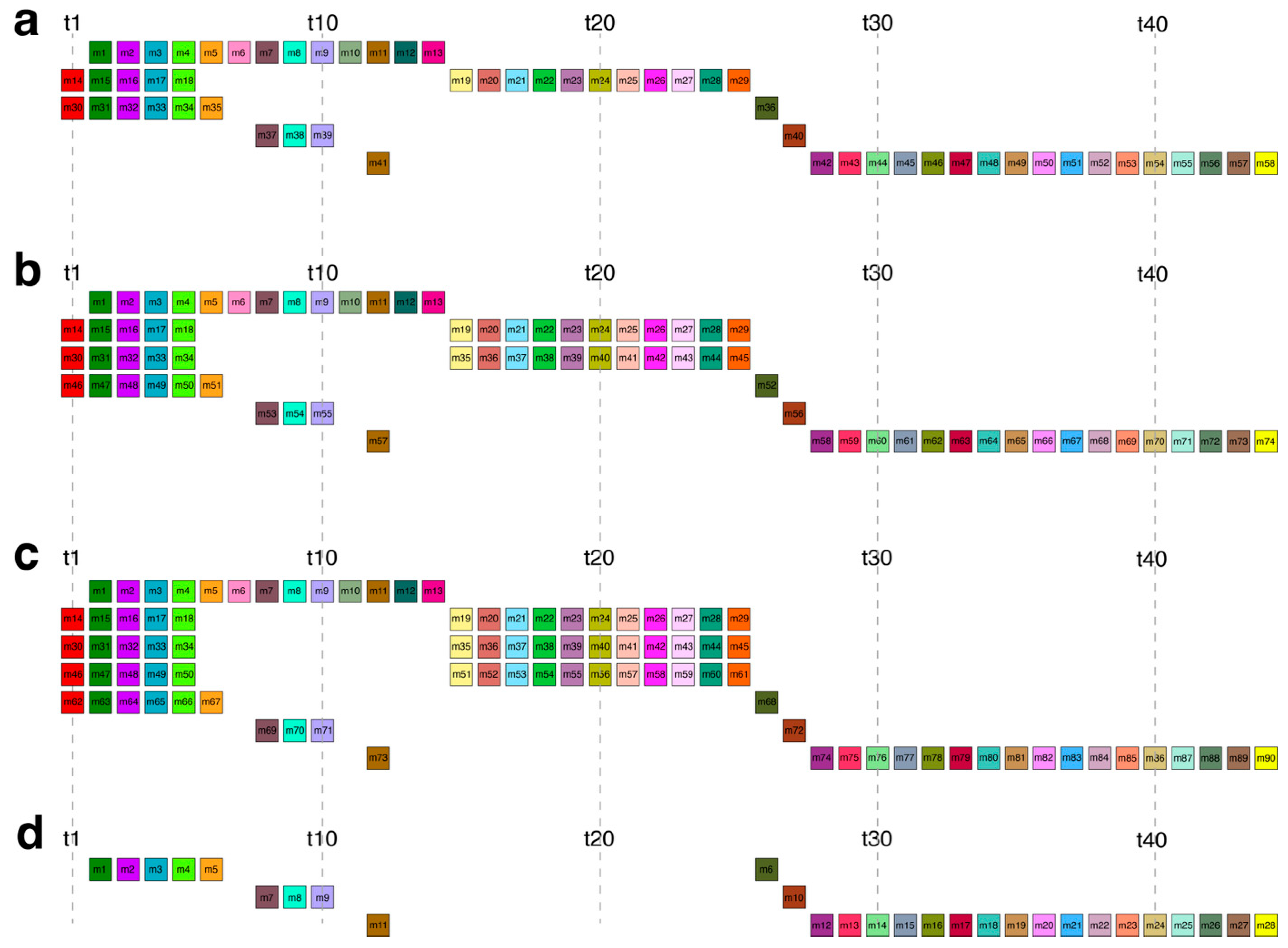
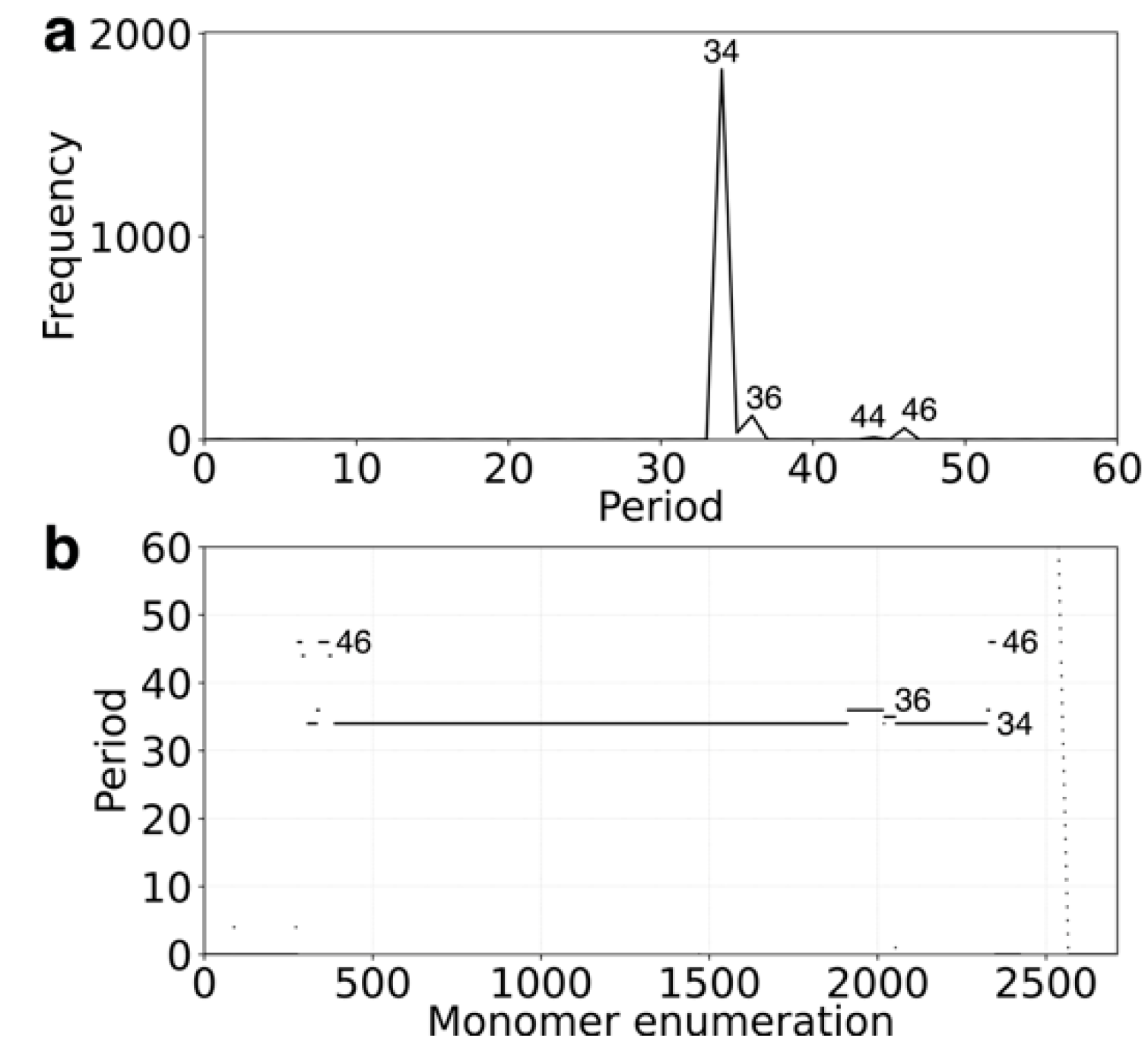
2.1. GRM and MD Diagrams for Orangutan Y Chromosome
| HOR Monomeric Scheme | No. of Monomers in 7 Successive Rows | No. of Monomeric Rows in HOR | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| C58 | 13 | 16 | – | – | 7 | 4 | 18 | 5 |
| V74 | 13 | 16 | 16 | – | 7 | 4 | 18 | 6 |
| V90 | 13 | 16 | 16 | 16 | 7 | 4 | 18 | 7 |
| V28 | – | – | – | – | 6 | 4 | 18 | 3 |
| V87 | 13 | 16 | 16 | 13 | 7 | 4 | 18 | 7 |
| V59 | 14 | 16 | – | – | 7 | 4 | 18 | 5 |
| V57 | 13 | 15 | – | – | 7 | 4 | 18 | 5 |
| V55 | 13 | 16 | – | – | 7 | 4 | 15 | 5 |
| V42 | 13 | – | – | – | 7 | 4 | 18 | 4 |
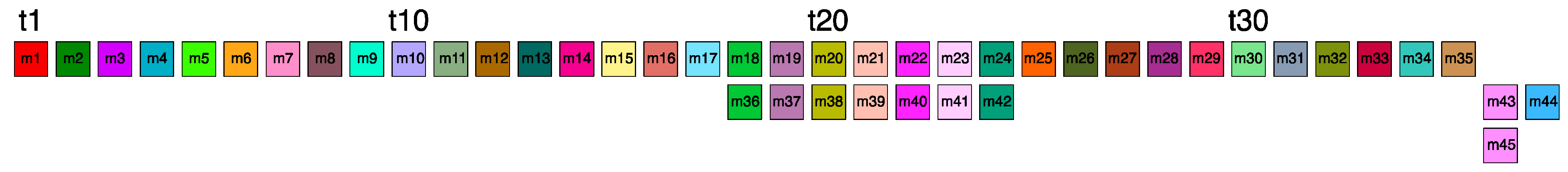
2.2. Novel Cascading 45mer HOR
2.3. MD-Frequency Table for Orangutan Chromosome Y
2.4. Cascading 18mer HOR in Gorilla and Willard’s Type 28mer HOR in Chimpanzee Chromosome Y
2.5. Comparison of Alpha Satellite HORs in Human Y Chromosome Assemblies GCA_018873775.2 and T2T_CHM13v2.0
3. Discussion
3.1. Comparative Overview of Y-Centromere HOR Architectures
3.2. Observations Versus Interpretations
3.3. Functional Implications for Kinetochore and Chromatin
3.4. Evolutionary Dynamics and Concerted Evolution
3.5. Linking Alpha-Satellite HOR Architecture to NBPF HORs and Functional Context
3.6. Limitations
4. Materials and Methods
Step-by-Step GRMhor Pipeline
- Monomer extraction—MonFinder identifies alpha satellite monomers by aligning the target genomic sequence to a 171 bp consensus alpha satellite monomer, searching in both forward and reverse complement orientations. Matches with ≥95% identity are retained as monomers.
- Monomer type classification—Each monomer is assigned to a specific monomer type if its sequence differs by less than 5% from other members of that type. Once monomer types are determined, the structure of the HOR array is established by vertically aligning monomers of the same type (i.e., those mutually differing by <5%) into columns. This process produces the aligned HOR schematic, in which each column contains monomers of a single type positioned consistently across HOR copies. Such vertical alignment not only enables a clear visual representation of the HOR structure but also ensures that monomer type assignment is consistent with the positional architecture of the array. This approach improves robustness against misclassification that could arise if only pairwise divergence was considered, by integrating sequence similarity with structural context in the final HOR diagram.
- Treatment of gaps—In the general case, undefined bases (‘N’) in the genomic assembly are ignored. Any monomer overlapping such a region is excluded from analysis to prevent erroneous HOR detection. In this study, however, all organisms were analyzed using complete assemblies without gaps in the targeted centromeric regions (orangutan, human, chimpanzee, gorilla), so this filtering step was not required.
- GRMhor analysis—The monomer list is analyzed to compute the GRM diagram (repeat periods vs. frequency) and the MD diagram (monomer positions vs. repeat period), using default parameters (max period = 90). No additional smoothing or pre-filtering was applied.
- Canonical and variant HOR definition—Within each detected HOR array, the canonical HOR is defined as the most frequent, complete n-mer unit. HOR copies that deviate from this dominant structure—through insertions, deletions, or internal duplications of one or more monomers—are classified as variant HORs. GRMhor assigns these labels by aligning all HOR copies monomer-by-monomer to the canonical reference and quantifying structural concordance; variant types are explicitly annotated in the aligned HOR schemes presented in the Results. This definition ensures that structural comparisons are made relative to a clearly established reference within each array.
- Output—For each HOR family, GRMhor outputs graphical representations (GRM, MD, aligned HOR scheme) and tabulated monomer composition, enabling full reconstruction of HOR organization.
- -
- Orangutan (NHGRI_mPonAbe1-v2.0_pri, GCF_028885655.2, National Human Genome Research Institute, National Institutes of Health, 5 January 2024)
- -
- Human (T2T-CHM13v2.0, GCF_009914755.1, T2T Consortium, 24 January 2022) and (HG01243v3.0, GCA_018873775.2, Johns Hopkins University, 27 September 2021)
- -
- Chimpanzee (NHGRI_mPanTro3-v2.0_pri, GCF_028858775.2, National Human Genome Research Institute, National Institutes of Health, 8 January 2024)
- -
- Gorilla (NHGRI_mGorGor1-v2.0_pri, GCF_029281585.2, National Human Genome Research Institute, National Institutes of Health, 8 January 2024)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Logsdon, G.A.; Rozanski, A.N.; Ryabov, F.; Potapova, T.; Shepelev, V.A.; Catacchio, C.R.; Porubsky, D.; Mao, Y.; Yoo, D.; Rautiainen, M.; et al. The variation and evolution of complete human centromeres. Nature 2024, 629, 136–145. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef]
- Cechova, M.; Miga, K.H. Comprehensive variant discovery in the era of complete human reference genomes. Nat. Methods 2023, 20, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Altemose, N.; Logsdon, G.A.; Bzikadze, A.V.; Sidhwani, P.; Langley, S.A.; Caldas, G.V.; Hoyt, S.J.; Uralsky, L.; Ryabov, F.D.; Shew, C.J.; et al. Complete genomic and epigenetic maps of human centromeres. Science 2022, 376, eabl4178. [Google Scholar] [CrossRef] [PubMed]
- Willard, H.F. Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 1985, 37, 524–532. [Google Scholar] [PubMed]
- Waye, J.S.; Willard, H.F. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: Evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol. Cell Biol. 1986, 6, 3156–3165. [Google Scholar] [CrossRef]
- Tyler-Smith, C.; Brown, W.R. Structure of the major block of alphoid satellite DNA on the human Y chromosome. J. Mol. Biol. 1987, 195, 457–470. [Google Scholar] [CrossRef]
- Choo, K.H.; Vissel, B.; Nagy, A.; Earle, E.; Kalitsis, P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991, 19, 1179–1182. [Google Scholar] [CrossRef]
- Warburton, P.E.; Willard, H.F. Evolution of centromeric alpha satellite DNA: Molecular organisation within and between human primate chromosomes. In Human Genome Evolution; BIOS Scientific Publisher: Milton Park, UK, 1996; pp. 121–145. [Google Scholar]
- Alexandrov, I.; Kazakov, A.; Tumeneva, I.; Shepelev, V.; Yurov, Y. Alpha-satellite DNA of primates: Old and new families. Chromosoma 2001, 110, 253–266. [Google Scholar] [CrossRef]
- Schueler, M.G.; Higgins, A.W.; Rudd, M.K.; Gustashaw, K.; Willard, H.F. Genomic and genetic definition of a functional human centromere. Science 2001, 294, 109–115. [Google Scholar] [CrossRef]
- Alkan, C.; Eichler, E.E.; Bailey, J.A.; Sahinalp, S.C.; Tuzun, E. The role of unequal crossover in alpha-satellite DNA evolution: A computational analysis. J. Comput. Biol. 2004, 11, 933–944. [Google Scholar] [CrossRef]
- Rudd, M.K.; Wray, G.A.; Willard, H.F. The evolutionary dynamics of alpha-satellite. Genome Res. 2006, 16, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Paar, V.; Gluncic, M.; Rosandic, M.; Basar, I.; Vlahovic, I. Intragene higher order repeats in neuroblastoma breakpoint family genes distinguish humans from chimpanzees. Mol. Biol. Evol. 2011, 28, 1877–1892. [Google Scholar] [CrossRef]
- Hayden, K.E.; Strome, E.D.; Merrett, S.L.; Lee, H.R.; Rudd, M.K.; Willard, H.F. Sequences associated with centromere competency in the human genome. Mol. Cell Biol. 2013, 33, 763–772. [Google Scholar] [CrossRef]
- Aldrup-Macdonald, M.E.; Sullivan, B.A. The past, present, and future of human centromere genomics. Genes 2014, 5, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Shepelev, V.A.; Uralsky, L.I.; Alexandrov, A.A.; Yurov, Y.B.; Rogaev, E.I.; Alexandrov, I.A. Annotation of suprachromosomal families reveals uncommon types of alpha satellite organization in pericentromeric regions of hg38 human genome assembly. Genom. Data 2015, 5, 139–146. [Google Scholar] [CrossRef]
- Sullivan, L.L.; Chew, K.; Sullivan, B.A. alpha satellite DNA variation and function of the human centromere. Nucleus 2017, 8, 331–339. [Google Scholar] [CrossRef]
- Uralsky, L.I.; Shepelev, V.A.; Alexandrov, A.A.; Yurov, Y.B.; Rogaev, E.I.; Alexandrov, I.A. Classification and monomer-by-monomer annotation dataset of suprachromosomal family 1 alpha satellite higher-order repeats in hg38 human genome assembly. Data Brief. 2019, 24, 103708. [Google Scholar] [CrossRef]
- Baldini, A.; Miller, D.A.; Shridhar, V.; Rocchi, M.; Miller, O.J.; Ward, D.C. Comparative mapping of a gorilla-derived alpha satellite DNA clone on great ape and human chromosomes. Chromosoma 1991, 101, 109–114. [Google Scholar] [CrossRef]
- Baldini, A.; Ried, T.; Shridhar, V.; Ogura, K.; D’Aiuto, L.; Rocchi, M.; Ward, D.C. An alphoid DNA sequence conserved in all human and great ape chromosomes: Evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum. Genet. 1993, 90, 577–583. [Google Scholar] [CrossRef] [PubMed]
- D’Aiuto, L.; Antonacci, R.; Marzella, R.; Archidiacono, N.; Rocchi, M. Cloning and comparative mapping of a human chromosome 4-specific alpha satellite DNA sequence. Genomics 1993, 18, 230–235. [Google Scholar] [CrossRef]
- Luke, S.; Verma, R.S. Human (Homo sapiens) and chimpanzee (Pan troglodytes) share similar ancestral centromeric alpha satellite DNA sequences but other fractions of heterochromatin differ considerably. Am. J. Phys. Anthropol. 1995, 96, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Haaf, T.; Mater, A.G.; Wienberg, J.; Ward, D.C. Presence and abundance of CENP-B box sequences in great ape subsets of primate-specific alpha-satellite DNA. J. Mol. Evol. 1995, 41, 487–491. [Google Scholar] [CrossRef]
- Haaf, T.; Willard, H.F. Orangutan alpha-satellite monomers are closely related to the human consensus sequence. Mamm. Genome 1998, 9, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Samonte, R.V.; Ramesh, K.H.; Verma, R.S. Comparative mapping of human alphoid satellite DNA repeat sequences in the great apes. Genetica 1997, 101, 97–104. [Google Scholar] [CrossRef]
- Gluncic, M.; Vlahovic, I.; Rosandic, M.; Paar, V. Novel Cascade Alpha Satellite HORs in Orangutan Chromosome 13 Assembly: Discovery of the 59mer HOR-The largest Unit in Primates-And the Missing Triplet 45/27/18 HOR in Human T2T-CHM13v2.0 Assembly. Int. J. Mol. Sci. 2024, 25, 7596. [Google Scholar] [CrossRef]
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Masumoto, H.; Masukata, H.; Muro, Y.; Nozaki, N.; Okazaki, T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 1989, 109, 1963–1973. [Google Scholar] [CrossRef]
- Jain, M.; Olsen, H.E.; Turner, D.J.; Stoddart, D.; Bulazel, K.V.; Paten, B.; Haussler, D.; Willard, H.F.; Akeson, M.; Miga, K.H. Linear assembly of a human centromere on the Y chromosome. Nat. Biotechnol. 2018, 36, 321–323. [Google Scholar] [CrossRef]
- Dover, G. Molecular drive: A cohesive mode of species evolution. Nature 1982, 299, 111–117. [Google Scholar] [CrossRef]
- Gluncic, M.; Vlahovic, I.; Rosandic, M.; Paar, V. Tandemly repeated NBPF HOR copies (Olduvai triplets): Possible impact on human brain evolution. Life Sci. Alliance 2023, 6, e202101306. [Google Scholar] [CrossRef] [PubMed]
- Gluncic, M.; Vlahovic, I.; Rosandic, M.; Paar, V. Tandem NBPF 3mer HORs (Olduvai triplets) in Neanderthal and two novel HOR tandem arrays in human chromosome 1 T2T-CHM13 assembly. Sci. Rep. 2023, 13, 14420. [Google Scholar] [CrossRef] [PubMed]
- Gluncic, M.; Baric, D.; Paar, V. Efficient genome monomer higher-order structure annotation and identification using the GRMhor algorithm. Bioinform. Adv. 2024, 4, vbae191. [Google Scholar] [CrossRef] [PubMed]
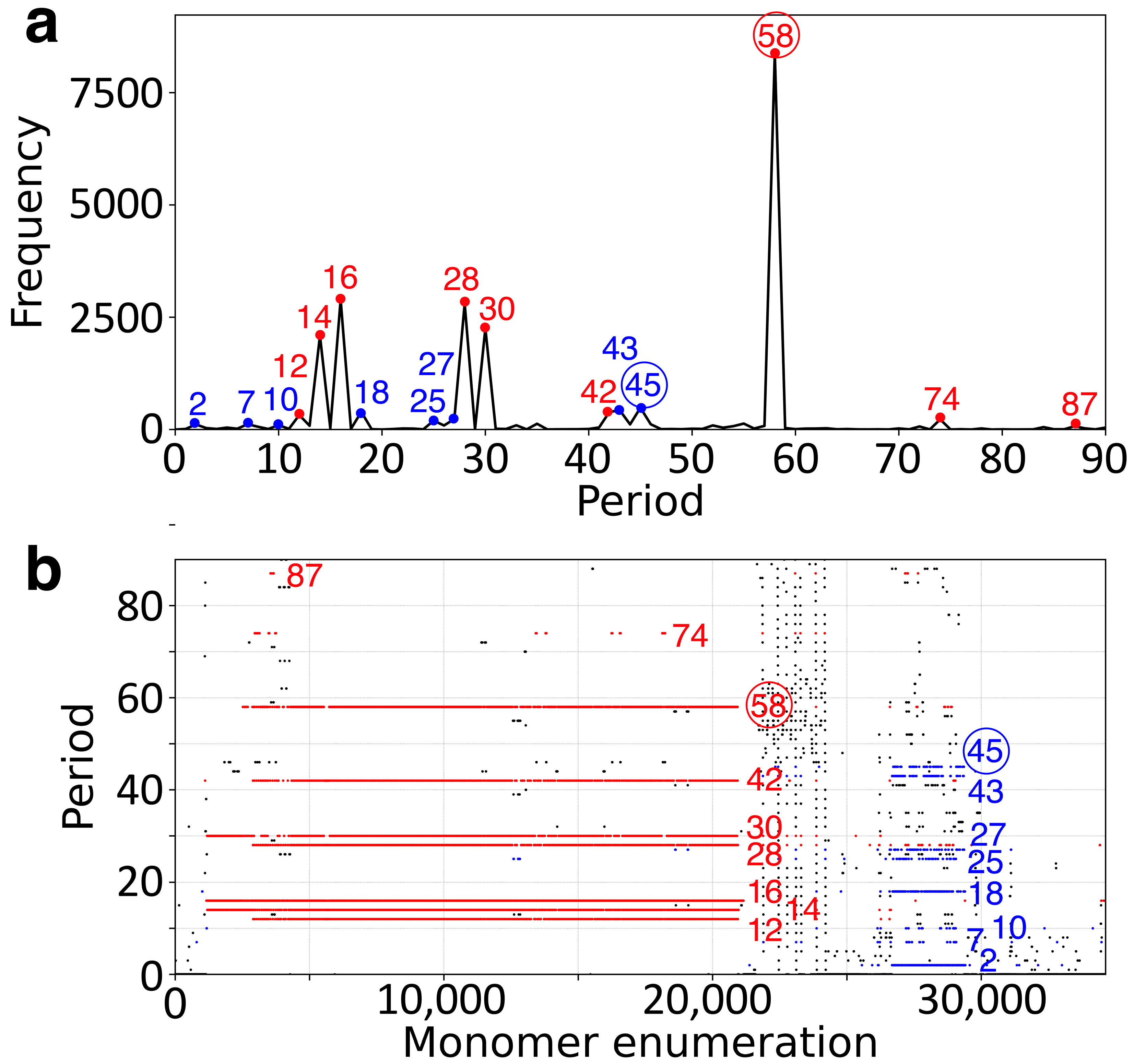
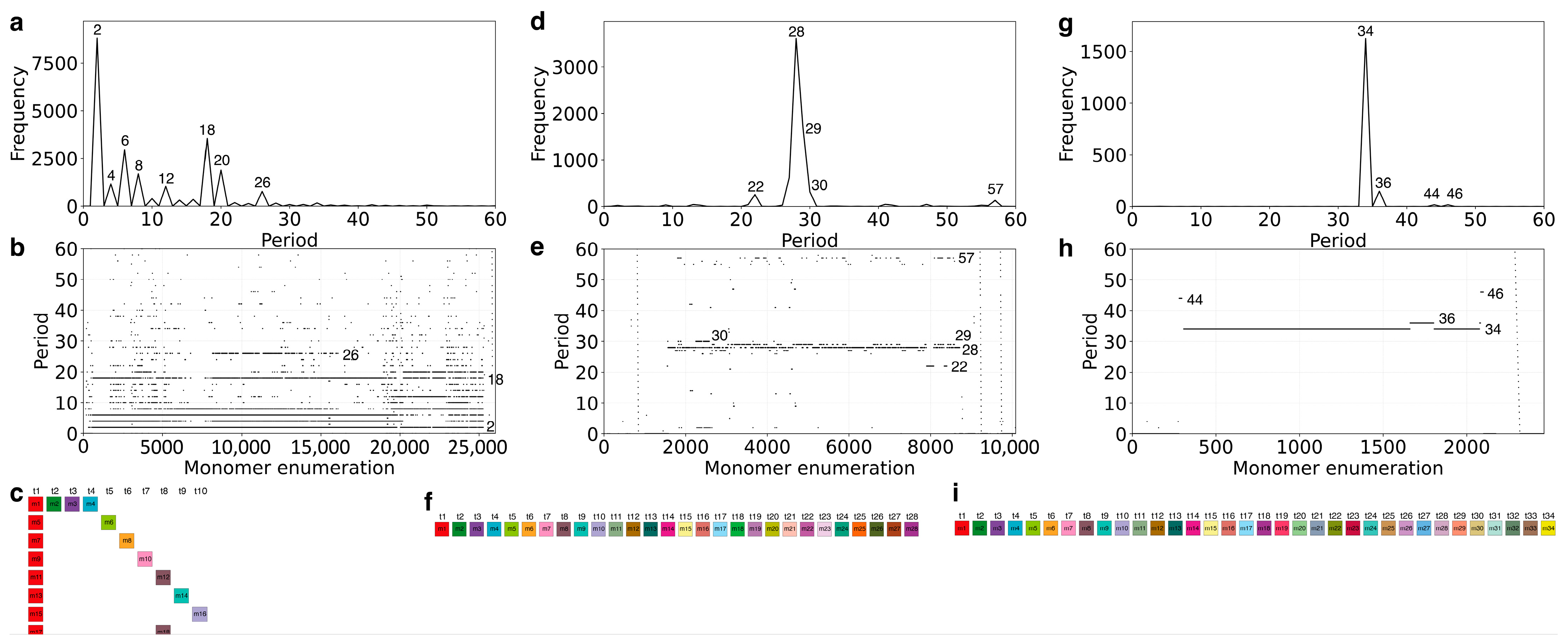
| Period | 2 | 7 | 10 | 12 | 14 | 16 | 18 | 25 | 27 | 28 | 30 | 33 | 35 | 42 | 43 | 44 | 45 | 46 | 52 | 55 | 58 | 74 |
| Freq. | 113 | 118 | 97 | 322 | 2089 | 2891 | 393 | 197 | 233 | 2803 | 2254 | 94 | 128 | 391 | 428 | 114 | 484 | 117 | 91 | 132 | 8389 | 225 |
| Human | Chimpanzee | Gorilla | Orangutan | |
|---|---|---|---|---|
| nmer HOR | 34mer | 28mer | 18mer | 58mer |
| No. HOR copies | 54 | 259 | 779 | 310 |
| No. Canonical HOR copies | 49 | 237 | 405 | 258 |
| HOR-type | Willard’s | Willard’s | Cascading | Cascading |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glunčić, M.; Vlahović, I.; Rosandić, M.; Paar, V. Cascading 58mer Alpha Satellite superHOR in Complete Orangutan Y Chromosome. Int. J. Mol. Sci. 2025, 26, 8122. https://doi.org/10.3390/ijms26178122
Glunčić M, Vlahović I, Rosandić M, Paar V. Cascading 58mer Alpha Satellite superHOR in Complete Orangutan Y Chromosome. International Journal of Molecular Sciences. 2025; 26(17):8122. https://doi.org/10.3390/ijms26178122
Chicago/Turabian StyleGlunčić, Matko, Ines Vlahović, Marija Rosandić, and Vladimir Paar. 2025. "Cascading 58mer Alpha Satellite superHOR in Complete Orangutan Y Chromosome" International Journal of Molecular Sciences 26, no. 17: 8122. https://doi.org/10.3390/ijms26178122
APA StyleGlunčić, M., Vlahović, I., Rosandić, M., & Paar, V. (2025). Cascading 58mer Alpha Satellite superHOR in Complete Orangutan Y Chromosome. International Journal of Molecular Sciences, 26(17), 8122. https://doi.org/10.3390/ijms26178122







