Comparative Metabolomics Reveals Phosphine-Induced Metabolic Disruptions in Planococcus citri (Risso)
Abstract
1. Introduction
2. Results
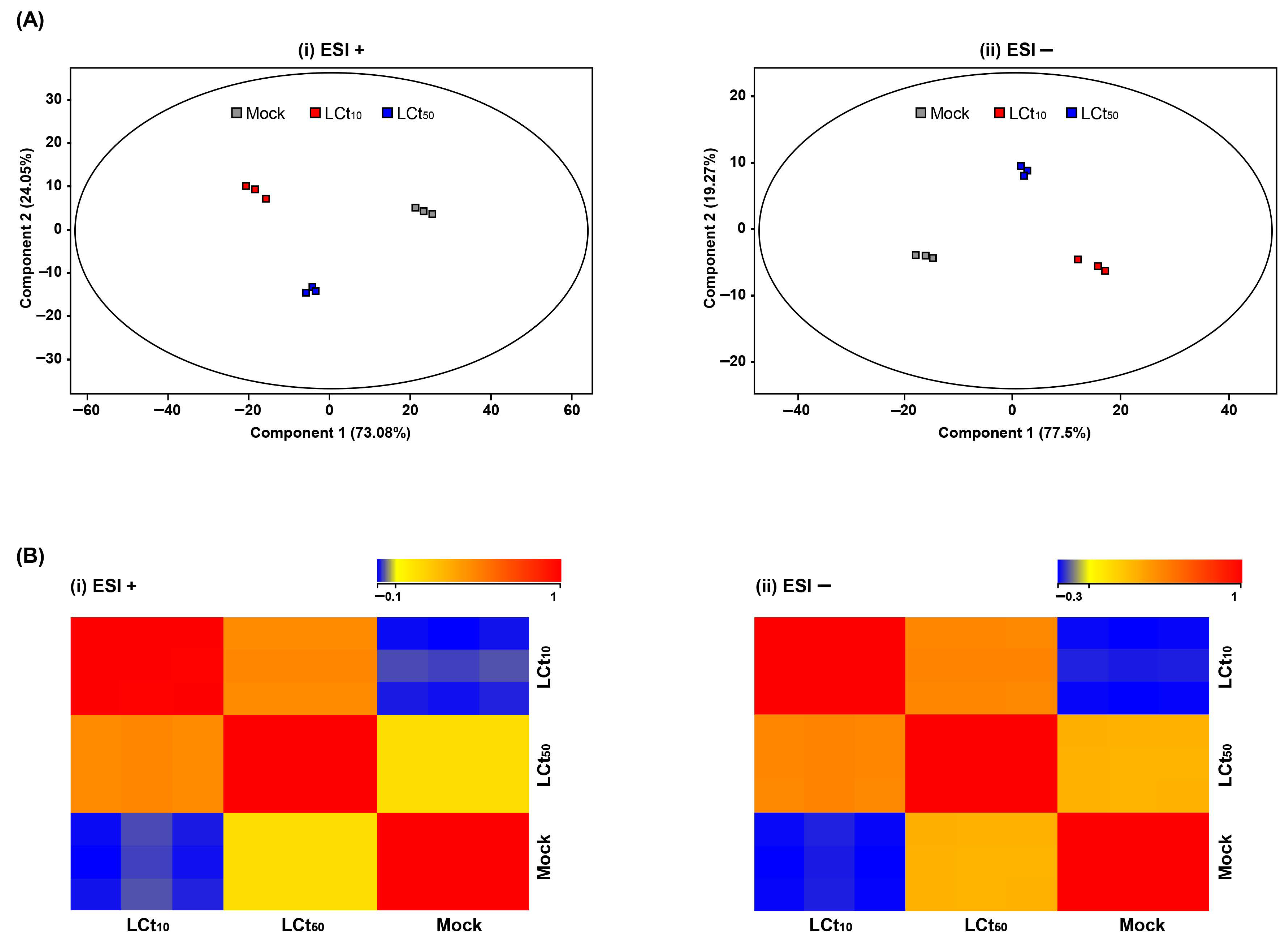
2.1. Comparative Metabolic Profiling by PH3 Exposure
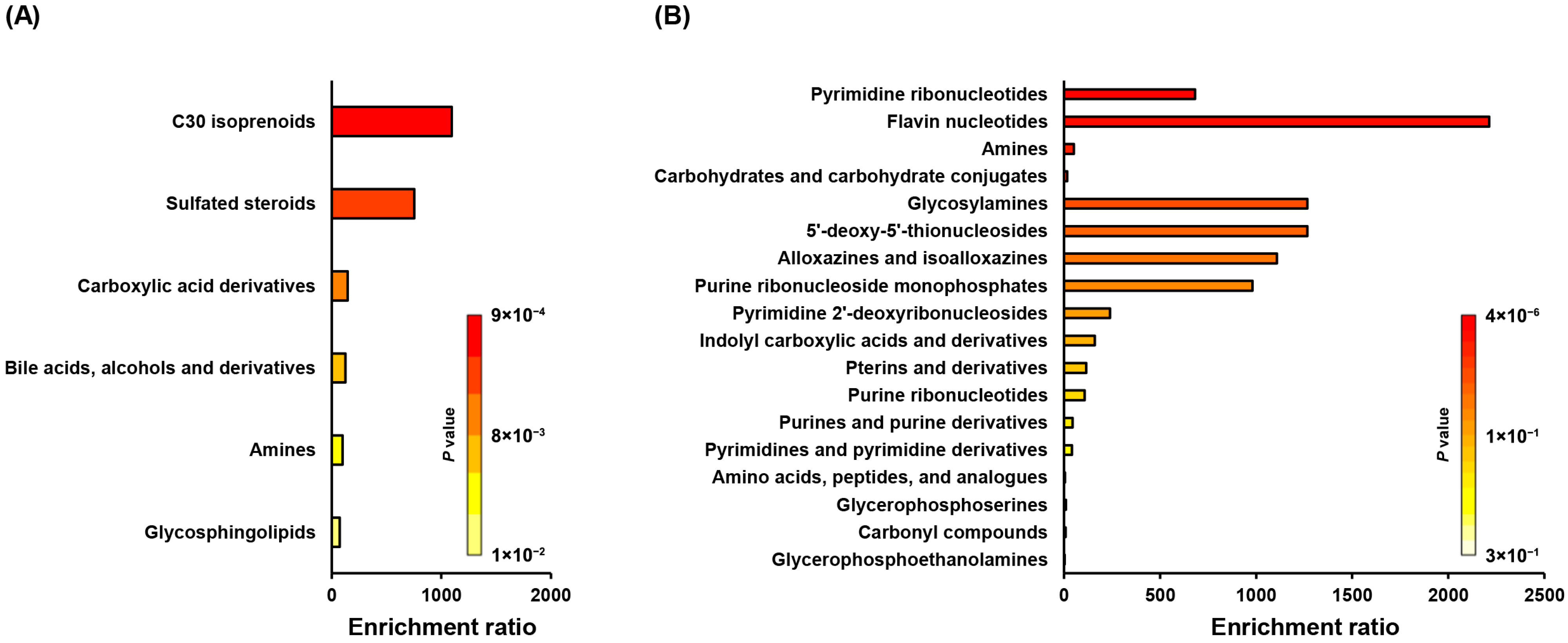
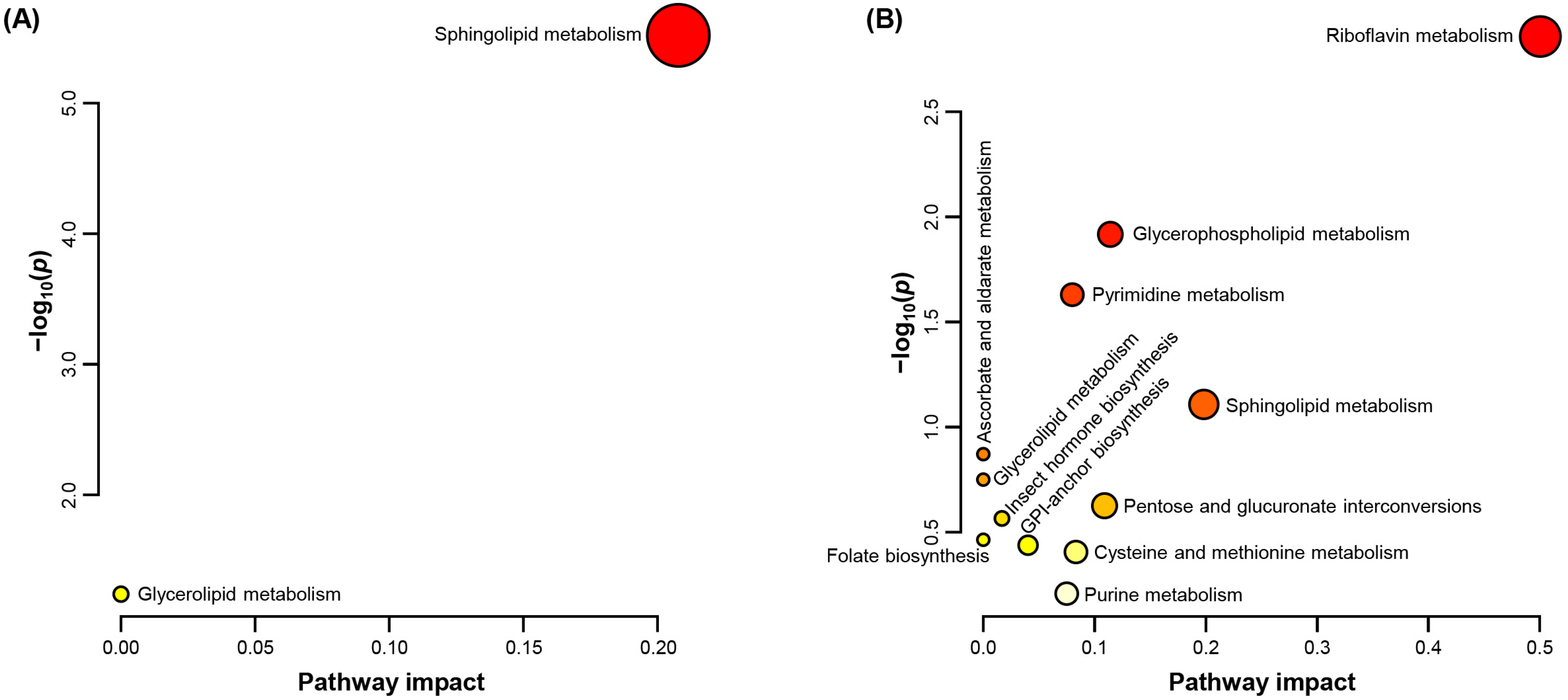
2.2. Enrichment and Pathway Impact of Altered Metabolites
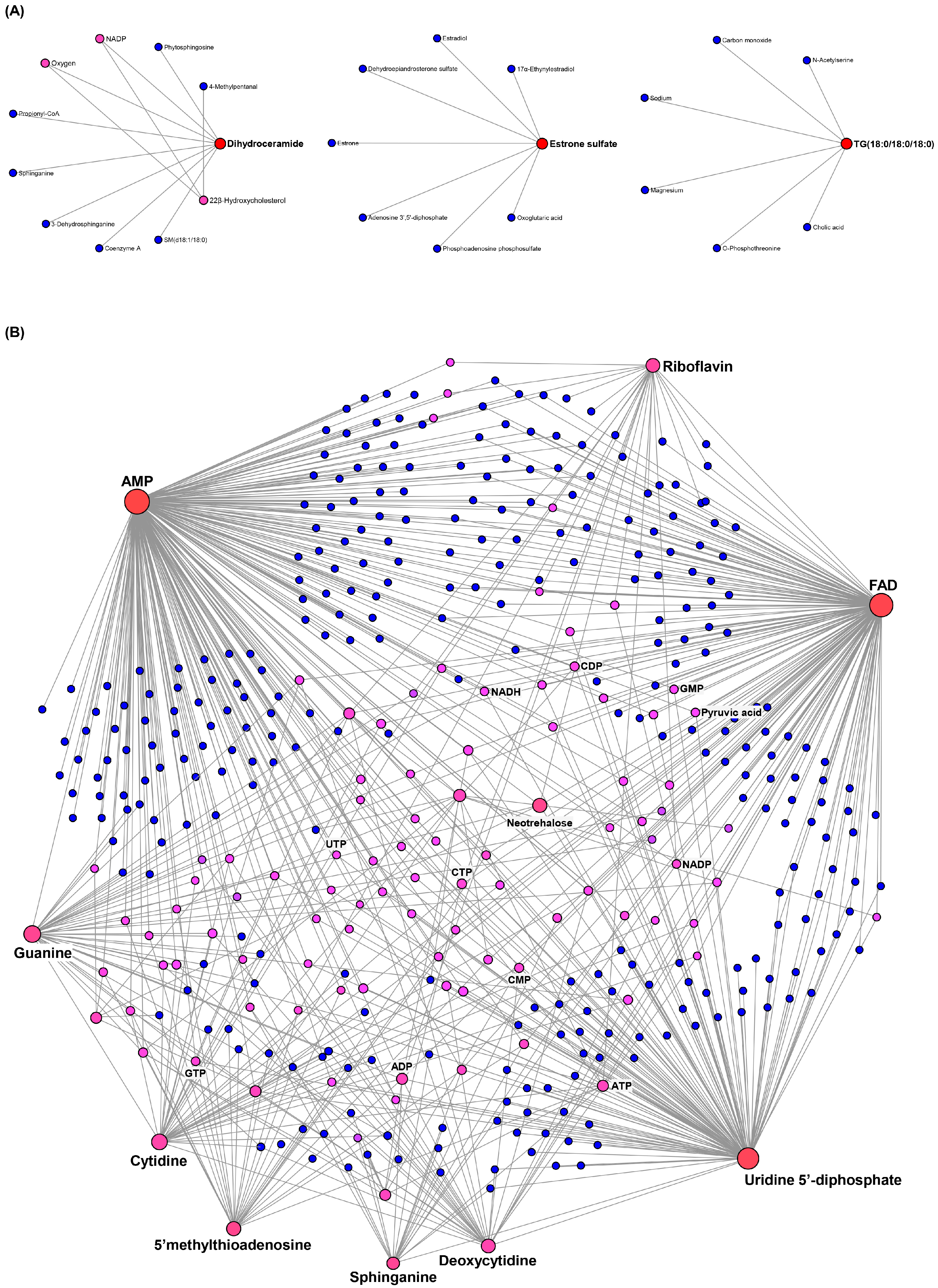
2.3. Metabolite–Metabolite Interaction Network
2.4. Lipid Metabolism Disorders by PH3 Stress
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. Phosphine and Thermal Treatment
4.3. Metabolite Extraction
4.4. Lipid Extraction
4.5. Metabolomics
4.6. Data Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ren, Y.; Lee, B.; Padovan, B. Penetration of methyl bromide, sulfuryl fluoride, ethanedinitrile and phosphine into timber blocks and the sorption rate of the fumigants. J. Stored Prod. Res. 2011, 47, 63–68. [Google Scholar] [CrossRef]
- Ridley, A.W.; Burrill, P.R.; Cook, C.C.; Daglish, G.J. Phosphine fumigation of silo bags. J. Stored Prod. Res. 2011, 47, 349–356. [Google Scholar] [CrossRef]
- Ryan, R.F.; De Lima, C.F. Phosphine-an overview of a unique 80 year fumigant. Gen. Appl. Entomol. J. Entomol. Soc. New South Wales 2014, 42, 31–42. [Google Scholar]
- Cichón, L.; Garrido, S.; Gómez, R.; Fernández, D.; Argañaraz, L.; Gastaminza, G. Evaluation of phosphine gas as a quarantine treatment for obscure mealybug for export markets. In Proceedings of the XI International Pear Symposium 909, General Roca, Argentina, 13–26 November 2010; pp. 479–484. [Google Scholar]
- Jamieson, L.; Page-Weir, N.; Chhagan, A.; Brash, D.; Klementz, D.; Bycroft, B.; Connolly, P.; Waddell, B.; Gilbertson, R.; Bollen, F. Phosphine fumigation to disinfest kiwifruit. New Zealand Plant Prot. 2012, 65, 35–43. [Google Scholar] [CrossRef]
- Kim, B.S.; Park, C.G.; Moon, Y.M.; Sung, B.K.; Ren, Y.; Wylie, S.J.; Lee, B.H. Quarantine treatments of imported nursery plants and exported cut flowers by phosphine gas (PH3) as methyl bromide alternative. J. Econ. Entomol. 2016, 109, 2334–2340. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-B. Effects of packing methods and fumigation of phosphine for control of rice weevil (Sitophilus oryzae). Appl. Biol. Chem. 1989, 32, 265–269. [Google Scholar]
- Horn, P.; Horn, F.; Tumambing, J.; Rogers, M. Studies and commercial application of Vaporph3os phosphine fumigant for disinfestation of exported fruits and vegetables in South America. In Proceedings of the International Symposium Postharvest Pacifica 2009-Pathways to Quality: V International Symposium on Managing Quality in 880, Napier, New Zealand, 15–19 November 2009; pp. 407–414. [Google Scholar]
- Tumambing, J.; Depalo, M.; Garnier, J.P.; Mallari, R. Eco2Fume and Vaporph3OS® phosphine fumigants-global application updates. In Proceedings of the 9th International Conference on Controlled Atmosphere and Fumigation in Stored Products, Antalya, Turkey, 15–19 October 2012. [Google Scholar]
- Williams, P.; Ryan, R. Eco2fume for the postharvest disinfestation of horticulture produce. In Proceedings of International Conference on Controlled Atmosphere and Fumigation in Stored Products, Fresno, CA, USA, 29 October–3 November 2000; pp. 365–371. [Google Scholar]
- Blumberg, D.; Klein, M.; Mendel, Z. Response by encapsulation of four mealybug species (Homoptera: Pseudococcidae) to parasitization by Anagyrus pseudococci. Phytoparasitica 1995, 23, 157–163. [Google Scholar] [CrossRef]
- Demirci, F.; Muştu, M.; Bora Kaydan, M.; Ülgentürk, S. Laboratory evaluation of the effectiveness of the entomopathogen; Isaria farinosa, on citrus mealybug, Planococcus citri. J. Pest Sci. 2011, 84, 337–342. [Google Scholar] [CrossRef]
- Franco, J.C.; Suma, P.; da Silva, E.B.; Blumberg, D.; Mendel, Z. Management strategies of mealybug pests of citrus in Mediterranean countries. Phytoparasitica 2004, 32, 507–522. [Google Scholar] [CrossRef]
- Franco, J.C.; Zada, A.; Mendel, Z. Novel approaches for the management of mealybug pests. Biorational Control. Arthropod Pests Appl. Resist. Manag. 2009, 233–278. [Google Scholar]
- Dhami, M.K.; Weir, B.S.; Taylor, M.W.; Beggs, J.R. Diverse honeydew-consuming fungal communities associated with scale insects. PLoS ONE 2013, 8, e70316. [Google Scholar] [CrossRef]
- Walton, V.; Pringle, K. Effects of pesticides used on table grapes on the mealybug parasitoid Coccidoxenoides peregrinus (Timberlake)(Hymenoptera: Encyrtidae). South Afr. J. Enol. Vitic. 1999, 20, 31–34. [Google Scholar] [CrossRef][Green Version]
- Gullan, P.J.; Kosztarab, M. Adaptations in scale insects. Annu. Rev. Entomol. 1997, 42, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Ulusoy, S.; Kahya, D.; Bilgin, M.G.; Apalak, A. The effectiveness of wax secretion on chemical control in some mealybug species. J. Asia-Pac. Entomol. 2022, 25, 101954. [Google Scholar] [CrossRef]
- Chaudhry, M. Review a review of the mechanisms involved in the action of phosphine as an insecticide and phosphine resistance in stored-product insects. Pestic. Sci. 1997, 49, 213–228. [Google Scholar] [CrossRef]
- Kim, B.S.; Shin, E.-M.; Park, Y.J.; Yang, J.O. Susceptibility of the cigarette beetle Lasioderma serricorne (Fabricius) to phosphine, ethyl formate and their combination, and the sorption and desorption of fumigants on cured tobacco leaves. Insects 2020, 11, 599. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Song, J.-E.; Park, J.S.; Park, Y.J.; Shin, E.-M.; Yang, J.O. Insecticidal effects of fumigants (EF, MB, and PH3) towards phosphine-susceptible and-resistant Sitophilus oryzae (Coleoptera: Curculionidae). Insects 2019, 10, 327. [Google Scholar] [CrossRef]
- Kim, K.; Park, M.-G.; Lee, Y.H.; Jeon, H.-J.; Kwon, T.H.; Kim, C.; Park, J.; Lee, B.-H.; Yang, J.O.; Lee, S.-E. Synergistic effects and toxic mechanism of phosphine with ethyl formate against citrus mealybug (Planococcus citri). Appl. Sci. 2021, 11, 9877. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Powers, H.J. Riboflavin (vitamin B-2) and health. Am. J. Clin. Nutr. 2003, 77, 1352–1360. [Google Scholar] [CrossRef]
- Voet, D.; Voet, J.G. Biochemistry; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Lu, S.C. Glutathione synthesis. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 3143–3153. [Google Scholar] [CrossRef]
- Stover, P.J.; Field, M.S. Trafficking of intracellular folates. Adv. Nutr. 2011, 2, 325–331. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef]
- Jadhav, U.; Mundhe, S.; Kumar, Y.; Jogaiah, S.; Upadhyay, A.; Gupta, V.S.; Kadoo, N.Y. Gibberellic acid induces unique molecular responses in ‘Thompson Seedless’ grapes as revealed by non-targeted metabolomics. J. Plant Growth Regul. 2021, 40, 293–304. [Google Scholar] [CrossRef]
- Yang, J.O.; Park, Y.; Hyun, I.-H.; Kim, G.-H.; Kim, B.-S.; Lee, B.-H.; Ren, Y. A combination treatment using ethyl formate and phosphine to control Planococcus citri (Hemiptera: Pseudococcidae) on pineapples. J. Econ. Entomol. 2016, 109, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Aittokallio, T.; Schwikowski, B. Graph-based methods for analysing networks in cell biology. Brief. Bioinform. 2006, 7, 243–255. [Google Scholar] [CrossRef]
- Serrato-Salas, J.; Gendrin, M. Involvement of microbiota in insect physiology: Focus on B vitamins. MBio 2023, 14, e02225-22. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect P450 enzymes. Annu. Rev. Entomol. 1999, 44, 507–533. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Abdollahi, M.; Ranjbar, A.; Shadnia, S.; Nikfar, S.; Rezaie, A. Pesticides and oxidative stress: A review. Med. Sci. Monit. 2004, 10, 141–147. [Google Scholar]
- Suwannasom, N.; Kao, I.; Pruß, A.; Georgieva, R.; Bäumler, H. Riboflavin: The health benefits of a forgotten natural vitamin. Int. J. Mol. Sci. 2020, 21, 950. [Google Scholar] [CrossRef] [PubMed]
- Bolter, C.J.; Chefurka, W. Extramitochondrial release of hydrogen peroxide from insect and mouse liver mitochondria using the respiratory inhibitors phosphine, myxothiazol, and antimycin and spectral analysis of inhibited cytochromes. Arch. Biochem. Biophys. 1990, 278, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Nath, N.S.; Bhattacharya, I.; Tuck, A.G.; Schlipalius, D.I.; Ebert, P.R. Mechanisms of phosphine toxicity. J. Toxicol. 2011, 2011, 494168. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.-K.; Jeon, J.-C.; Seok, S.-J.; Kim, G.-H.; Koo, H.-N.; Lee, D.-W. Metabolite changes by combined treatment, ethyl formate and low temperature, in Drosophila suzukii. Sci. Rep. 2024, 14, 25948. [Google Scholar] [CrossRef]
- Felton, G.W.; Summers, C.B. Antioxidant systems in insects. Arch. Insect Biochem. Physiol. 1995, 29, 187–197. [Google Scholar] [CrossRef]
- Casani, S.; Gómez-Pastor, R.; Matallana, E.; Paricio, N. Antioxidant compound supplementation prevents oxidative damage in a Drosophila model of Parkinson’s disease. Free Radic. Biol. Med. 2013, 61, 151–160. [Google Scholar] [CrossRef]
- Kanzok, S.M.; Fechner, A.; Bauer, H.; Ulschmid, J.K.; Müller, H.-M.; Botella-Munoz, J.; Schneuwly, S.; Schirmer, R.H.; Becker, K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science 2001, 291, 643–646. [Google Scholar] [CrossRef]
- Gřešková, A.; Petřivalský, M. Thioredoxin System in Insects: Uncovering the Roles of Thioredoxins and Thioredoxin Reductase beyond the Antioxidant Defences. Insects 2024, 15, 797. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Bauer, H.; Massey, V.; Arscott, L.D.; Schirmer, R.H.; Ballou, D.P.; Williams, C.H. The mechanism of high M r thioredoxin reductase from Drosophila melanogaster. J. Biol. Chem. 2003, 278, 33020–33028. [Google Scholar] [CrossRef]
- Gao, Z.; Batool, R.; Xie, W.; Huang, X.; Wang, Z. Transcriptome and metabolome analysis reveals the importance of amino-acid metabolism in Spodoptera frugiperda exposed to spinetoram. Insects 2022, 13, 852. [Google Scholar] [CrossRef]
- Chen, L.; He, T.; Ding, L.; Lan, X.; Sun, J.; Xu, X.; Wu, H.; Zhou, D.; Huang, Z.; Zhou, T. Effect of Spinetoram Stress on Midgut Detoxification Enzyme and Gene Expression of Apis cerana cerana Fabricius. Insects 2025, 16, 492. [Google Scholar] [CrossRef]
- Stevens, R.; Page, D.; Gouble, B.; Garchery, C.; Zamir, D.; Causse, M. Tomato fruit ascorbic acid content is linked with monodehydroascorbate reductase activity and tolerance to chilling stress. Plant Cell Environ. 2008, 31, 1086–1096. [Google Scholar] [CrossRef]
- Lv, N.; Ma, K.; Li, R.; Liang, P.; Liang, P.; Gao, X. Sublethal and lethal effects of the imidacloprid on the metabolic characteristics based on high-throughput non-targeted metabolomics in Aphis gossypii Glover. Ecotoxicol. Environ. Saf. 2021, 212, 111969. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, S.M.; Ebert, P.R. Pesticidal toxicity of phosphine and its interaction with other pest control treatments. Curr. Issues Mol. Biol. 2023, 45, 2461–2473. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xie, N.; Illes, P.; Di Virgilio, F.; Ulrich, H.; Semyanov, A.; Verkhratsky, A.; Sperlagh, B.; Yu, S.-G.; Huang, C. From purines to purinergic signalling: Molecular functions and human diseases. Signal Transduct. Target. Ther. 2021, 6, 162. [Google Scholar] [CrossRef]
- Friedman, D.L. Role of cyclic nucleotides in cell growth and differentiation. Physiol. Rev. 1976, 56, 652–708. [Google Scholar] [CrossRef]
- Petitgas, C.; Seugnet, L.; Dulac, A.; Matassi, G.; Mteyrek, A.; Fima, R.; Strehaiano, M.; Dagorret, J.; Chérif-Zahar, B.; Marie, S. Metabolic and neurobehavioral disturbances induced by purine recycling deficiency in Drosophila. Elife 2024, 12, RP88510. [Google Scholar] [CrossRef]
- Kim, D.; Lee, S.-E. Proteomic evaluation of pathways associated with phosphine-induced mitochondrial dysfunction and resistance mechanisms in Tribolium castaneum against phosphine fumigation: Whole and partial proteome identification. Ecotoxicol. Environ. Saf. 2025, 289, 117652. [Google Scholar] [CrossRef]
- Koo, H.-N.; Seok, S.J.; Kim, H.K.; Kim, G.-H.; Yang, J.O. Comparative Proteomics Analysis of Phosphine-Resistant and Phosphine-Susceptible Sitophilus oryzae (Coleoptera: Curculionidae). Appl. Sci. 2021, 11, 4163. [Google Scholar] [CrossRef]
- Lee, J.; Kim, C.-H.; Jang, H.A.; Kim, J.K.; Kotaki, T.; Shinoda, T.; Shinada, T.; Yoo, J.-W.; Lee, B.L. Burkholderia gut symbiont modulates titer of specific juvenile hormone in the bean bug Riptortus pedestris. Dev. Comp. Immunol. 2019, 99, 103399. [Google Scholar] [CrossRef]
- Alnajim, I.; Aldosary, N.; Agarwal, M.; Liu, T.; Du, X.; Ren, Y. Role of lipids in phosphine resistant stored-grain insect pests Tribolium castaneum and Rhyzopertha dominica. Insects 2022, 13, 798. [Google Scholar] [CrossRef] [PubMed]
- Arrese, E.L.; Soulages, J.L. Insect fat body: Energy, metabolism, and regulation. Annu. Rev. Entomol. 2010, 55, 207–225. [Google Scholar] [CrossRef] [PubMed]
- Downer, R.; Matthews, J. Patterns of lipid distribution and utilisation in insects. Am. Zool. 1976, 16, 733–745. [Google Scholar] [CrossRef]
- Bohdanowicz, M.; Grinstein, S. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol. Rev. 2013, 93, 69–106. [Google Scholar] [CrossRef]
- Bieberich, E. Sphingolipids and lipid rafts: Novel concepts and methods of analysis. Chem. Phys. Lipids 2018, 216, 114–131. [Google Scholar] [CrossRef]
- Rajendran, L.; Simons, K. Lipid rafts and membrane dynamics. J. Cell Sci. 2005, 118, 1099–1102. [Google Scholar] [CrossRef]
- van Meer, G.; Lisman, Q. Sphingolipid transport: Rafts and translocators. J. Biol. Chem. 2002, 277, 25855–25858. [Google Scholar] [CrossRef]
- Borges, A.R.; Link, F.; Engstler, M.; Jones, N.G. The glycosylphosphatidylinositol anchor: A linchpin for cell surface versatility of trypanosomatids. Front. Cell Dev. Biol. 2021, 9, 720536. [Google Scholar] [CrossRef]
- Kinoshita, T. Glycosylphosphatidylinositol (GPI) anchors: Biochemistry and cell biology: Introduction to a thematic review series. J. Lipid Res. 2016, 57, 4–5. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.-K.; Jeon, J.-C.; Seok, S.-J.; Kim, G.-H.; Koo, H.-N.; Lee, D.-W. Comparative metabolic profiling in Drosophila suzukii by combined treatment of fumigant phosphine and low temperature. Metabolites 2024, 14, 526. [Google Scholar] [CrossRef]
- Beenakkers, A.M.T.; Van der Horst, D.J.; Van Marrewijk, W.J.A. Insect lipids and lipoproteins, and their role in physiological processes. Prog. Lipid Res. 1985, 24, 19–67. [Google Scholar] [CrossRef] [PubMed]
- Jutsum, A.; Goldsworthy, G. Fuels for flight in Locusta. J. Insect Physiol. 1976, 22, 243–249. [Google Scholar] [CrossRef]
- Finney, D. Probit Analysis. Cambridge University Press: Cambridge, UK, 1971. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
| Stage | Number Treated | LCt10 (95% CI) | LCt50 (95% CI) | Slope ± SE | df | χ2 |
|---|---|---|---|---|---|---|
| Adult | 450 | 0.161 (0.137–0.174) | 0.216 (0.205–0.232) | 5.17 ± 1.93 | 13 | 17.03 |
| KEGG ID | Compound | Fold Change (vs. [Mock]) | Related Pathway | ||
|---|---|---|---|---|---|
| [LCt10] | [LCt50] | ||||
| C00422 | TG(12:0/12:0/12:0) | 8.765 | 5.578 | api00561 | Glycerolipid metabolism |
| C02960 | Cer(d18:1/18:0) | 12.796 | 3.470 | api00600 | Sphingolipid metabolism |
| C00319 | Sphingosine | 1.709 | N.D. | api00600 | Sphingolipid metabolism |
| C06126 | Galabiosylceramide (d18:1/22:0) | 339,388 | N.D. | api00600 | Sphingolipid metabolism |
| C12126 | Dihydroceramide | 7,290,115 | N.D. | api00600 | Sphingolipid metabolism |
| C03997 | 5-Hydroxymethyldeoxycytidylate | 213,974 | N.D. | api00240 | Pyrimidine metabolism |
| C02538 | Estrone 3-sulfate | 56,747 | N.D. | api01100 | Metabolic pathways |
| C05502 | 22R-hydroxycholesterol | N.D. | 129,099 | api01100 | Metabolic pathways |
| C08972 | Quillaic acid | N.D. | 242,641 | api01100 | Metabolic pathways |
| C11472 | D-glycero-D-manno-heptose 1,7-bisphosphate | N.D. | 115,917 | api01250 | Biosynthesis of nucleotide sugars |
| C00350 | PE(17:2(9Z,12Z)/20:1(11Z)) | −3.167 | −1.173 | api00563 | GPI-anchor biosynthesis |
| api00564 | Glycerophospholipid metabolism | ||||
| C00319 | D-erythro-sphingosine C-17 | −2.251 | −1.868 | api00600 | Sphingolipid metabolism |
| C00836 | C17 sphinganine | −2.648 | −2.127 | api00600 | Sphingolipid metabolism |
| C03033 | Epinephrine glucuronide | −3.985 | −1.869 | api00040 | Pentose and glucuronate interconversions |
| api00053 | Ascorbate and aldarate metabolism | ||||
| C00255 | Riboflavin (vitamin B2) | −3.677 | −1.833 | api00740 | Riboflavin metabolism |
| api02010 | ABC transporters | ||||
| C00242 | Guanine | −3.125 | −1.762 | api00230 | Purine metabolism |
| C00881 | Deoxycytidine | −8.308 | −2.426 | api00240 | Pyrimidine metabolism |
| api02010 | ABC transporters | ||||
| C16582 | 2-Hydroxyfelbamate | −3.179 | −1.727 | api00982 | Drug metabolism: cytochrome P450 |
| C14876 | S-(2-Hydroxyethyl)-N-acetyl-L-cysteine | −6.718 | −2.825 | api00980 | Metabolism of xenobiotics by cytochrome P450 |
| C00475 | Cytidine | −7.167 | −2.400 | api00240 | Pyrimidine metabolism |
| api02010 | ABC transporters | ||||
| C06193 | Guanosine 3’-phosphate | −3.831 | −2.315 | api00230 | Purine metabolism |
| C00331 | Indolepyruvate | −3.874 | −1.644 | api00380 | Tryptophan metabolism |
| C02237 | 5-Oxo-D-proline | −4.788 | −2.178 | api01100 | Metabolic pathways |
| C00020 | Adenosine 5’-monophosphate (AMP) | −3.620 | −1.854 | api00230 | Purine metabolism |
| C02989 | L-Methionine S-oxide | −5.030 | −2.178 | api00270 | Cysteine and methionine metabolism |
| C06156 | D-Glucosamine 1-phosphate | −4.036 | −1.826 | api01250 | Biosynthesis of nucleotide sugars |
| C00015 | Uridine diphosphate (UDP) | −2.803 | −1.853 | api00240 | Pyrimidine metabolism |
| C00570 | CDP-ethanolamine | −3.028 | −1.651 | api00564 | Glycerophospholipid metabolism |
| C10556 | Deoxypodophyllotoxin | −10.196 | −2.280 | api01100 | Metabolic pathways |
| C02737 | PS(18:0/0:0) | −4.339 | −3.272 | api00564 | Glycerophospholipid metabolism |
| C06542 | Ajmaline | −6.059 | −1.892 | api01100 | Metabolic pathways |
| C19564 | 4-(Nitrosoamino)-1-(3-pyridinyl)-1-butanone | −2.978 | −1.793 | api00980 | Metabolism of xenobiotics by cytochrome P450 |
| C11680 | Cathenamine | N.D. | N.D. | api01100 | Metabolic pathways |
| C00422 | TG(16:1(9Z)/14:0/16:1(9Z))[iso3] | N.D. | N.D. | api00561 | Glycerolipid metabolism |
| C16692 | Mannopine | N.D. | N.D. | api02010 | ABC transporters |
| C01152 | 3-Methyl-L-histidine | N.D. | N.D. | api00340 | Histidine metabolism |
| C16505 | (10S)-Juvenile hormone III diol | N.D. | N.D. | api00981 | Insect hormone biosynthesis |
| C04874 | 7,8-Dihydroneopterin | N.D. | N.D. | api00790 | Folate biosynthesis |
| C19606 | NNAL-N-glucuronide | N.D. | N.D. | api00980 | Metabolism of xenobiotics by cytochrome P450 |
| C00350 | PE(12:0/19:1(9Z)) | N.D. | −1.260 | api00563 | GPI-anchor biosynthesis |
| api00564 | Glycerophospholipid metabolism | ||||
| C00350 | PE(19:1(9Z)/0:0) | N.D. | 1.079 | api00563 | GPI-anchor biosynthesis |
| api00564 | Glycerophospholipid metabolism | ||||
| C16365 | 5-Acetylamino-6-formylamino-3-methyluracil | N.D. | −1.010 | api00232 | Caffeine metabolism |
| C00016 | Flavin adenine dinucleotide (FAD) | N.D. | −1.875 | api00740 | Riboflavin metabolism |
| api04977 | Vitamin digestion and absorption | ||||
| C00170 | 5’-Deoxy-5’-(methylthio)adenosine | −4.264 | N.D. | api00270 | Cysteine and methionine metabolism |
| Log2 Fold Change (vs. [Mock]) | LMP ID | Category | Main Class | Compound | |
|---|---|---|---|---|---|
| [LCt10] | [LCt50] | ||||
| −16.17 | −16.17 | LMSP02010018 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:0/13:0) |
| −1.03 | −0.87 | LMSP02020015 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:0/18:1(9Z)) |
| −22.06 | −0.38 | LMSP02010004 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:1/16:0) |
| −18.80 | −0.54 | LMSP02010006 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:1/18:0) |
| −1.20 | −1.06 | LMSP02010003 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:1/18:1(9Z)) |
| 0.00 | 15.00 | LMSP02010007 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:1/20:0) |
| −1.36 | −1.48 | LMSP02010008 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:1/22:0) |
| −1.16 | −1.11 | LMSP02010026 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:2/20:0) |
| −1.39 | −1.04 | LMSP02010027 | Sphingolipids [SP] | Ceramides [SP02] | Cer(d18:2/20:1) |
| −17.13 | −17.13 | LMSP02050007 | Sphingolipids [SP] | Ceramides [SP02] | CerP(d18:1/24:1(15Z)) |
| 2.29 | 2.10 | LMSP0501AA31 | Sphingolipids [SP] | Neutral glycosphingolipids [SP05] | GlcCer(d16:1/23:0) |
| 1.76 | 1.73 | LMSP0501AA32 | Sphingolipids [SP] | Neutral glycosphingolipids [SP05] | GlcCer(d18:1/23:0) |
| 1.97 | 1.50 | LMSP0501AA10 | Sphingolipids [SP] | Neutral glycosphingolipids [SP05] | GlcCer(d18:1/26:1(17Z)) |
| 1.17 | 0.77 | LMGL02010095 | Glycerolipids [GL] | Diradylglycerols [GL02] | DG(16:1(9Z)/21:0/0:0)[iso2] |
| 18.29 | 18.12 | LMGL02010034 | Glycerolipids [GL] | Diradylglycerols [GL02] | DG(17:0/18:2(9Z,12Z)/0:0)[iso2] |
| 1.25 | 0.87 | LMGL02010068 | Glycerolipids [GL] | Diradylglycerols [GL02] | DG(17:0/20:2(11Z,14Z)/0:0)[iso2] |
| 1.46 | 1.22 | LMGL02010088 | Glycerolipids [GL] | Diradylglycerols [GL02] | DG(18:3(9Z,12Z,15Z)/19:0/0:0)[iso2] |
| 1.16 | 0.23 | LMGL02010190 | Glycerolipids [GL] | Diradylglycerols [GL02] | DG(19:0/22:0/0:0)[iso2] |
| 0.00 | 19.84 | LMGL02010250 | Glycerolipids [GL] | Diradylglycerols [GL02] | DG(19:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)/0:0)[iso2] |
| 16.28 | 16.15 | LMGL02010296 | Glycerolipids [GL] | Diradylglycerols [GL02] | DG(22:3(10Z,13Z,16Z)/22:5(7Z,10Z,13Z,16Z,19Z)/0:0)[iso2] |
| −18.49 | −18.49 | LMGL03010017 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(16:0/16:0/16:1(9Z))[iso3] |
| −0.70 | −1.06 | LMGL03010970 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(16:0/20:2(11Z,14Z)/22:1(13Z))[iso6] |
| −15.83 | −15.83 | LMGL03010303 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(16:1(9Z)/18:2(9Z,12Z)/20:1(11Z))[iso6] |
| −16.51 | −16.51 | LMGL03010948 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(16:1(9Z)/20:4(5Z,8Z,11Z,14Z)/20:5(5Z,8Z,11Z,14Z,17Z))[iso6] |
| −16.53 | −16.53 | LMGL03010177 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(17:0/17:1(9Z)/20:0)[iso6] |
| 2.87 | 2.20 | LMGL03010554 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(17:0/18:3(9Z,12Z,15Z)/20:4(5Z,8Z,11Z,14Z))[iso6] |
| 15.02 | 14.48 | LMGL03010196 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(17:2(9Z,12Z)/17:2(9Z,12Z)/18:3(9Z,12Z,15Z))[iso3] |
| 2.03 | 1.42 | LMGL03011824 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(17:2(9Z,12Z)/20:5(5Z,8Z,11Z,14Z,17Z)/22:5(7Z,10Z,13Z,16Z,19Z))[iso6] |
| 1.76 | 2.06 | LMGL03010748 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(18:1(9Z)/18:1(9Z)/20:5(5Z,8Z,11Z,14Z,17Z))[iso3] |
| 17.21 | 17.33 | LMGL03011321 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(18:1(9Z)/20:4(5Z,8Z,11Z,14Z)/20:5(5Z,8Z,11Z,14Z,17Z))[iso6] |
| −18.36 | −18.36 | LMGL03011398 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(18:2(9Z,12Z)/20:4(5Z,8Z,11Z,14Z)/20:5(5Z,8Z,11Z,14Z,17Z))[iso6] |
| 2.05 | 1.56 | LMGL03011018 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(18:3(9Z,12Z,15Z)/19:0/20:4(5Z,8Z,11Z,14Z))[iso6] |
| −14.22 | −14.22 | LMGL03012401 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(18:3(9Z,12Z,15Z)/22:4(7Z,10Z,13Z,16Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z))[iso6] |
| −18.42 | −18.42 | LMGL03011840 | Glycerolipids [GL] | Triradylglycerols [GL03] | TG(20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z)) |
| −17.36 | −17.36 | LMGP02010101 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(10:0/10:0) |
| 16.43 | 15.77 | LMGP02010232 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(11:0/14:0)[U] |
| −22.56 | −22.56 | LMGP02010230 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(11:0/16:0)[U] |
| −21.89 | −21.89 | LMGP02010371 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(12:0/19:1(9Z)) |
| −21.95 | −21.95 | LMGP02010397 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(13:0/20:2(11Z,14Z)) |
| −22.61 | −22.61 | LMGP02010062 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(14:0/15:0)[U] |
| 0.00 | 21.68 | LMGP02010419 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(14:0/22:1(11Z)) |
| −16.58 | −16.58 | LMGP02010430 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(14:1(9Z)/17:1(9Z)) |
| 22.52 | 0.00 | LMGP02010441 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(14:1(9Z)/20:1(11Z)) |
| 0.58 | −20.36 | LMGP02010456 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(15:0/17:1(9Z)) |
| −23.50 | −23.50 | LMGP02010458 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(15:0/18:1(9Z)) |
| −23.79 | −23.79 | LMGP02010041 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(16:0/18:3(9Z,12Z,15Z)) |
| −25.08 | −25.08 | LMGP02010509 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(16:0/19:1(9Z)) |
| −21.21 | −21.21 | LMGP02010356 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(16:1(5Z)/16:1(5Z)) |
| −26.03 | −26.03 | LMGP02010528 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(16:1(9Z)/19:1(9Z)) |
| 22.34 | 22.19 | LMGP02010531 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(16:1(9Z)/20:2(11Z,14Z)) |
| −21.36 | −21.36 | LMGP02010251 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(17:0/14:0)[U] |
| 0.00 | 23.89 | LMGP02010580 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(17:1(9Z)/20:1(11Z)) |
| −16.42 | −16.42 | LMGP02010630 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(18:0/19:0) |
| −22.82 | −22.82 | LMGP02011202 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(18:0/20:3(8Z,11Z,14Z)) |
| −16.27 | −16.27 | LMGP20020004 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(18:0/22:6(4Z,7Z,10Z,12E,16Z,19Z)(14OH)) |
| 0.00 | 19.69 | LMGP02010768 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(18:4(6Z,9Z,12Z,15Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) |
| −19.63 | −19.63 | LMGP02010779 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(19:0/18:4(6Z,9Z,12Z,15Z)) |
| −19.78 | −19.78 | LMGP02010816 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(19:1(9Z)/20:4(5Z,8Z,11Z,14Z)) |
| −28.16 | −28.16 | LMGP02010851 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(20:1(11Z)/17:2(9Z,12Z)) |
| −20.02 | −20.02 | LMGP02010906 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(20:3(8Z,11Z,14Z)/15:0) |
| −18.03 | −18.03 | LMGP02010955 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(20:4(5Z,8Z,11Z,14Z)/20:5(5Z,8Z,11Z,14Z,17Z)) |
| −16.70 | −16.70 | LMGP02011167 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(21:0/18:0) |
| 15.71 | 15.54 | LMGP02050025 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(22:0/0:0) |
| −16.39 | −0.16 | LMGP02010292 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(22:0/24:1(15Z)) |
| −18.97 | −18.97 | LMGP02011083 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(22:2(13Z,16Z)/18:1(9Z)) |
| 19.91 | 0.00 | LMGP02020034 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(O-16:0/21:0) |
| 20.15 | 19.95 | LMGP02020048 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(O-18:0/18:2(9Z,12Z)) |
| −17.57 | −17.57 | LMGP02020070 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE(O-20:0/17:2(9Z,12Z)) |
| −22.94 | −22.94 | LMGP02010347 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE-NMe(16:0/16:0)[U] |
| −24.09 | −24.09 | LMGP02010326 | Glycerophospholipids [GP] | Glycerophosphoethanolamines [GP02] | PE-NMe2(18:1(9Z)/18:1(9Z)) |
| 15.77 | 15.40 | LMGP03050009 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(14:0/0:0) |
| 18.94 | 18.83 | LMGP03010921 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(14:0/22:0) |
| −24.05 | −24.05 | LMGP03010135 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(14:1(9Z)/22:1(11Z)) |
| −21.60 | −21.60 | LMGP03010238 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(17:0/19:1(9Z)) |
| 15.20 | 15.06 | LMGP03050003 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(18:0/0:0)[U] |
| 1.00 | 0.79 | LMGP03010961 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(18:0/20:1(11Z)) |
| −17.84 | −17.84 | LMGP03010960 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(18:0/20:2(11Z,14Z)) |
| 16.47 | 16.26 | LMGP03050011 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(18:2(9Z,12Z)/0:0) |
| −18.20 | −18.20 | LMGP03010458 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(19:0/16:1(9Z)) |
| 18.98 | 0.00 | LMGP03010868 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(19:0/19:0) |
| 18.05 | 17.70 | LMGP03010536 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(20:1(11Z)/17:0) |
| −19.65 | 0.30 | LMGP03010537 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(20:1(11Z)/17:1(9Z)) |
| 18.39 | 18.34 | LMGP03010614 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(20:3(8Z,11Z,14Z)/21:0) |
| −22.02 | −22.02 | LMGP03010733 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(22:1(11Z)/17:0) |
| 1.11 | 0.79 | LMGP03010768 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(22:2(13Z,16Z)/18:1(9Z)) |
| −15.14 | −15.14 | LMGP03010840 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/20:5(5Z,8Z,11Z,14Z,17Z)) |
| −14.47 | −14.47 | LMGP03030030 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(P-18:0/14:0) |
| −15.69 | −15.69 | LMGP03030053 | Glycerophospholipids [GP] | Glycerophosphoserines [GP03] | PS(P-18:0/22:1(11Z)) |
| −18.04 | −0.57 | LMGP04010123 | Glycerophospholipids [GP] | Glycerophosphoglycerols [GP04] | PG(14:1(9Z)/18:3(9Z,12Z,15Z)) |
| 1.29 | 0.85 | LMGP04010256 | Glycerophospholipids [GP] | Glycerophosphoglycerols [GP04] | PG(17:1(9Z)/17:0) |
| 0.00 | 15.39 | LMGP04010004 | Glycerophospholipids [GP] | Glycerophosphoglycerols [GP04] | PG(21:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) |
| −15.58 | −15.58 | LMGP04020088 | Glycerophospholipids [GP] | Glycerophosphoglycerols [GP04] | PG(O-16:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) |
| −15.83 | −15.83 | LMGP04020037 | Glycerophospholipids [GP] | Glycerophosphoglycerols [GP04] | PG(O-18:0/21:0) |
| −20.74 | −20.74 | LMGP04060001 | Glycerophospholipids [GP] | Glycerophosphoglycerols [GP04] | PG(O-20:0/0:0) |
| 16.64 | 15.98 | LMGP04020065 | Glycerophospholipids [GP] | Glycerophosphoglycerols [GP04] | PG(O-20:0/21:0) |
| −18.54 | −18.54 | LMGP06010167 | Glycerophospholipids [GP] | Glycerophosphoinositols [GP06] | PI(16:0/22:1(11Z)) |
| −17.98 | −17.98 | LMGP06010168 | Glycerophospholipids [GP] | Glycerophosphoinositols [GP06] | PI(16:0/22:2(13Z,16Z)) |
| 0.00 | 22.05 | LMGP06010185 | Glycerophospholipids [GP] | Glycerophosphoinositols [GP06] | PI(16:1(9Z)/20:1(11Z)) |
| 0.39 | −22.33 | LMGP06010283 | Glycerophospholipids [GP] | Glycerophosphoinositols [GP06] | PI(18:0/18:3(6Z,9Z,12Z)) |
| −21.93 | −21.93 | LMGP06010428 | Glycerophospholipids [GP] | Glycerophosphoinositols [GP06] | PI(19:0/17:1(9Z)) |
| −18.01 | −18.01 | LMGP06010611 | Glycerophospholipids [GP] | Glycerophosphoinositols [GP06] | PI(20:4(5Z,8Z,11Z,14Z)/21:0) |
| −15.77 | −15.77 | LMGP10010921 | Glycerophospholipids [GP] | Glycerophosphates [GP10] | PA(14:1(9Z)/14:1(9Z)) |
| −16.64 | −16.64 | LMGP10050024 | Glycerophospholipids [GP] | Glycerophosphates [GP10] | PA(18:4(6Z,9Z,12Z,15Z)/0:0) |
| −17.23 | 2.22 | LMGP10010853 | Glycerophospholipids [GP] | Glycerophosphates [GP10] | PA(21:0/13:0) |
| 16.05 | 15.57 | LMGP10010004 | Glycerophospholipids [GP] | Glycerophosphates [GP10] | PA(21:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) |
| −17.53 | −17.53 | LMGP10020015 | Glycerophospholipids [GP] | Glycerophosphates [GP10] | PA(O-16:0/21:0) |
| −15.26 | −15.26 | LMGP10020043 | Glycerophospholipids [GP] | Glycerophosphates [GP10] | PA(O-20:0/13:0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Suh, S.-J.; Kim, B.-S.; Lee, D.-W. Comparative Metabolomics Reveals Phosphine-Induced Metabolic Disruptions in Planococcus citri (Risso). Int. J. Mol. Sci. 2025, 26, 8020. https://doi.org/10.3390/ijms26168020
Lee J, Suh S-J, Kim B-S, Lee D-W. Comparative Metabolomics Reveals Phosphine-Induced Metabolic Disruptions in Planococcus citri (Risso). International Journal of Molecular Sciences. 2025; 26(16):8020. https://doi.org/10.3390/ijms26168020
Chicago/Turabian StyleLee, Junbeom, Soo-Jung Suh, Bong-Su Kim, and Dae-Weon Lee. 2025. "Comparative Metabolomics Reveals Phosphine-Induced Metabolic Disruptions in Planococcus citri (Risso)" International Journal of Molecular Sciences 26, no. 16: 8020. https://doi.org/10.3390/ijms26168020
APA StyleLee, J., Suh, S.-J., Kim, B.-S., & Lee, D.-W. (2025). Comparative Metabolomics Reveals Phosphine-Induced Metabolic Disruptions in Planococcus citri (Risso). International Journal of Molecular Sciences, 26(16), 8020. https://doi.org/10.3390/ijms26168020







