Hijacking the Host Cell for Replication: Pro-Viral Host Factors Involved in EVA71 Infection
Abstract
1. Introduction
2. Pro-Viral Host Factors Facilitating Viral Entry
3. Pro-Viral Host Factors Regulating Viral RNA Dynamics
4. Pro-Viral Host Factors Modulating Viral Protein
5. Host Metabolic Pro-Viral Factors
6. Pro-Viral Host Factors Orchestrating Immune Evasion
7. Pro-Viral Host Factors Mediating Virion Egress and Dissemination
8. Other Pro-Viral Factors
9. Conclusions
10. Perspectives
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EVA71 | Enterovirus A71 |
| HFMD | Hand, foot, and mouth disease |
| UTR | Untranslated region |
| SLs | Stem loops |
| IRES | Internal ribosome entry site |
| ITAFs | IRES transfactors |
| HSP | Heat shock protein |
| SCARB2 | Scavenger receptor class B member 2 |
| SLC35B2 | Solute carrier family 35 member B2 |
| B3GAT3 | Beta-1, 3-glucuronyltransferase 3 |
| HS | Heparan sulfate |
| ROs | Replication organelles |
| PI4KB | Phosphatidylinositol-4-kinase IIIβ |
| PI4P | Phosphatidylinositol 4-phosphate |
| ARF1 | ADP-ribosylation factor 1 |
| GTPase | Guanosine triphosphatase |
| GBF1 | Golgi Brefeldin A-resistant guanine nucleotide exchange factor 1 |
| ACBD3 | Acyl-coenzyme A-binding domain containing 3 |
| NAT | N-terminal acetyltransferase |
| ANXA2 | Annexin A2 |
| RNP | Ribonucleoprotein |
| DDX | DEAD-box helicase |
| 4E-T | eIF4E-transporter |
| METTL3 | Methyltransferase-like 3 |
| NAT10 | N-acetyltransferase 10 |
| PCBP2 | Poly(rC)-binding protein 2 |
| DDX3X | DEAD-box helicase 3 X-linked |
| GADD34 | DNA damage-inducible protein 34 |
| eIF2α | Eukaryotic translation initiation factor 2α |
| hnRNP A1 | heterogeneous nuclear ribonucleoprotein A1 |
| NSUN2 | NOP2/Sun RNA methyltransferase family 2 |
| USP | Ubiquitin-specific protease |
| ABPs | Activity-based probes |
| PTB | Polypyrimidine tract-binding protein 1 |
| ER | Endoplasmic reticulum |
| ATG | Autophagy-related protein |
| RAB11A | RAS-associated protein 11A |
| TCA | Tricarboxylic acid cycle |
| PI3K | Phosphoinositide 3-kinase |
| AKT | Protein kinase B |
| SLC38A8 | Solute carrier family 38 member 8 |
| p70S6K1 | Ribosomal protein S6 kinase beta-1 |
| rpS6 | Ribosomal protein S6 |
| G3BP2 | Stress granule assembly factor 2 |
| OSBP | Oxysterol-binding protein |
| VDAC3 | Voltage-dependent anion channel 3 |
| ROS | Reactive oxygen species |
| RIG-I | Retinoic acid-inducible gene I |
| MAVS | Mitochondrial antiviral-signaling protein |
| IFN | Interferon |
| IRF | IFN response factor |
| G3BP1 | Ras GTPase-activating protein-binding protein |
| LRRC25 | Leucine-rich repeat-containing 25 |
| YTHDF2 | YTH N6-methyladenosine RNA binding protein F2 |
| GTPBP4 | GTP-binding protein 4 |
| PTEN | Phosphatase and tensin homolog |
| TRAF | Tumor necrosis factor receptor-associated factor |
| EVs | Extracellular vesicles |
| MVs | Microvesicles |
| BBB | Blood–brain barrier |
| CNS | Central nervous system |
| ZO-1 | zonula occludens-1 |
| VPS | Vacuolar protein sorting |
| PVRL1 | Poliovirus receptor-related 1 |
| AJs | Adherens Junctions |
| BECLIN1 | BCL-2 interacting protein 1 |
| HMGB1 | High mobility group box 1 |
| HSBP1 | Heat shock factor-binding protein 1 |
| ULK | Kinase unc-51-like autophagy-activating kinase |
| HSV-1 | Herpes simplex virus 1 |
| OAS | 2′-5′-oligoadenylate synthetase |
| CVA16 | Coxsackievirus 16 |
References
- Nayak, G.; Bhuyan, S.K.; Bhuyan, R.; Sahu, A.; Kar, D.; Kuanar, A. Global emergence of Enterovirus 71: A systematic review. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 78. [Google Scholar] [CrossRef]
- Yi, E.-J.; Shin, Y.-J.; Kim, J.-H.; Kim, T.-G.; Chang, S.-Y. Enterovirus 71 infection and vaccines. Clin. Exp. Vaccine Res. 2017, 6, 4–14. [Google Scholar] [CrossRef]
- Mao, Q.-y.; Wang, Y.; Bian, L.; Xu, M.; Liang, Z. EV71 vaccine, a new tool to control outbreaks of hand, foot and mouth disease (HFMD). Expert Rev. Vaccines 2016, 15, 599–606. [Google Scholar] [CrossRef]
- Baggen, J.; Thibaut, H.J.; Strating, J.R.P.M.; van Kuppeveld, F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018, 16, 368–381. [Google Scholar] [CrossRef] [PubMed]
- Davila-Calderon, J.; Patwardhan, N.N.; Chiu, L.-Y.; Sugarman, A.; Cai, Z.; Penutmutchu, S.R.; Li, M.-L.; Brewer, G.; Hargrove, A.E.; Tolbert, B.S. IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex. Nat. Commun. 2020, 11, 4775. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.R.; Sarnow, P. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology 2003, 315, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Shih, S.-R.; Stollar, V.; Li, M.-L. Host factors in enterovirus 71 replication. J. Virol. 2011, 85, 9658–9666. [Google Scholar] [CrossRef]
- Lozano, G.; Martínez-Salas, E. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015, 12, 113–120. [Google Scholar] [CrossRef]
- Rossmann, M.G.; Johnson, J.E. Icosahedral RNA virus structure. Annu. Rev. Biochem. 1989, 58, 533–573. [Google Scholar] [CrossRef]
- Tan, Y.W.; Hong, W.J.; Chu, J.J. Inhibition of enterovirus VP4 myristoylation is a potential antiviral strategy for hand, foot and mouth disease. Antivir. Res. 2016, 133, 191–195. [Google Scholar] [CrossRef]
- Dang, M.; Wang, X.; Wang, Q.; Wang, Y.; Lin, J.; Sun, Y.; Li, X.; Zhang, L.; Lou, Z.; Wang, J.; et al. Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71. Protein Cell 2014, 5, 692–703. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Yamashita, Y.; Li, J.; Hanagata, N.; Minowa, T.; Takemura, T.; Koike, S. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 2009, 15, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Shimojima, M.; Tano, Y.; Miyamura, T.; Wakita, T.; Shimizu, H. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 2009, 15, 794–797. [Google Scholar] [CrossRef]
- De Sena, J.; Mandel, B. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology 1977, 78, 554–566. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wang, X.; Hu, Z.; Gao, Q.; Sun, Y.; Li, X.; Porta, C.; Walter, T.S.; Gilbert, R.J.; Zhao, Y.; et al. Picornavirus uncoating intermediate captured in atomic detail. Nat. Commun. 2013, 4, 1929. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Ye, H.Q.; Deng, C.L.; Li, R.; Zhang, B.; Gong, P. A nucleobase-binding pocket in a viral RNA-dependent RNA polymerase contributes to elongation complex stability. Nucleic Acids Res. 2020, 48, 1392–1405. [Google Scholar] [CrossRef]
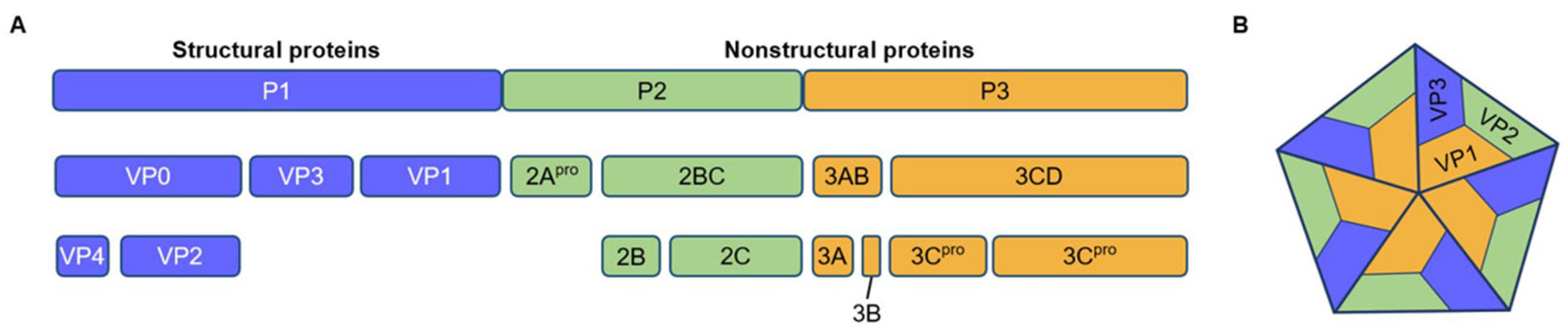
- Yuan, J.; Shen, L.; Wu, J.; Zou, X.; Gu, J.; Chen, J.; Mao, L. Enterovirus A71 Proteins: Structure and function. Front. Microbiol. 2018, 9, 286. [Google Scholar] [CrossRef]
- Wang, R.Y.; Kuo, R.L.; Ma, W.C.; Huang, H.I.; Yu, J.S.; Yen, S.M.; Huang, C.R.; Shih, S.R. Heat shock protein-90-beta facilitates enterovirus 71 viral particles assembly. Virology 2013, 443, 236–247. [Google Scholar] [CrossRef]
- Tsou, Y.L.; Lin, Y.W.; Chang, H.W.; Lin, H.Y.; Shao, H.Y.; Yu, S.L.; Liu, C.C.; Chitra, E.; Sia, C.; Chow, Y.H. Heat shock protein 90: Role in enterovirus 71 entry and assembly and potential target for therapy. PLoS ONE 2013, 8, e77133. [Google Scholar] [CrossRef]
- Gu, J.; Wu, J.; Fang, D.; Qiu, Y.; Zou, X.; Jia, X.; Yin, Y.; Shen, L.; Mao, L. Exosomes cloak the virion to transmit Enterovirus 71 non-lytically. Virulence 2019, 11, 32–38. [Google Scholar] [CrossRef]
- Guo, D.; Yu, X.; Wang, D.; Li, Z.; Zhou, Y.; Xu, G.; Yuan, B.; Qin, Y.; Chen, M. SLC35B2 acts in a dual role in the host sulfation required for EV71 infection. J. Virol. 2022, 96, e0204221. [Google Scholar] [CrossRef]
- Kobayashi, K.; Mizuta, K.; Koike, S. Heparan sulfate attachment receptor is a major selection factor for attenuated enterovirus 71 mutants during cell culture adaptation. PLoS Pathog. 2020, 16, e1008428. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, M.; Cheng, A.; Wen, X.; Ou, X.; Mao, S.; Gao, Q.; Sun, D.; Jia, R.; Yang, Q.; et al. Enterovirus replication organelles and inhibitors of their formation. Front. Microbiol. 2020, 11, 1817. [Google Scholar] [CrossRef] [PubMed]
- den Boon, J.A.; Nishikiori, M.; Zhan, H.; Ahlquist, P. Positive-strand RNA virus genome replication organelles: Structure, assembly, control. Trends Genet. 2024, 40, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-Y.; Dyakov, B.J.A.; Zhang, J.; Knight, J.D.R.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.-C. Properties of stress granule and P-body proteomes. Mol. Cell 2019, 76, 286–294. [Google Scholar] [CrossRef]
- Hsu, N.-Y.; Ilnytska, O.; Belov, G.; Santiana, M.; Chen, Y.-H.; Takvorian, P.M.; Pau, C.; van der Schaar, H.; Kaushik-Basu, N.; Balla, T.; et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 2010, 141, 799–811. [Google Scholar] [CrossRef]
- Xiao, X.; Lei, X.; Zhang, Z.; Ma, Y.; Qi, J.; Wu, C.; Xiao, Y.; Li, L.; He, B.; Wang, J.; et al. Enterovirus 3A facilitates viral replication by promoting phosphatidylinositol 4-Kinase IIIβ–ACBD3 interaction. J. Virol. 2017, 91, e00791-17. [Google Scholar] [CrossRef]
- Lyoo, H.; van der Schaar, H.M.; Dorobantu, C.M.; Rabouw, H.H.; Strating, J.R.P.M.; van Kuppeveld, F.J.M.; Racaniello, V.R. ACBD3 is an essential pan-enterovirus host factor that mediates the interaction between viral 3A protein and cellular protein PI4KB. mBio 2019, 10, e02742-18. [Google Scholar] [CrossRef]
- Yang, H.; Fan, T.; Xun, M.; Wu, B.; Guo, S.; Li, X.; Zhao, X.; Yao, H.; Wang, H. N-terminal acetyltransferase 6 facilitates enterovirus 71 replication by regulating PI4KB expression and replication organelle biogenesis. J. Virol. 2024, 98, e0174923. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, S.; Lei, P.; Li, Z.; Chen, F.; Chen, Q.; Wang, Y.; Gong, J.; Tang, Q.; Liu, X.; et al. ANXA2 facilitates Enterovirus 71 infection by interacting with 3D polymerase and PI4KB to assist the assembly of replication organelles. Virol. Sin. 2021, 36, 1387–1399. [Google Scholar] [CrossRef]
- Hubstenberger, A.; Courel, M.; Bénard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.B.; Munier, A.; Fradet, M.; et al. P-Body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 2017, 68, 144–157.e145. [Google Scholar] [CrossRef]
- Chahar, H.S.; Chen, S.; Manjunath, N. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 2013, 436, 1–7. [Google Scholar] [CrossRef]
- Ariumi, Y.; Kuroki, M.; Kushima, Y.; Osugi, K.; Hijikata, M.; Maki, M.; Ikeda, M.; Kato, N. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 2011, 85, 6882–6892. [Google Scholar] [CrossRef]
- Dougherty, J.D.; White, J.P.; Lloyd, R.E. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 2011, 85, 64–75. [Google Scholar] [CrossRef]
- Fan, S.; Xu, Z.; Liu, P.; Qin, Y.; Chen, M.; López, S. Enterovirus 71 2A protease inhibits P-Body formation to promote viral RNA synthesis. J. Virol. 2021, 95, e0092221. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Hao, S.; Chen, H.; Chen, Z.; Zhang, Y.; Wang, J.; Wang, H.; Zhang, B.; Qiu, J.; Deng, F.; et al. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 2019, 47, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, Z.; Shu, B.; Meng, J.; Zhang, Y.; Zheng, C.; Ke, X.; Gong, P.; Hu, Q.; Wang, H. SUMO modification stabilizes Enterovirus 71 polymerase 3D to facilitate viral replication. J. Virol. 2016, 90, 10472–10485. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Liu, W.; Miao, Y.; Ma, L.; Yu, B.; Liu, L.; Yang, C.; Zhang, K.; Chen, Z.; Yang, J.; et al. N4-acetylcytidine regulates the replication and pathogenicity of enterovirus 71. Nucleic Acids Res. 2022, 50, 9339–9354. [Google Scholar] [CrossRef]
- Beckham, S.A.; Matak, M.Y.; Belousoff, M.J.; Venugopal, H.; Shah, N.; Vankadari, N.; Elmlund, H.; Nguyen, J.H.C.; Semler, B.L.; Wilce, M.C.J.; et al. Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES. Nucleic Acids Res. 2020, 48, 8006–8021. [Google Scholar] [CrossRef]
- Su, Y.-S.; Tsai, A.-H.; Ho, Y.-F.; Huang, S.-Y.; Liu, Y.-C.; Hwang, L.-H. Stimulation of the internal ribosome entry site (IRES)-dependent translation of Enterovirus 71 by DDX3X RNA helicase and viral 2A and 3C proteases. Front. Microbiol. 2018, 9, 1324. [Google Scholar] [CrossRef]
- Li, H.; Li, W.; Zhang, S.; Qiu, M.; Li, Z.; Lin, Y.; Tan, J.; Qiao, W. Enterovirus 71 activates GADD34 via precursor 3CD to promote IRES-mediated viral translation. Microbiol. Spectr. 2022, 10, e0138821. [Google Scholar] [CrossRef]
- Dan, X.; Wan, Q.; Yi, L.; Lu, J.; Jiao, Y.; Li, H.; Song, D.; Chen, Y.; Xu, H.; He, M.L. Hsp27 responds to and facilitates Enterovirus A71 replication by enhancing viral internal ribosome entry site-mediated translation. J. Virol. 2019, 93, e02322-18. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Sharma, B.; Tiwari, P.K. The small heat shock protein Hsp27: Present understanding and future prospects. J. Therm. Biol. 2017, 69, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Ye, F.; Wang, G.; Han, W.; Wei, Z.; Yin, B.; Yuan, J.; Qiang, B.; Peng, X. Polypyrimidine tract-binding protein regulates enterovirus 71 translation through interaction with the internal ribosomal entry site. Virol. Sin. 2019, 34, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, Z.; Zhang, K.; Hao, H.; Ma, L.; Liu, H.; Yu, B.; Ding, S.; Zhang, X.; Zhu, M.; et al. NSUN2 mediates distinct pathways to regulate enterovirus 71 replication. Virol. Sin. 2024, 39, 574–586. [Google Scholar] [CrossRef]
- Schumann, U.; Zhang, H.-N.; Sibbritt, T.; Pan, A.; Horvath, A.; Gross, S.; Clark, S.J.; Yang, L.; Preiss, T. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 2020, 18, 40. [Google Scholar] [CrossRef]
- Duan, H.; Zhu, M.; Xiong, Q.; Wang, Y.; Xu, C.; Sun, J.; Wang, C.; Zhang, H.; Xu, P.; Peng, Y. Regulation of enterovirus 2A protease-associated viral IRES activities by the cell’s ERK signaling cascade: Implicating ERK as an efficiently antiviral target. Antivir. Res. 2017, 143, 13–21. [Google Scholar] [CrossRef]
- Li, H.; Chen, Y.; Zhang, J.; Lin, Y.; Yang, Z.; Tan, J.; Qiao, W. Identification of the internal ribosome entry sites in the 5′-untranslated region of the c-fos gene. Int. J. Mol. Med. 2021, 47, 56. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, H.; Yang, H.; Liu, Y.; Xun, M.; Li, X.; Fan, T.; Wu, B.; Guo, S.; Wang, H. N-acetyltransferase 8 promotes viral replication by increasing the stability of Enterovirus 71 nonstructural proteins. J. Virol. 2022, 96, e0011922. [Google Scholar] [CrossRef]
- Yang, X.; Tang, M.; Zang, L.; Hao, P.; Chen, Y.; Yuan, Y.; Miao, Y.; Zuo, Y.; Wu, Z.; Che, Z.; et al. Ubiquitin-specific protease 21 aggravates enterovirus 71 (EV71) infection by restricting Lys48-linked ubiquitination of EV71-2A protease. Int. J. Biol. Macromol. 2025, 314, 144202. [Google Scholar] [CrossRef]
- Sun, Y.; Zheng, Q.; Wang, Y.; Pang, Z.; Liu, J.; Yin, Z.; Lou, Z.; Pfeiffer, J.K. Activity-based protein profiling identifies ATG4B as a key host factor for Enterovirus 71 proliferation. J. Virol. 2019, 93, e01092-19. [Google Scholar] [CrossRef]
- Ng, Q.Y.; Mahendran, V.; Lim, Z.Q.; Tan, J.H.Y.; Wong, J.J.F.; Chu, J.J.H.; Chow, V.T.K.; Sze, N.S.K.; Alonso, S. Enterovirus-A71 exploits RAB11 to recruit chaperones for virus morphogenesis. J. Biomed. Sci. 2024, 31, 65. [Google Scholar] [CrossRef]
- Moreno-Altamirano, M.M.B.; Kolstoe, S.E.; Sánchez-García, F.J. Virus control of cell metabolism for replication and evasion of host immune responses. Front. Cell. Infect. Microbiol. 2019, 9, 95. [Google Scholar] [CrossRef]
- Eisenreich, W.; Rudel, T.; Heesemann, J.; Goebel, W. How viral and intracellular bacterial pathogens reprogram the metabolism of host cells to allow their intracellular replication. Front. Cell. Infect. Microbiol. 2019, 9, 42. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, W.; Xiao, M.Z.X.; Zheng, Y.; Liang, Q. The interplay between lipid droplets and virus infection. J. Med. Virol. 2023, 95, e28967. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Liu, S.; Tan, Z.; Yin, L.; Zeng, L.; Liu, T.; Zhang, S.; Zhang, L. Proteomic and metabonomic analysis uncovering Enterovirus A71 reprogramming host cell metabolic pathway. Proteomics 2022, 23, 2200362. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Dang, D.; Zhou, G.; Tao, L.; Sun, T.; Li, D.; Cheng, C.; Feng, H.; Long, J.; Chen, S.; et al. Metabolomic analysis reveals an important role of sphingosine 1-phosphate in the development of HFMD due to EV-A71 infection. Antimicrob. Agents Chemother. 2025, 69, e0127224. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, Z.; Xue, Q.; Yang, F.; Cao, W.; Xue, Z.; Liu, X.; Zheng, H. Picornavirus infection enhances aspartate by the SLC38A8 transporter to promote viral replication. PLoS Pathog. 2023, 19, e1011126. [Google Scholar] [CrossRef]
- Zou, L.; Zhao, H.; Wang, D.; Wang, M.; Zhang, C.; Xiao, F. Expression and purification of a functional recombinant aspartate aminotransferase (AST) from Escherichia coli. J. Microbiol. Biotechnol. 2014, 24, 998–1003. [Google Scholar] [CrossRef]
- Cheng, M.L.; Chien, K.Y.; Lai, C.H.; Li, G.J.; Lin, J.F.; Ho, H.Y. Metabolic reprogramming of host cells in response to enteroviral infection. Cells 2020, 9, 473. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Lai, R.-H.; Chow, Y.-H.; Lin, Y.-W.; Chung, N.-H.; Nien, S.-W.; Juang, J.-L. Hyperglycemia facilitates EV71 replication: Insights into miR-206-mediated regulation of G3BP2 promoting EV71 IRES activity. Theranostics 2024, 14, 2706–2718. [Google Scholar] [CrossRef]
- Jin, G.; Zhang, Z.; Wan, J.; Wu, X.; Liu, X.; Zhang, W. G3BP2: Structure and function. Pharmacol Res 2022, 186, 106548. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, J.; Lu, Z.; Huang, S.; Zhang, S.; Cai, J.; Zhou, Y.; Cao, G.; Yu, J.; Qin, Z.; et al. Enterovirus A71 utilizes host cell lipid β-oxidation to promote its replication. Front. Microbiol. 2022, 13, 961942. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Arita, M.; Sakai, S.; Kojima, H.; Senda, M.; Senda, T.; Hanada, K.; Kato, R. Ligand recognition by the lipid transfer domain of human OSBP is important for enterovirus replication. ACS Infect. Dis. 2022, 8, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-L.; Wu, C.-H.; Chien, K.-Y.; Lai, C.-H.; Li, G.-J.; Liu, Y.-Y.; Lin, G.; Ho, H.-Y. Enteroviral 2B interacts with VDAC3 to regulate reactive oxygen species generation that is essential to viral replication. Viruses 2022, 14, 1717. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, J.; Ouyang, W.; Yao, R.S.Y.; Cao, W.; Li, J.; Zou, R.; Fang, C.; Zeng, F.; Yang, F.; et al. Suppression of innate and acquired immunity in severe hand foot and mouth disease caused by EV71 infections in children. Clin. Immunol. 2023, 248, 109260. [Google Scholar] [CrossRef]
- Wei, J.; Lv, L.; Wang, T.; Gu, W.; Luo, Y.; Feng, H. Recent progress in innate immune responses to Enterovirus A71 and viral evasion strategies. Int. J. Mol. Sci. 2024, 25, 5688. [Google Scholar] [CrossRef]
- Gu, Z.; Shi, W.; Zhang, L.; Hu, Z.; Xu, C. USP19 suppresses cellular type I interferon signaling by targeting TRAF3 for deubiquitination. Future Microbiol. 2017, 12, 767–779. [Google Scholar] [CrossRef]
- Zhao, X.; Di, Q.; Yu, J.; Quan, J.; Xiao, Y.; Zhu, H.; Li, H.; Ling, J.; Chen, W. USP19 (ubiquitin specific peptidase 19) promotes TBK1 (TANK-binding kinase 1) degradation via chaperone-mediated autophagy. Autophagy 2022, 18, 891–908. [Google Scholar] [CrossRef]
- Jayabalan, A.K.; Griffin, D.E.; Leung, A.K.L. Pro-viral and anti-viral roles of the RNA-binding protein G3BP1. Viruses 2023, 15, 449. [Google Scholar] [CrossRef] [PubMed]
- Visser, L.J.; Langereis, M.A.; Rabouw, H.H.; Wahedi, M.; Muntjewerff, E.M.; de Groot, R.J.; van Kuppeveld, F.J.M.; Gallagher, T. Essential role of Enterovirus 2A protease in counteracting stress granule Formation and the Induction of Type I Interferon. J. Virol. 2019, 93, e00222-19. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, Y.; Lin, L.; Si, X.; Wang, T.; Zhong, X.; Tong, L.; Luan, Y.; Chen, Y.; Li, X.; et al. Protease 2A induces stress granule formation during coxsackievirus B3 and enterovirus 71 infections. Virol. J. 2014, 11, 192. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, L.; Xu, X.; Han, H.; Li, P.; Zou, D.; Li, X.; Zheng, L.; Cheng, L.; Shen, Y.; et al. Enterovirus 71 inhibits cytoplasmic stress granule formation during the late stage of infection. Virus Res. 2018, 255, 55–67. [Google Scholar] [CrossRef]
- Yang, W.; Li, D.; Ru, Y.; Bai, J.; Ren, J.; Zhang, J.; Li, L.; Liu, X.; Zheng, H.; Williams, B.R.G. Foot-and-Mouth Disease virus 3A protein causes upregulation of autophagy-related protein LRRC25 to inhibit the G3BP1-mediated RIG-Like helicase-signaling pathway. J. Virol. 2020, 94, e02086-19. [Google Scholar] [CrossRef]
- Du, Y.; Duan, T.; Feng, Y.; Liu, Q.; Lin, M.; Cui, J.; Wang, R.F. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 2018, 37, 351–366. [Google Scholar] [CrossRef]
- Liu, H.; Xue, Q.; Yang, F.; Cao, W.; Liu, P.; Liu, X.; Zhu, Z.; Zheng, H. Foot-and-mouth disease virus VP1 degrades YTHDF2 through autophagy to regulate IRF3 activity for viral replication. Autophagy 2024, 20, 1597–1615. [Google Scholar] [CrossRef]
- Sun, Y.; Feng, L.; Li, J.; Xu, H.; Mei, X.; Feng, L.; Sun, H.; Gao, J.; Zhang, X. miR-545 promoted enterovirus 71 replication via directly targeting phosphatase and tensin homolog and tumor necrosis factor receptor-associated factor 6. J. Cell. Physiol. 2019, 234, 15686–15697. [Google Scholar] [CrossRef]
- Li, D.; Chen, S.; Zhang, W.; Zhang, C.; Sun, T.; Du, Y.; Ding, R.; Gao, Y.; Jin, Y.; Duan, G. MicroRNA-628-5p facilitates Enterovirus 71 infection by suppressing TRAF3 signaling. Cell. Mol. Immunol. 2020, 18, 1320–1322. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Gu, J.; Zhao, Y.; Wu, J.; Chen, Y.; Yin, Y.; Jia, X.; Mao, L. Enterovirus-71 utilizes small extracellular vesicles to cross the blood–brain barrier for infecting the central nervous system via transcytosis. J. Med. Virol. 2022, 95. [Google Scholar] [CrossRef]
- Tian, X.; Liu, B.; Li, L.; Yuan, M.; You, Q.; Zhang, R.; Chen, D.; Cheng, M.; Zheng, N.; He, M.; et al. Microvesicles carrying EV71 virions cross BBB through endocytic pathway to induce brain injury. Cell Commun. Signal. 2025, 23, 183. [Google Scholar] [CrossRef]
- Ruan, Z.; Liang, Y.; Chen, Z.; Yin, J.; Li, C.; Pan, P.; Zhang, Q.; Wu, J.; Luo, Z. Enterovirus 71 non-structural protein 3A hijacks vacuolar protein sorting 25 to boost exosome biogenesis to facilitate viral replication. Front. Microbiol. 2022, 13, 1024899. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Kong, Y.; Rao, M.; Zhou, X.; Li, C.; Meng, Y.; Chen, Y.; Li, H.; Luo, Z. Inhibition of ESCRT-independent extracellular vesicles biogenesis suppresses enterovirus 71 replication and pathogenesis in mice. Int. J. Biol. Macromol. 2024, 267, 131453. [Google Scholar] [CrossRef] [PubMed]
- Hierro, A.; Sun, J.; Rusnak, A.S.; Kim, J.; Prag, G.; Emr, S.D.; Hurley, J.H. Structure of the ESCRT-II endosomal trafficking complex. Nature 2004, 431, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Song, J.; Liu, L.; Zhang, Y.; Wang, L.; Li, Q. microRNA-4516 contributes to different functions of epithelial permeability barrier by targeting poliovirus receptor related protein 1 in Enterovirus 71 and Coxsackievirus A16 infections. Front. Cell. Infect. Microbiol. 2018, 8, 110. [Google Scholar] [CrossRef]
- Samanta, D.; Almo, S.C. Nectin family of cell-adhesion molecules: Structural and molecular aspects of function and specificity. Cell. Mol. Life Sci. 2015, 72, 645–658. [Google Scholar] [CrossRef]
- Chen, T.; Tu, S.; Ding, L.; Jin, M.; Chen, H.; Zhou, H. The role of autophagy in viral infections. J. Biomed. Sci. 2023, 30, 5. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Chaumorcel, M.; Lussignol, M.; Mouna, L.; Cavignac, Y.; Fahie, K.; Cotte-Laffitte, J.; Geballe, A.; Brune, W.; Beau, I.; Codogno, P.; et al. The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1. J. Virol. 2012, 86, 2571–2584. [Google Scholar] [CrossRef]
- Gannagé, M.; Dormann, D.; Albrecht, R.; Dengjel, J.; Torossi, T.; Rämer, P.C.; Lee, M.; Strowig, T.; Arrey, F.; Conenello, G.; et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 2009, 6, 367–380. [Google Scholar] [CrossRef]
- Gladue, D.P.; O’Donnell, V.; Baker-Branstetter, R.; Holinka, L.G.; Pacheco, J.M.; Fernandez-Sainz, I.; Lu, Z.; Brocchi, E.; Baxt, B.; Piccone, M.E.; et al. Foot-and-mouth disease virus nonstructural protein 2C interacts with Beclin1, modulating virus replication. J. Virol. 2012, 86, 12080–12090. [Google Scholar] [CrossRef]
- Hernaez, B.; Cabezas, M.; Muñoz-Moreno, R.; Galindo, I.; Cuesta-Geijo, M.A.; Alonso, C. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr. Mol. Med. 2013, 13, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Kyei, G.B.; Dinkins, C.; Davis, A.S.; Roberts, E.; Singh, S.B.; Dong, C.; Wu, L.; Kominami, E.; Ueno, T.; Yamamoto, A.; et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009, 186, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Orvedahl, A.; Alexander, D.; Tallóczy, Z.; Sun, Q.; Wei, Y.; Zhang, W.; Burns, D.; Leib, D.A.; Levine, B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 2007, 1, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Wan, P.; Yang, G.; Huang, S.; Qin, M.; Yang, H.; Luo, Z.; Wu, K.; Wu, J. Beclin1 binds to Enterovirus 71 3D protein to promote the virus replication. Viruses 2020, 12, 756. [Google Scholar] [CrossRef]
- Li, J.; Ma, J.; Cao, R.; Zhang, Q.; Li, M.; Wang, W.; Wang, Y.; Li, W.; Zhu, Y.; Leng, L. A skin organoid-based infection platform identifies an inhibitor specific for HFMD. Nat. Commun. 2025, 16, 2513. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, J.; Zi, R.; Ji, L.; Hu, J.; Wu, Z.; Fu, Y. Enterovirus 71-induced autophagosome fusion with multivesicular bodies facilitates viral RNA packaging into exosomes. Microb. Pathog. 2022, 173, 105875. [Google Scholar] [CrossRef]
- Wilson, A.; McCormick, C. Reticulophagy and viral infection. Autophagy 2025, 21, 3–20. [Google Scholar] [CrossRef]
- Kuo, R.L.; Kung, S.H.; Hsu, Y.Y.; Liu, W.T. Infection with enterovirus 71 or expression of its 2A protease induces apoptotic cell death. J. Gen. Virol. 2002, 83, 1367–1376. [Google Scholar] [CrossRef]
- Li, M.L.; Hsu, T.A.; Chen, T.C.; Chang, S.C.; Lee, J.C.; Chen, C.C.; Stollar, V.; Shih, S.R. The 3C protease activity of enterovirus 71 induces human neural cell apoptosis. Virology 2002, 293, 386–395. [Google Scholar] [CrossRef]
- Jan, E.; Li, M.-L.; Lin, J.-Y.; Chen, B.-S.; Weng, K.-F.; Shih, S.-R.; Calderon, J.D.; Tolbert, B.S.; Brewer, G. EV71 3C protease induces apoptosis by cleavage of hnRNP A1 to promote apaf-1 translation. PLoS ONE 2019, 14, e0221048. [Google Scholar] [CrossRef]
- Li, J.; Yao, Y.; Chen, Y.; Xu, X.; Lin, Y.; Yang, Z.; Qiao, W.; Tan, J.; López, S. Enterovirus 71 3C promotes apoptosis through cleavage of PinX1, a telomere binding protein. J. Virol. 2017, 91, e02016-16. [Google Scholar] [CrossRef]
- Cong, H.; Du, N.; Yang, Y.; Song, L.; Zhang, W.; Tien, P.; López, S. Enterovirus 71 2B induces cell apoptosis by directly inducing the conformational activation of the proapoptotic protein bax. J. Virol. 2016, 90, 9862–9877. [Google Scholar] [CrossRef]
- Mauthe, M.; Dinesh Kumar, N.; Verlhac, P.; van de Beek, N.; Reggiori, F. HSBP1 is a novel interactor of FIP200 and ATG13 that promotes autophagy initiation and picornavirus replication. Front. Cell. Infect. Microbiol. 2021, 11, 745640. [Google Scholar] [CrossRef]
- Mauthe, M.; Langereis, M.; Jung, J.; Zhou, X.; Jones, A.; Omta, W.; Tooze, S.A.; Stork, B.; Paludan, S.R.; Ahola, T.; et al. An siRNA screen for ATG protein depletion reveals the extent of the unconventional functions of the autophagy proteome in virus replication. J. Cell Biol. 2016, 214, 619–635. [Google Scholar] [CrossRef] [PubMed]
- Mathew, S.S.; Della Selva, M.P.; Burch, A.D. Modification and reorganization of the cytoprotective cellular chaperone Hsp27 during herpes simplex virus type 1 infection. J. Virol. 2009, 83, 9304–9312. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.W.; Yang, Y.X.; Hu, H.D.; An, X.; Ye, F.; Ren, H.; Li, S.L.; Zhang, D.Z. HSPB1 is an intracellular antiviral factor against hepatitis B virus. J. Cell Biochem. 2013, 114, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Sarnow, P.; Tan, R.; Zhang, Y.; Huang, M.; Chen, H.; Liu, Z.; Wang, Z.; Li, X.; Wang, T.; Wang, Z. EV-D68 cleaves LARP1 and PABPC1 by 3Cpro to redirect host mRNA translation machinery toward its genomic RNA. PLoS Pathog. 2025, 21, e1013098. [Google Scholar] [CrossRef]
- Hellen, C.U.; Witherell, G.W.; Schmid, M.; Shin, S.H.; Pestova, T.V.; Gil, A.; Wimmer, E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 1993, 90, 7642–7646. [Google Scholar] [CrossRef]
- Zhang, R.; Cheng, M.; Liu, B.; Yuan, M.; Chen, D.; Wang, Y.; Wu, Z. DEAD-Box helicase DDX6 facilitated RIG-I-mediated type-I interferon response to EV71 infection. Front. Cell. Infect. Microbiol. 2021, 11, 725392. [Google Scholar] [CrossRef]
- Li, P.; Yang, S.; Hu, D.; Wei, D.; Lu, J.; Zheng, H.; Nie, S.; Liu, G.; Yang, H. Enterovirus 71 VP1 promotes mouse Schwann cell autophagy via ER stress-mediated PMP22 upregulation. Int. J. Mol. Med. 2019, 44, 759–767. [Google Scholar] [CrossRef]
- Too, I.H.; Yeo, H.; Sessions, O.M.; Yan, B.; Libau, E.A.; Howe, J.L.; Lim, Z.Q.; Suku-Maran, S.; Ong, W.Y.; Chua, K.B.; et al. Enterovirus 71 infection of motor neuron-like NSC-34 cells undergoes a non-lytic exit pathway. Sci. Rep. 2016, 6, 36983. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.C.; Chang, C.L.; Wang, P.S.; Tsai, Y.; Liu, H.S. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J. Med. Virol. 2009, 81, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Wu, J.; Lyu, R.; Cai, Y.; Jiang, N.; Liu, Y.; Zhang, F.; He, Y.; Chen, D.; Wu, Z. 6-thioguanine inhibits EV71 replication by reducing BIRC3-mediated autophagy. BMC Microbiol. 2025, 25, 53. [Google Scholar] [CrossRef] [PubMed]
- Battaglioni, S.; Benjamin, D.; Wälchli, M.; Maier, T.; Hall, M.N. mTOR substrate phosphorylation in growth control. Cell 2022, 185, 1814–1836. [Google Scholar] [CrossRef]
- Liu, Z.-W.; Zhuang, Z.-C.; Chen, R.; Wang, X.-R.; Zhang, H.-L.; Li, S.-H.; Wang, Z.-Y.; Wen, H.-L. Enterovirus 71 VP1 protein regulates viral replication in SH-SY5Y cells via the mTOR autophagy signaling pathway. Viruses 2019, 12, 11. [Google Scholar] [CrossRef]
- Zhan, Y.; Yu, S.; Yang, S.; Qiu, X.; Meng, C.; Tan, L.; Song, C.; Liao, Y.; Liu, W.; Sun, Y.; et al. Newcastle Disease virus infection activates PI3K/Akt/mTOR and p38 MAPK/Mnk1 pathways to benefit viral mRNA translation via interaction of the viral NP protein and host eIF4E. PLoS Pathog. 2020, 16, e1008610. [Google Scholar] [CrossRef]
- Peterson, R.T.; Schreiber, S.L. Translation control: Connecting mitogens and the ribosome. Curr. Biol. 1998, 8, R248–R250. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Overeem, N.J.; van der Vries, E.; Huskens, J. A Dynamic, Supramolecular view on the multivalent interaction between influenza virus and host cell. Small 2021, 17, e2007214. [Google Scholar] [CrossRef] [PubMed]
- Leonard, R.A.; Burke, K.N.; Spreng, R.L.; Macintyre, A.N.; Tam, Y.; Alameh, M.G.; Weissman, D.; Heaton, N.S. Improved influenza vaccine responses after expression of multiple viral glycoproteins from a single mRNA. Nat. Commun. 2024, 15, 8712. [Google Scholar] [CrossRef]
- Cai, Z.; Ni, W.; Li, W.; Wu, Z.; Yao, X.; Zheng, Y.; Zhao, Y.; Yuan, W.; Liang, S.; Wang, Q.; et al. SARS-CoV-2 S protein disrupts the formation of ISGF3 complex through conserved S2 subunit to antagonize type I interferon response. J. Virol. 2025, 99, e0151624. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, W.; Ren, L.; Ju, X.; Gong, M.; Rao, J.; Sun, L.; Li, P.; Ding, Q.; Wang, J.; et al. Comparison of viral RNA–host protein interactomes across pathogenic RNA viruses informs rapid antiviral drug discovery for SARS-CoV-2. Cell Res. 2021, 32, 9–23. [Google Scholar] [CrossRef]
- Soriaga, L.B.; Balce, D.R.; Bartha, I.; Park, A.; Wong, E.; McAllaster, M.; Mueller, E.A.; Barauskas, O.; Carabajal, E.; Kowalski, B.; et al. Shared host genetic landscape of respiratory viral infection. Proc. Natl. Acad. Sci. USA 2025, 122, e2414202122. [Google Scholar] [CrossRef]
- Girault, V.; Stukalov, A.; Carter-Timofte, M.E.; Hertzog, J.; Verin, M.; Austen, K.; Haas, D.A.; Oubraham, L.; Piras, A.; Maidl, S.; et al. Multi-proteomic profiling of the varicella-zoster virus-host interface reveals host susceptibilities to severe infection. Nat. Microbiol. 2025, 10, 2048–2072. [Google Scholar] [CrossRef]
- Hoang, M.T.V.; Nguyen, T.A.; Tran, T.T.; Vu, T.T.H.; Le, N.T.N.; Nguyen, T.H.N.; Le, T.H.N.; Nguyen, T.T.H.; Nguyen, T.H.; Le, N.T.N.; et al. Clinical and aetiological study of hand, foot and mouth disease in southern Vietnam, 2013–2015: Inpatients and outpatients. Int. J. Infect. Dis. 2019, 80, 1–9. [Google Scholar] [CrossRef]
- Xie, J.; Yang, X.H.; Hu, S.Q.; Zhan, W.L.; Zhang, C.B.; Liu, H.; Zhao, H.Y.; Chai, H.Y.; Chen, K.Y.; Du, Q.Y.; et al. Co-circulation of coxsackieviruses A-6, A-10, and A-16 causes hand, foot, and mouth disease in Guangzhou city, China. BMC Infect. Dis. 2020, 20, 271. [Google Scholar] [CrossRef]
- Duan, X.; Zhang, C.; Wang, X.; Ren, X.; Peng, H.; Tang, X.; Zhang, L.; Chen, Z.; Ye, Y.; Zheng, M.; et al. Molecular epidemiology and clinical features of hand, foot and mouth disease requiring hospitalization after the use of enterovirus A71 inactivated vaccine in chengdu, China, 2017-2022: A descriptive study. Emerg. Microbes Infect. 2022, 11, 2510–2519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Dai, W.; Zhang, C.; Zhou, Y.; Xiong, P.; Wang, S.; Ye, X.; Liu, Q.; Zhou, D.; Huang, Z. A virus-like particle-based tetravalent vaccine for hand, foot, and mouth disease elicits broad and balanced protective immunity. Emerg. Microbes Infect. 2018, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shen, C.; Zhang, C.; Zhang, W.; Wang, L.; Lan, K.; Liu, Q.; Huang, Z. Yeast-produced recombinant virus-like particles of coxsackievirus A6 elicited protective antibodies in mice. Antivir. Res. 2016, 132, 165–169. [Google Scholar] [CrossRef]
- Zheng, X.; Jin, G. Progress in research and development of preventive vaccines for children in China. Front. Pediatr. 2024, 12, 1414177. [Google Scholar] [CrossRef]
- Zeng, S.; Meng, X.; Huang, Q.; Lei, N.; Zeng, L.; Jiang, X.; Guo, X. Spiramycin and azithromycin, safe for administration to children, exert antiviral activity against enterovirus A71 in vitro and in vivo. Int. J. Antimicrob. Agents 2019, 53, 362–369. [Google Scholar] [CrossRef]
- Kang, H.; Kim, C.; Kim, D.E.; Song, J.H.; Choi, M.; Choi, K.; Kang, M.; Lee, K.; Kim, H.S.; Shin, J.S.; et al. Synergistic antiviral activity of gemcitabine and ribavirin against enteroviruses. Antivir. Res. 2015, 124, 1–10. [Google Scholar] [CrossRef]
- Bustos-Hamdan, A.; Bracho-Gallardo, J.I.; Hamdan-Partida, A.; Bustos-Martínez, J. Repositioning of antibiotics in the treatment of viral infections. Curr. Microbiol. 2024, 81, 427. [Google Scholar] [CrossRef]
- Wang, S.; Pang, Z.; Fan, H.; Tong, Y. Advances in anti-EV-A71 drug development research. J. Adv. Res. 2024, 56, 137–156. [Google Scholar] [CrossRef]
 indicates sulfation and
indicates sulfation and  indicates phosphorylation.
indicates phosphorylation.
 indicates sulfation and
indicates sulfation and  indicates phosphorylation.
indicates phosphorylation.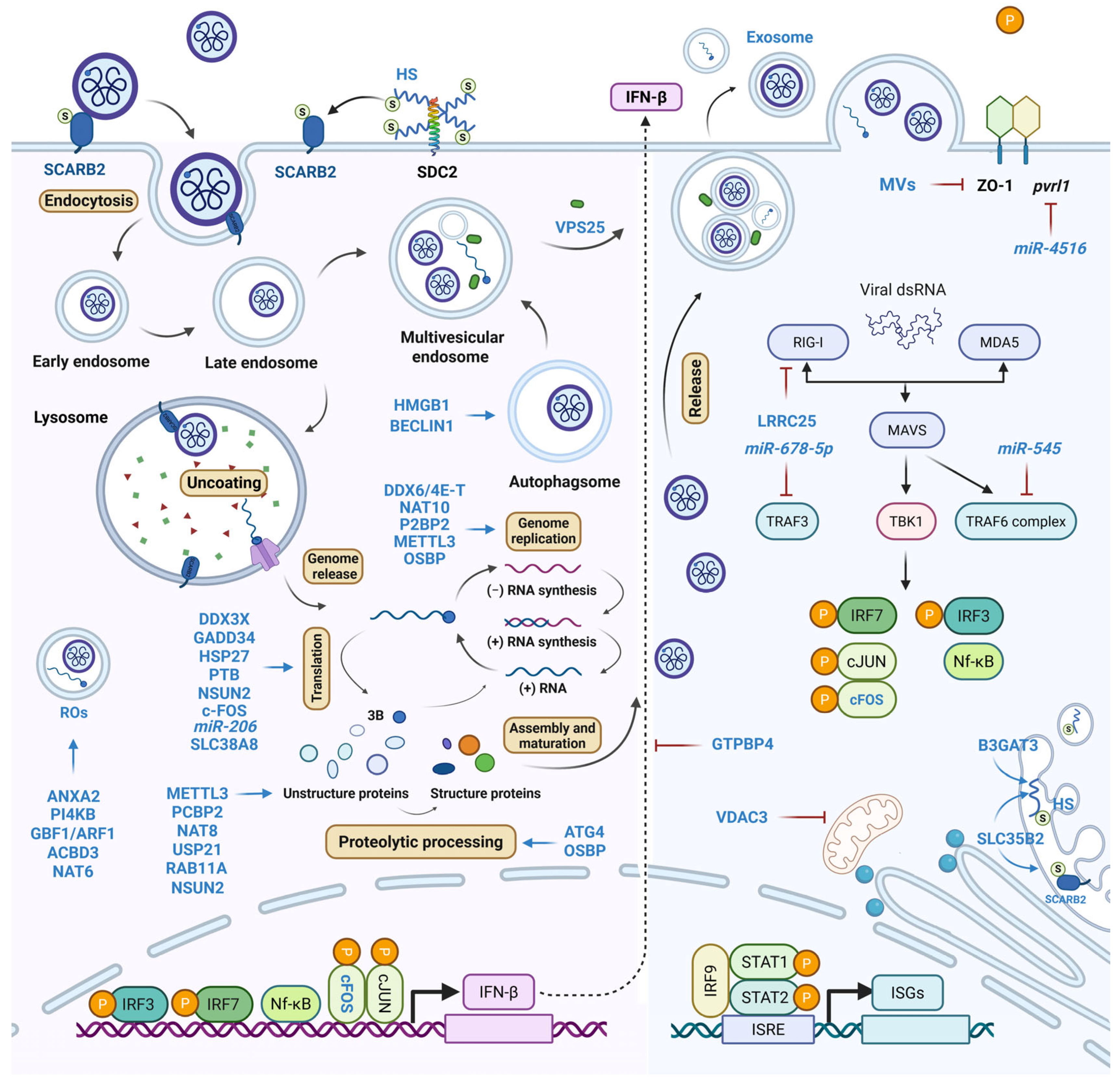
| Step | Factor | Function | Refs. |
|---|---|---|---|
| Viral entry | SCARB2 | Receptor | [21] |
| SLC35B2 | Tyrosine sulfation of receptors | [21] | |
| B3GAT3 | HS backbone biosynthesis | [21] | |
| Viral RNA | PI4KB | ROs formation | [26,27,28] |
| GBF1/ARF1 | PI4KB recruitment | [28] | |
| ACBD3 | PI4KB recruitment | [28] | |
| NAT6 | Stabilizing ACBD3 | [29] | |
| ANXA2 | PI4P induction and viral 3D recruitment | [30] | |
| DDX6/4E-T | Viral RNA binding | [35] | |
| NAT10 | ac4C modification of viral 5′ UTR | [38] | |
| PCBP2 | Stabilizing viral RNA-3D interaction | [39] | |
| Viral protein | DDX3X | Facilitating ribosome entry | [40] |
| GADD34 | Promoting IRES activity | [41] | |
| HSP27 | Activating IRES-mediated translation | [42] | |
| PTB | Activating IRES-mediated translation | [44] | |
| c-FOS | Activating MEK/ERK signaling | [48] | |
| NAT8 | Lysine acetylation of 2B, 3AB, and 3C | [49] | |
| USP21 | Deubiquitinating 2Apro | [50] | |
| ATG4 | 3Cpro protease-like activity | [51] | |
| RAB11A | Acting as scaffold of viral proteins | [52] | |
| SLC38A8 | Activating mTOR/p70S6K1 signaling | [58,60] | |
| miR-206 | Repressing G3BP2 expression | [62] | |
| Viral RNA and protein | METTL3 | Sumoylation and ubiquitination of 3D | [36] |
| NSUN2 | m5C modifications of EVA71 RNAs; stabilizing VP1 | [45] | |
| OSBP | Promoting 3AB cleavage and plus-strand RNA synthesis | [65] | |
| LRRC25 | Degradation of G3BP1 and RIG-I to inhibit stress granule formation and innate immunity | [76] | |
| Immune evasion | GTPBP4 | Negative regulator of IFNβ | [77] |
| miR-545 | Inhibiting IFN signaling via PENT and TRAF6 | [78] | |
| miR-628-5p | Targeting TRAF3 to impair innate immunity | [79] | |
| VPS25 | Promoting exosome biogenesis and secretion | [83] | |
| Packaging and dissemination | miR-4516 | Reducing pvrl1 mRNA to disrupt CAMs | [86] |
| BECLIN1 | ATG protein recruitment | [96] | |
| HMGB1 | Regulating autophagosome formation | [97] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Wu, X.; Liu, M.; Li, L.; Wang, Y.; He, Q.; Zhang, X.; Liang, Z.; Gao, F.; Ma, X. Hijacking the Host Cell for Replication: Pro-Viral Host Factors Involved in EVA71 Infection. Int. J. Mol. Sci. 2025, 26, 7992. https://doi.org/10.3390/ijms26167992
Wang Q, Wu X, Liu M, Li L, Wang Y, He Q, Zhang X, Liang Z, Gao F, Ma X. Hijacking the Host Cell for Replication: Pro-Viral Host Factors Involved in EVA71 Infection. International Journal of Molecular Sciences. 2025; 26(16):7992. https://doi.org/10.3390/ijms26167992
Chicago/Turabian StyleWang, Qian, Xing Wu, Mingchen Liu, Lu Li, Ying Wang, Qian He, Xuanxuan Zhang, Zhenglun Liang, Fan Gao, and Xiao Ma. 2025. "Hijacking the Host Cell for Replication: Pro-Viral Host Factors Involved in EVA71 Infection" International Journal of Molecular Sciences 26, no. 16: 7992. https://doi.org/10.3390/ijms26167992
APA StyleWang, Q., Wu, X., Liu, M., Li, L., Wang, Y., He, Q., Zhang, X., Liang, Z., Gao, F., & Ma, X. (2025). Hijacking the Host Cell for Replication: Pro-Viral Host Factors Involved in EVA71 Infection. International Journal of Molecular Sciences, 26(16), 7992. https://doi.org/10.3390/ijms26167992





