A Machine Learning Approach to Understanding the Genetic Role in COVID-19 Prognosis: The Influence of Gene Polymorphisms Related to Inflammation, Vitamin D, and ACE2
Abstract
1. Introduction
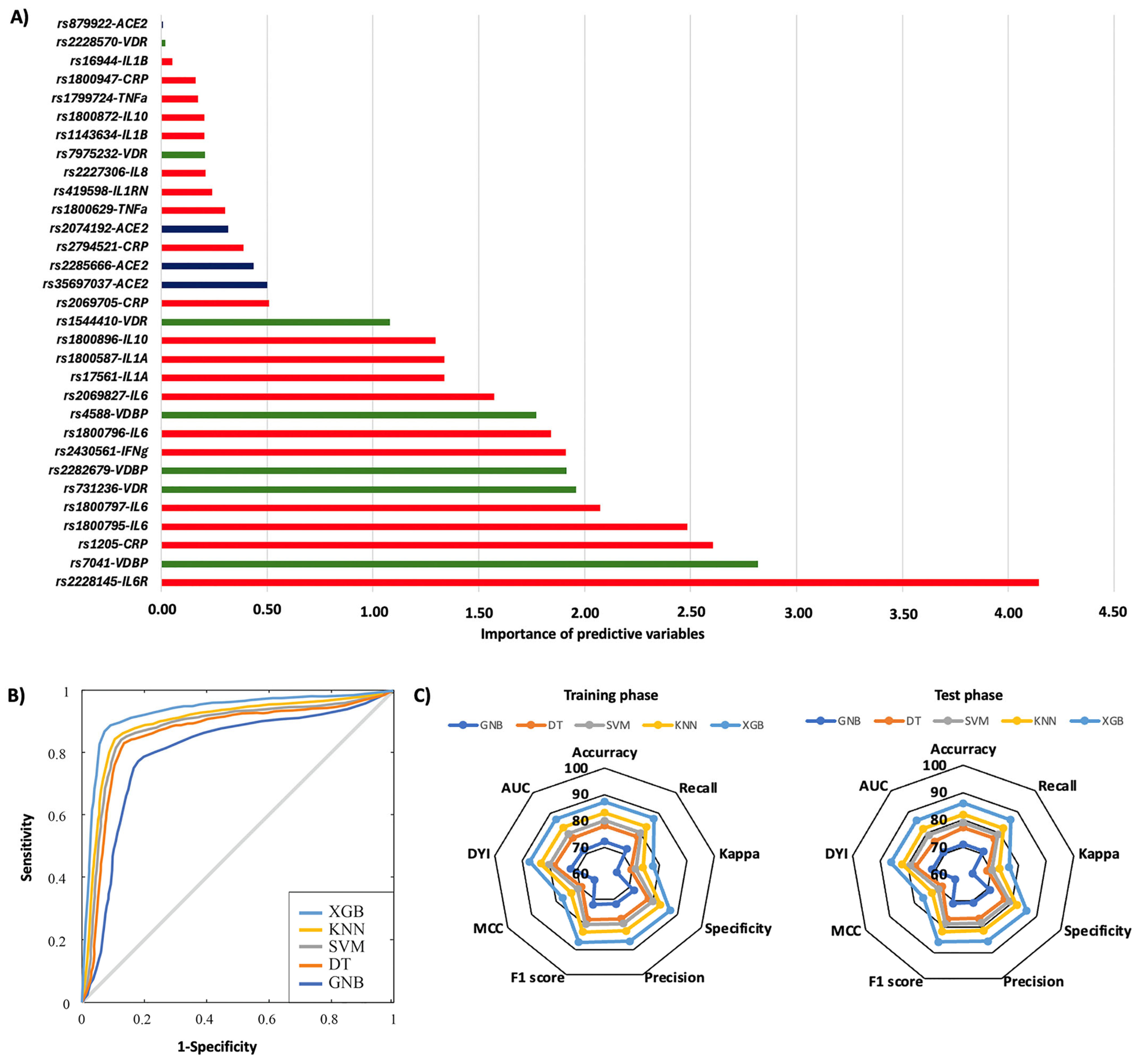
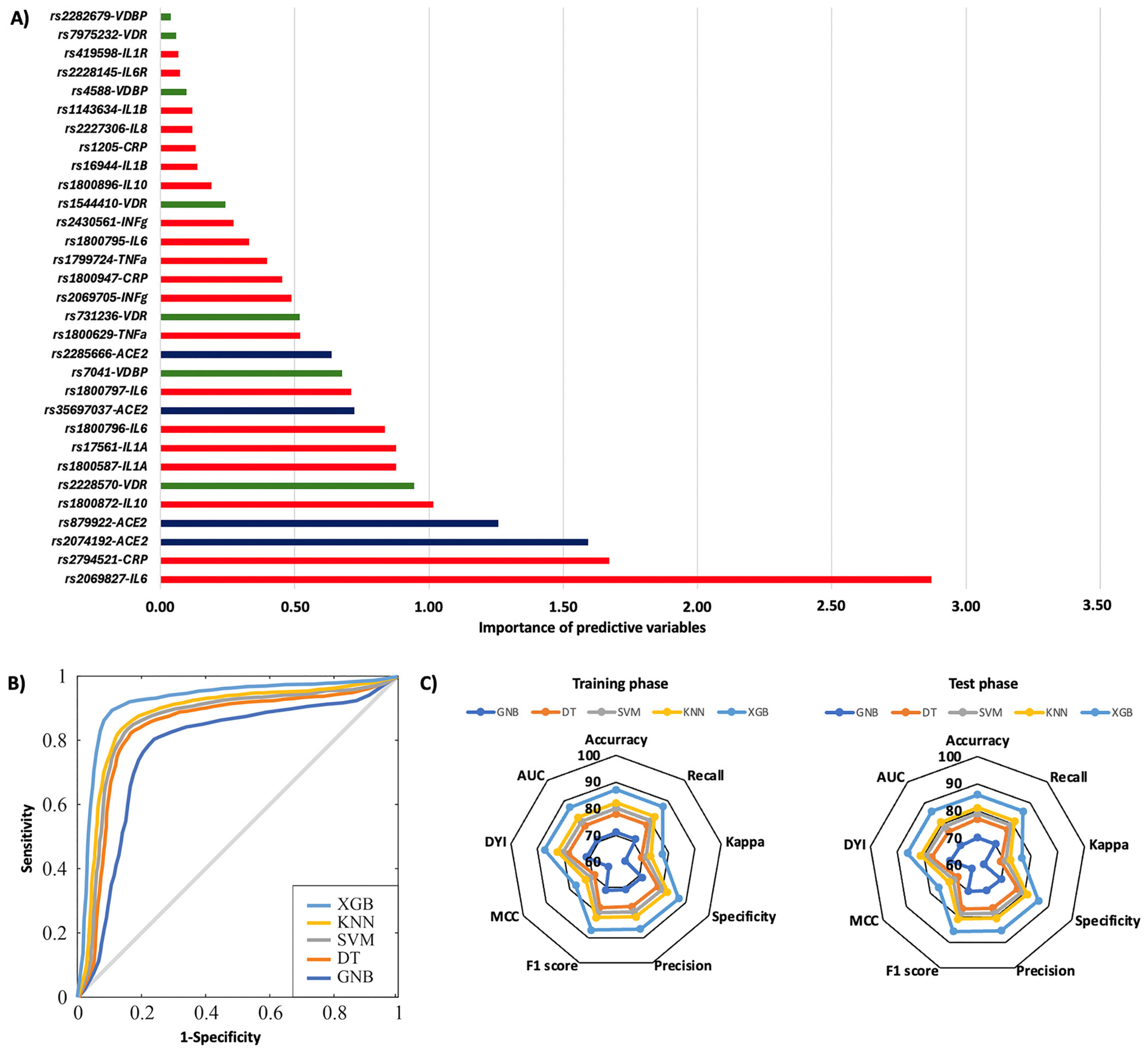
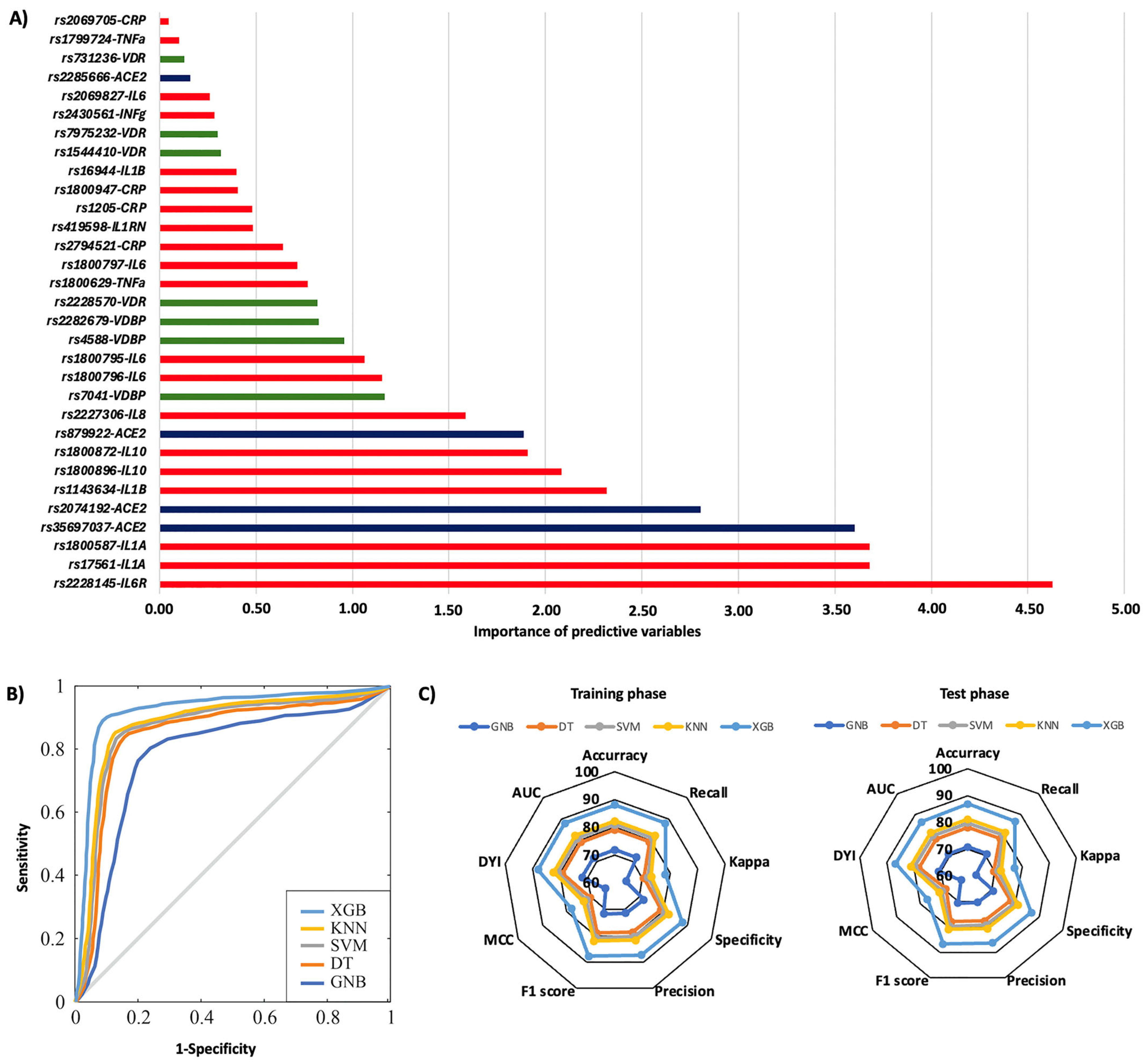
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. DNA Isolation and Polymorphism Genotyping
4.3. Machine Learning Analysis
4.4. Ethical Aspects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Gong, Z.; Song, T.; Hu, M.; Che, Q.; Guo, J.; Zhang, H.; Li, H.; Wang, Y.; Liu, B.; Shi, N. Natural and socio-environmental factors in the transmission of COVID-19: A comprehensive analysis of epidemiology and mechanisms. BMC Public Health 2024, 24, 2196. [Google Scholar] [CrossRef]
- Ingraham, N.E.; Barakat, A.G.; Reilkoff, R.; Bezdicek, T.; Schacker, T.; Chipman, J.G.; Tignanelli, C.J.; Puskarich, M.A. Understanding the renin-angiotensin-aldosterone-SARS-CoV axis: A comprehensive review. Eur. Respir. J. 2020, 56, 2000912. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436, 112–116. [Google Scholar] [CrossRef]
- Annoni, F.; Orbegozo, D.; Rahmania, L.; Irazabal, M.; Mendoza, M.; De Backer, D.; Taccone, F.S.; Creteur, J.; Vincent, J.-L. Angiotensin-converting enzymes in acute respiratory distress syndrome. Intensive Care Med. 2019, 45, 1159–1160. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Jose, R.J.; Manuel, A. COVID-19 cytokine storm: The interplay between inflammation and coagulation. Lancet Respir. Med. 2020, 8, e46–e47. [Google Scholar] [CrossRef]
- Gustine, J.N.; Jones, D. Immunopathology of Hyperinflammation in COVID-19. Am. J. Pathol. 2021, 191, 4–17. [Google Scholar] [CrossRef]
- Tan, L.Y.; Komarasamy, T.V.; Balasubramaniam, V.R. Hyperinflammatory Immune Response and COVID-19: A Double Edged Sword. Front. Immunol. 2021, 12, 742941. Available online: https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2021.742941/full (accessed on 24 April 2025). [CrossRef] [PubMed]
- Minton, K. Vitamin D shuts down T cell-mediated inflammation. Nat. Rev. Immunol. 2022, 22, 1. [Google Scholar] [CrossRef] [PubMed]
- Colotta, F.; Jansson, B.; Bonelli, F. Modulation of inflammatory and immune responses by vitamin D. J. Autoimmun. 2017, 85, 78–97. [Google Scholar] [CrossRef] [PubMed]
- Zdrenghea, M.T.; Makrinioti, H.; Bagacean, C.; Bush, A.; Johnston, S.L.; Stanciu, L.A. Vitamin D modulation of innate immune responses to respiratory viral infections. Rev. Med. Virol. 2017, 27, e1909. [Google Scholar] [CrossRef]
- Mohan, M.; Cherian, J.J.; Sharma, A. Exploring links between vitamin D deficiency and COVID-19. PLoS Pathog. 2020, 16, e1008874. [Google Scholar] [CrossRef]
- Weir, E.K.; Thenappan, T.; Bhargava, M.; Chen, Y. Does vitamin D deficiency increase the severity of COVID-19? Clin. Med. 2020, 20, e107–e108. [Google Scholar] [CrossRef]
- Radujkovic, A.; Hippchen, T.; Tiwari-Heckler, S.; Dreher, S.; Boxberger, M.; Merle, U. Vitamin D Deficiency and Outcome of COVID-19 Patients. Nutrients 2020, 12, 2757. [Google Scholar] [CrossRef]
- Fu, L.; Wang, B.; Yuan, T.; Chen, X.; Ao, Y.; Fitzpatrick, T.; Li, P.; Zhou, Y.; Lin, Y.; Duan, Q.; et al. Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: A systematic review and meta-analysis. J. Infect. 2020, 80, 656–665. [Google Scholar] [CrossRef]
- Camps-Vilaró, A.; Pinsach-Abuin, M.; Degano, I.R.; Ramos, R.; Martí-Lluch, R.; Elosua, R.; Subirana, I.; Solà-Richarte, C.; Puigmulé, M.; Pérez, A.; et al. Genetic characteristics involved in COVID-19 severity. The CARGENCORS case-control study and meta-analysis. J. Med. Virol. 2024, 96, e29404. [Google Scholar] [CrossRef]
- Velavan, T.P.; Pallerla, S.R.; Rüter, J.; Augustin, Y.; Kremsner, P.G.; Krishna, S.; Meyer, C.G. Host genetic factors determining COVID-19 susceptibility and severity. eBioMedicine 2021, 72, 103629. Available online: https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(21)00422-9/fulltext (accessed on 23 April 2025). [CrossRef]
- Niemi, M.E.K.; Daly, M.J.; Ganna, A. The human genetic epidemiology of COVID-19. Nat. Rev. Genet. 2022, 23, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Waring, J.; Lindvall, C.; Umeton, R. Automated machine learning: Review of the state-of-the-art and opportunities for healthcare. Artif. Intell. Med. 2020, 104, 101822. [Google Scholar] [CrossRef] [PubMed]
- Handelman, G.S.; Kok, H.K.; Chandra, R.V.; Razavi, A.H.; Lee, M.J.; Asadi, H. eDoctor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Abobaker, A.; Nagib, T.; Alsoufi, A. The impact of certain genetic variants (single nucleotide polymorphisms) on incidence and severity of COVID-19. J. Gene Med. 2021, 23, e3310. [Google Scholar] [CrossRef]
- Ren, H.; Lin, Y.; Huang, L.; Xu, W.; Luo, D.; Zhang, C. Association of genetic polymorphisms with COVID-19 infection and outcomes: An updated meta-analysis based on 62 studies. Heliyon 2024, 10, e23662. [Google Scholar] [CrossRef]
- Gómez, J.; Albaiceta, G.M.; García-Clemente, M.; López-Larrea, C.; Amado-Rodríguez, L.; Lopez-Alonso, I.; Hermida, T.; Enriquez, A.I.; Herrero, P.; Melón, S.; et al. Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene 2020, 762, 145102. [Google Scholar] [CrossRef]
- Saengsiwaritt, W.; Jittikoon, J.; Chaikledkaew, U.; Udomsinprasert, W. Genetic polymorphisms of ACE1, ACE2, and TMPRSS2 associated with COVID-19 severity: A systematic review with meta-analysis. Rev. Med. Virol. 2022, 32, e2323. [Google Scholar] [CrossRef]
- Sabater Molina, M.; Nicolás Rocamora, E.; Bendicho, A.I.; Vázquez, E.G.; Zorio, E.; Rodriguez, F.D.; Gil Ortuño, C.; Rodríguez, A.I.; Sánchez-López, A.J.; Jara Rubio, R.; et al. Polymorphisms in ACE, ACE2, AGTR1 genes and severity of COVID-19 disease. PLoS ONE 2022, 17, e0263140. [Google Scholar] [CrossRef]
- Merad, M.; Blish, C.A.; Sallusto, F.; Iwasaki, A. The immunology and immunopathology of COVID-19. Science 2022, 375, 1122–1127. [Google Scholar] [CrossRef]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 323, 1824–1836. [Google Scholar] [CrossRef]
- Darbeheshti, F.; Mahdiannasser, M.; Uhal, B.D.; Ogino, S.; Gupta, S.; Rezaei, N. Interindividual immunogenic variants: Susceptibility to coronavirus, respiratory syncytial virus and influenza virus. Rev. Med. Virol. 2021, 31, e2234. [Google Scholar] [CrossRef]
- Yip, J.Q.; Oo, A.; Ng, Y.L.; Chin, K.L.; Tan, K.-K.; Chu, J.J.H.; AbuBakar, S.; Zainal, N. The role of inflammatory gene polymorphisms in severe COVID-19: A review. Virol. J. 2024, 21, 327. [Google Scholar] [CrossRef] [PubMed]
- Vogi, V.; Haschka, D.; Forer, L.; Schwendinger, S.; Petzer, V.; Coassin, S.; Tancevski, I.; Sonnweber, T.; Löffler-Ragg, J.; Puchhammer-Stöckl, E.; et al. Severe COVID-19 disease is associated with genetic factors affecting plasma ACE2 receptor and CRP concentrations. Sci. Rep. 2025, 15, 4708. [Google Scholar] [CrossRef] [PubMed]
- Bischoff-Ferrari, H.A.; Dawson-Hughes, B.; Stöcklin, E.; Sidelnikov, E.; Willett, W.C.; Edel, J.O.; Stähelin, H.B.; Wolfram, S.; Jetter, A.; Schwager, J.; et al. Oral supplementation with 25(OH)D3 versus vitamin D3: Effects on 25(OH)D levels, lower extremity function, blood pressure, and markers of innate immunity. J. Bone Miner. Res. 2012, 27, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Malmberg, H.-R.; Hanel, A.; Taipale, M.; Heikkinen, S.; Carlberg, C. Vitamin D Treatment Sequence Is Critical for Transcriptome Modulation of Immune Challenged Primary Human Cells. Front. Immunol. 2021, 12, 754056. [Google Scholar] [CrossRef]
- Bouillon, R.; Bikle, D. Vitamin D Metabolism Revised: Fall of Dogmas. J. Bone Miner. Res. 2019, 34, 1985–1992. [Google Scholar] [CrossRef]
- Manousaki, D.; Dudding, T.; Haworth, S.; Hsu, Y.-H.; Liu, C.-T.; Medina-Gómez, C.; Voortman, T.; van der Velde, N.; Melhus, H.; Robinson-Cohen, C.; et al. Low-Frequency Synonymous Coding Variation in CYP2R1 Has Large Effects on Vitamin D Levels and Risk of Multiple Sclerosis. Am. J. Hum. Genet. 2017, 101, 227–238. [Google Scholar] [CrossRef]
- Scazzone, C.; Agnello, L.; Bivona, G.; Lo Sasso, B.; Ciaccio, M. Vitamin D and Genetic Susceptibility to Multiple Sclerosis. Biochem. Genet. 2021, 59, 1–30. [Google Scholar] [CrossRef]
- Cools, M.; Goemaere, S.; Baetens, D.; Raes, A.; Desloovere, A.; Kaufman, J.M.; De Schepper, J.; Jans, I.; Vanderschueren, D.; Billen, J.; et al. Calcium and bone homeostasis in heterozygous carriers of CYP24A1 mutations: A cross-sectional study. Bone 2015, 81, 89–96. [Google Scholar] [CrossRef]
- Kuan, V.; Martineau, A.R.; Griffiths, C.J.; Hyppönen, E.; Walton, R. DHCR7 mutations linked to higher vitamin D status allowed early human migration to Northern latitudes. BMC Evol. Biol. 2013, 13, 144. [Google Scholar] [CrossRef]
- Saccone, D.; Asani, F.; Bornman, L. Regulation of the vitamin D receptor gene by environment, genetics and epigenetics. Gene 2015, 561, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Karcıoğlu Batur, L.; Dokur, M.; Koç, S.; Karabay, M.; Akcay, Z.N.; Gunger, E.; Hekim, N. Investigation of the Relationship between Vitamin D Deficiency and Vitamin D-Binding Protein Polymorphisms in Severe COVID-19 Patients. Diagnostics 2024, 14, 1941. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chi, X.; Sun, Y.; Li, H. Vitamin D Binding Protein: A Potential Factor in Geriatric COVID-19 Acute Lung Injury. J. Inflamm. Res. 2024, 17, 4419–4429. [Google Scholar] [CrossRef] [PubMed]
- Dobrijevic, Z.; Robajac, D.; Gligorijevic, N.; Šunderic, M.; Penezic, A.; Miljuš, G.; Nedic, O. The association of ACE1, ACE2, TMPRSS2, IFITM3 and VDR polymorphisms with COVID-19 severity: A systematic review and meta-analysis. EXCLI J. 2022, 21, 818–839. [Google Scholar]
- Tentolouris, N.; Achilla, C.; Anastasiou, I.A.; Eleftheriadou, I.; Tentolouris, A.; Basoulis, D.; Kosta, O.; Lambropoulos, A.; Yavropoulou, M.P.; Chatzikyriakidou, A.; et al. The Association of Vitamin D Receptor Polymorphisms with COVID-19 Severity. Nutrients 2024, 16, 727. [Google Scholar] [CrossRef]
- Litmanovich, D.E.; Chung, M.; Kirkbride, R.R.; Kicska, G.; Kanne, J.P. Review of Chest Radiograph Findings of COVID-19 Pneumonia and Suggested Reporting Language. J. Thorac. Imaging 2020, 35, 354–360. [Google Scholar] [CrossRef]
- Schleinitz, D.; Distefano, J.K.; Kovacs, P. Targeted SNP genotyping using the TaqMan® assay. In Disease Gene Identification: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2011; Volume 700, pp. 77–87. [Google Scholar]
- Chen, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Cano, I.; Zhou, T. Xgboost: Extreme gradient boosting. Sci. Res. 2015, 1, 1–4. [Google Scholar]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Charbuty, B.; Abdulazeez, A. Classification Based on Decision Tree Algorithm for Machine Learning. J. Appl. Sci. Technol. Trends 2021, 2, 20–28. [Google Scholar] [CrossRef]
- Ampomah, E.K.; Nyame, G.; Qin, Z.; Addo, P.C.; Gyamfi, E.O.; Gyan, M. Stock Market Prediction with Gaussian Naïve Bayes Machine Learning Algorithm. Informatica 2021, 45, 3407. Available online: https://www.informatica.si/index.php/informatica/article/view/3407 (accessed on 23 April 2025). [CrossRef]
- Cubillos, M.; Wøhlk, S.; Wulff, J.N. A bi-objective k-nearest-neighbors-based imputation method for multilevel data. Expert Syst. Appl. 2022, 204, 117298. Available online: https://re.public.polimi.it/handle/11311/1252307 (accessed on 23 April 2025). [CrossRef]
- MATLAB Runtime—MATLAB Compiler. Available online: https://es.mathworks.com/products/compiler/matlab-runtime.html (accessed on 23 April 2025).
- Feurer, M.; Hutter, F. Hyperparameter Optimization. In Automated Machine Learning: Methods, Systems, Challenges; Hutter, F., Kotthoff, L., Vanschoren, J., Eds.; Springer International Publishing: Cham, Germany, 2019; pp. 3–33. ISBN 978-3-030-05318-5. [Google Scholar] [CrossRef]
- Chen, R.-C.; Dewi, C.; Huang, S.-W.; Caraka, R.E. Selecting critical features for data classification based on machine learning methods. J. Big Data 2020, 7, 52. [Google Scholar] [CrossRef]
- Herland, M.; Khoshgoftaar, T.M.; Wald, R. A review of data mining using big data in health informatics. J. Big Data 2014, 1, 2. [Google Scholar] [CrossRef]

| General Characteristics | Patients |
|---|---|
| Age, mean (SD) (years) | 73.26 (13.09) |
| Sex (male; female), n (%) | 182 (53.84); 156 (46.15) |
| Days from symptom onset to hospital admission, mean (SD) | 5.77 (5.45) |
| COVID-19 pneumonia, n (%) | 248 (73.3) |
| Days of admission, mean (SD) | 18.21 (22.75) |
| Death due to COVID-19, n (%) | 76 (22.48) |
| Dependency (yes; moderate; mild; independent), n (%) | 42 (12.42); 47 (13.90); 169 (50); 80 (83.67) |
| Rehospitalization in the first year, n (%) | 77 (29.3) |
| Smoking history (active; ex-smoker; never smoker), n (%) | 12 (3.56); 57 (16.91); 268 (79.52) |
| Dementia, n (%) | 41 (12.20) |
| Hypertension, n (%) | 196 (58.16) |
| Dyslipidemia n (%) | 133 (39.58) |
| Myocardial infarction (%) | 15 (4.45) |
| Heart failure, n (%) | 20 (5.97) |
| Cerebral ictus, n (%) | 18 (5.36) |
| Diabetes mellitus, n (%) | 68 (20.24) |
| COPD, n (%) | 8 (2.43) |
| Asthma, n (%) | 32 (9.46) |
| Obstructive sleep apnea, n (%) | 20 (5.93) |
| Chronic kidney disease (4–5), n (%) | 26 (7.15) |
| Tumor without metastasis, n (%) | 39 (11.57) |
| Tumor with metastasis, n (%) | 5 (1.48) |
| ACEi, n (%) | 82 (24.40) |
| ARBs, n (%) | 77 (22.91) |
| Statins, n (%) | 81 (24.11) |
| Metformin, n (%) | 36 (10.71) |
| DDP-4 inhibitors, n (%) | 30 (8.92) |
| Insulin, n (%) | 20 (5.95) |
| Inhaled corticosteroids, n (%) | 33 (9.82) |
| Corticosteroids, n (%) | 10 (2.97) |
| Immunosuppressors/immunomodulators, n (%) | 16 (4.76) |
| Test COVID-19 confirmation, n (%) | 338 (100) |
| Method | BA (%) | Recall | Precision | AUC | F1 Score | MCC | DYI | Kappa | |
|---|---|---|---|---|---|---|---|---|---|
| COVID-19 pneumonia | SVM | 79.09 | 79.18 | 78.53 | 0.79 | 78.85 | 70.18 | 79.09 | 70.41 |
| DT | 77.23 | 77.32 | 76.68 | 0.76 | 77.00 | 68.53 | 77.23 | 68.75 | |
| GNB | 71.16 | 71.25 | 70.65 | 0.71 | 70.95 | 63.14 | 71.16 | 63.35 | |
| KNN | 82.04 | 82.13 | 81.45 | 0.82 | 81.79 | 72.79 | 82.04 | 73.04 | |
| XGB | 86.10 | 86.20 | 85.48 | 0.86 | 85.84 | 76.40 | 86.10 | 76.65 | |
| Mortality | SVM | 79.20 | 79.29 | 78.64 | 0.79 | 78.96 | 70.28 | 79.20 | 70.51 |
| DT | 77.13 | 77.22 | 76.58 | 0.76 | 76.90 | 68.44 | 77.13 | 68.67 | |
| GNB | 70.18 | 70.26 | 69.68 | 0.70 | 69.97 | 62.27 | 70.18 | 62.48 | |
| KNN | 81.17 | 81.27 | 80.59 | 0.81 | 80.93 | 72.02 | 81.17 | 72.26 | |
| XGB | 86.00 | 86.10 | 85.39 | 0.86 | 85.74 | 76.31 | 86.00 | 76.56 | |
| Rehospitalization | SVM | 79.26 | 79.35 | 78.69 | 0.78 | 79.02 | 70.33 | 79.26 | 70.56 |
| DT | 77.48 | 77.58 | 76.93 | 0.77 | 77.25 | 68.75 | 77.48 | 68.98 | |
| GNB | 72.78 | 72.86 | 72.25 | 0.72 | 72.56 | 64.46 | 72.78 | 64.68 | |
| KNN | 81.94 | 82.04 | 81.36 | 0.81 | 81.69 | 72.71 | 81.94 | 72.95 | |
| XGB | 85.86 | 85.96 | 85.25 | 0.85 | 85.61 | 76.19 | 85.86 | 76.44 | |
| Mortality (rehospitalization) | SVM | 79.85 | 79.94 | 79.28 | 0.80 | 79.61 | 70.85 | 79.85 | 71.09 |
| DT | 78.12 | 78.21 | 77.56 | 0.78 | 77.88 | 69.31 | 78.12 | 69.54 | |
| GNB | 70.83 | 70.91 | 70.32 | 0.71 | 70.62 | 62.85 | 70.83 | 63.05 | |
| KNN | 81.17 | 81.27 | 80.59 | 0.81 | 80.93 | 72.02 | 81.17 | 72.26 | |
| XGB | 86.85 | 86.72 | 86.23 | 0.86 | 86.59 | 77.06 | 86.85 | 77.32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaurrieta-Largo, S.; Miramontes-González, J.P.; Corral-Gudino, L.; Gabella-Martín, M.; Pérez-Arroyo, S.; Torres, A.M.; Mateo, J.; Pérez-Castrillón, J.L.; Usategui-Martín, R. A Machine Learning Approach to Understanding the Genetic Role in COVID-19 Prognosis: The Influence of Gene Polymorphisms Related to Inflammation, Vitamin D, and ACE2. Int. J. Mol. Sci. 2025, 26, 7975. https://doi.org/10.3390/ijms26167975
Jaurrieta-Largo S, Miramontes-González JP, Corral-Gudino L, Gabella-Martín M, Pérez-Arroyo S, Torres AM, Mateo J, Pérez-Castrillón JL, Usategui-Martín R. A Machine Learning Approach to Understanding the Genetic Role in COVID-19 Prognosis: The Influence of Gene Polymorphisms Related to Inflammation, Vitamin D, and ACE2. International Journal of Molecular Sciences. 2025; 26(16):7975. https://doi.org/10.3390/ijms26167975
Chicago/Turabian StyleJaurrieta-Largo, Sofía, José Pablo Miramontes-González, Luis Corral-Gudino, Miriam Gabella-Martín, Sofía Pérez-Arroyo, Ana M. Torres, Jorge Mateo, José Luis Pérez-Castrillón, and Ricardo Usategui-Martín. 2025. "A Machine Learning Approach to Understanding the Genetic Role in COVID-19 Prognosis: The Influence of Gene Polymorphisms Related to Inflammation, Vitamin D, and ACE2" International Journal of Molecular Sciences 26, no. 16: 7975. https://doi.org/10.3390/ijms26167975
APA StyleJaurrieta-Largo, S., Miramontes-González, J. P., Corral-Gudino, L., Gabella-Martín, M., Pérez-Arroyo, S., Torres, A. M., Mateo, J., Pérez-Castrillón, J. L., & Usategui-Martín, R. (2025). A Machine Learning Approach to Understanding the Genetic Role in COVID-19 Prognosis: The Influence of Gene Polymorphisms Related to Inflammation, Vitamin D, and ACE2. International Journal of Molecular Sciences, 26(16), 7975. https://doi.org/10.3390/ijms26167975









