Construction and Analysis of Immune Infiltration and Competing Endogenous RNA Network in Moyamoya Disease
Abstract
1. Introduction
2. Results
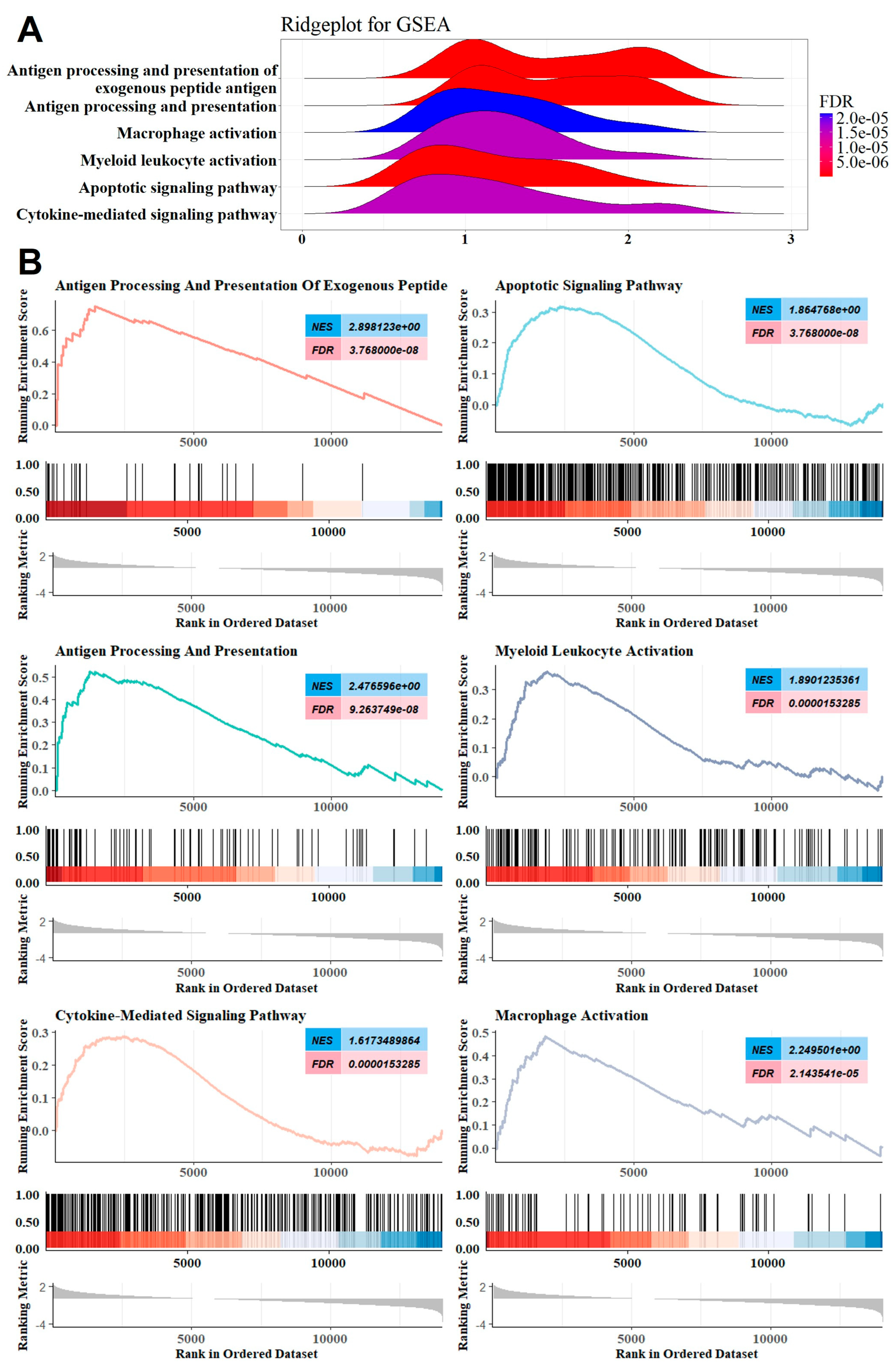
2.1. GSEA Revealed the Involvement of the Innate Immune System
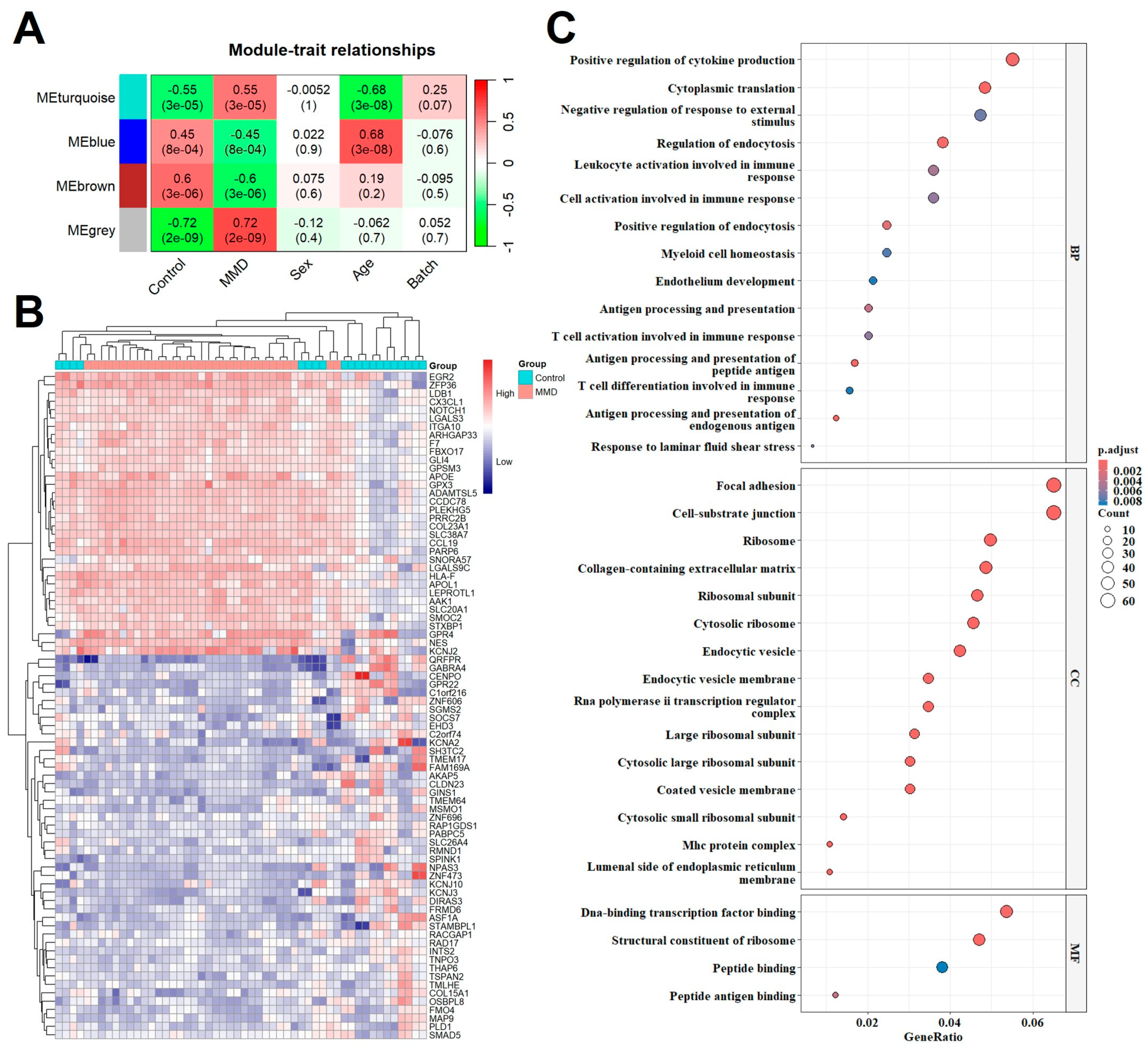
2.2. Co-Expression Modules and GO Analysis
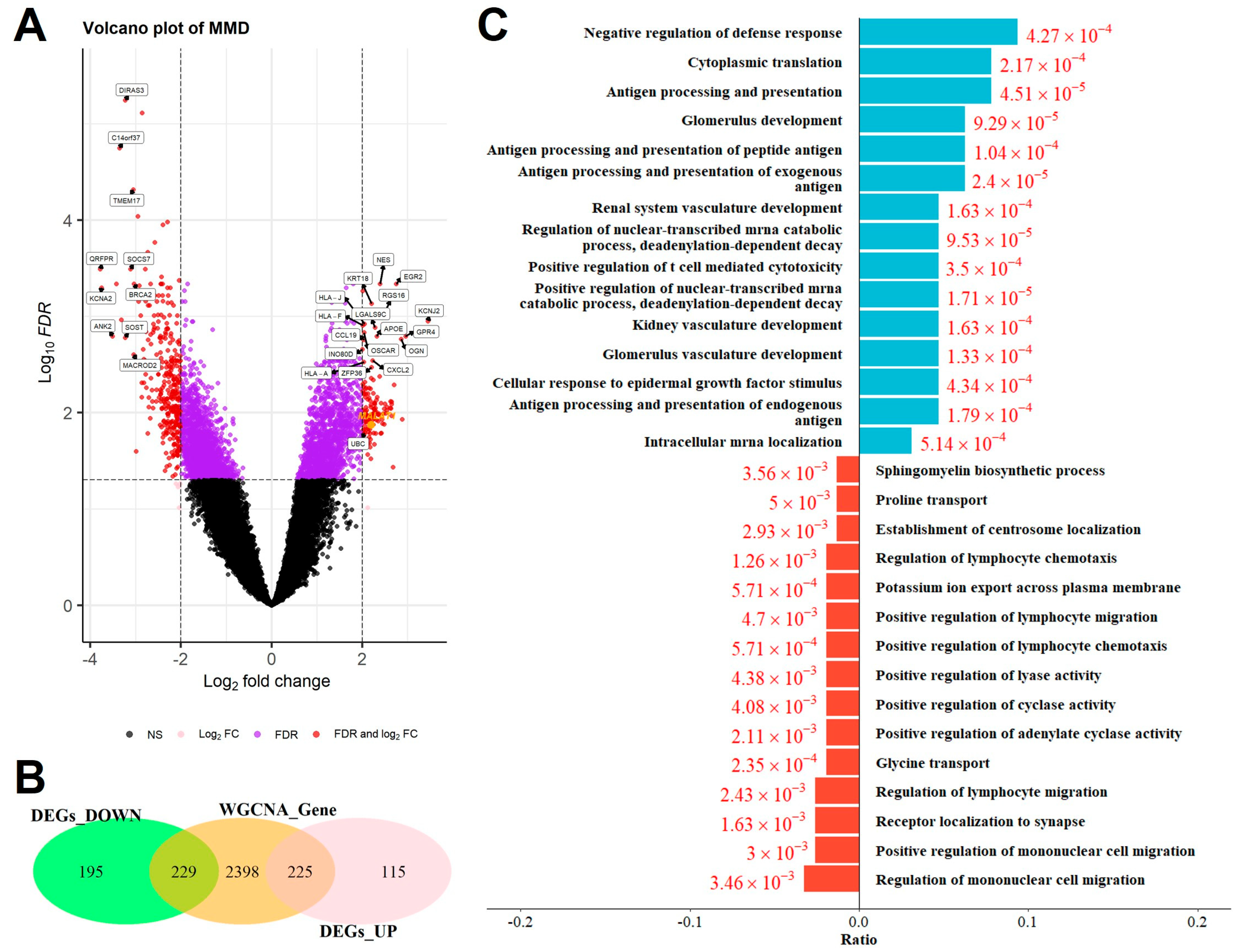
2.3. Identification of Overlapping DEGs and GO Analysis
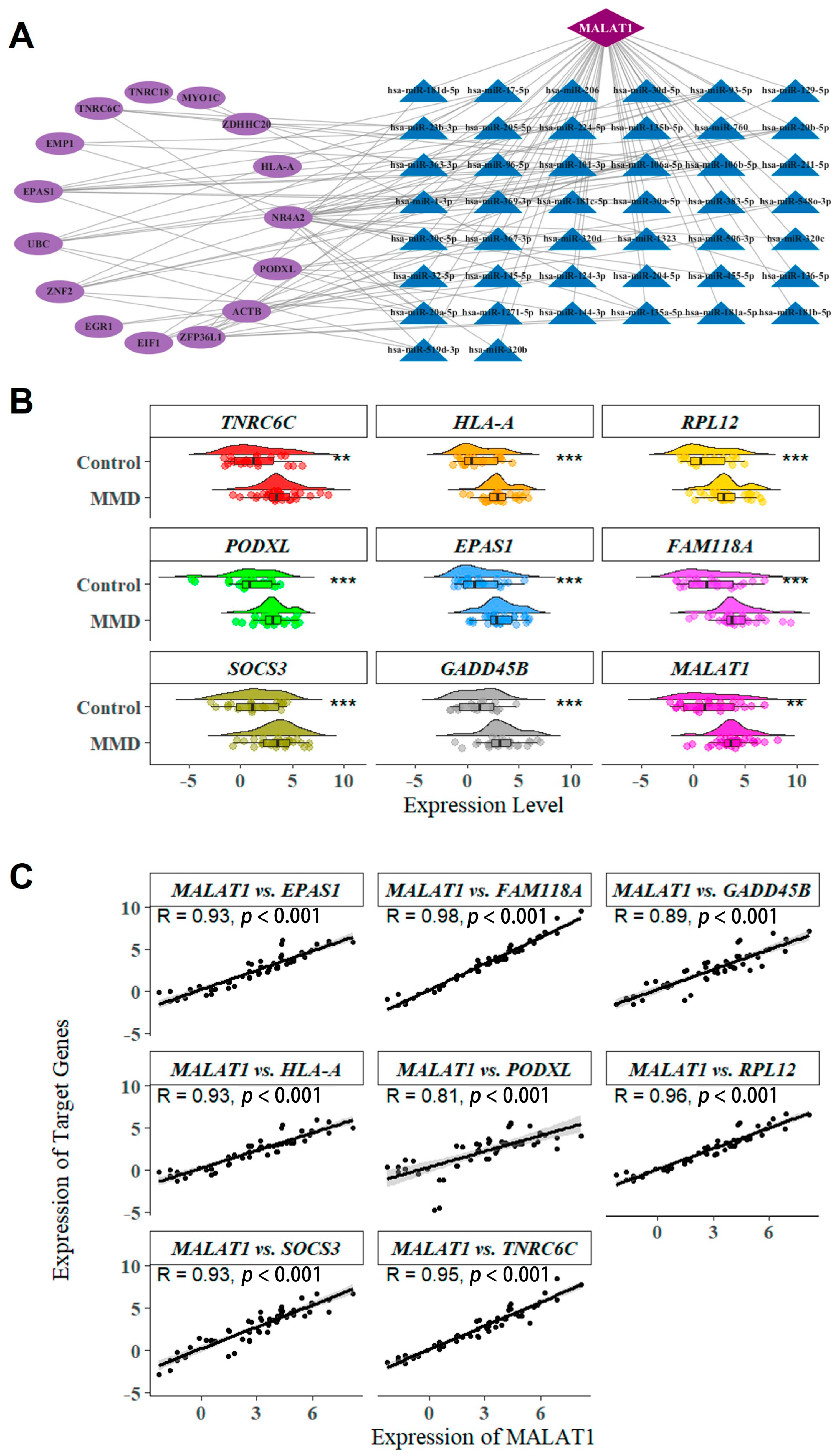
2.4. lncRNA-miRNA-mRNA ceRNA Network Construction
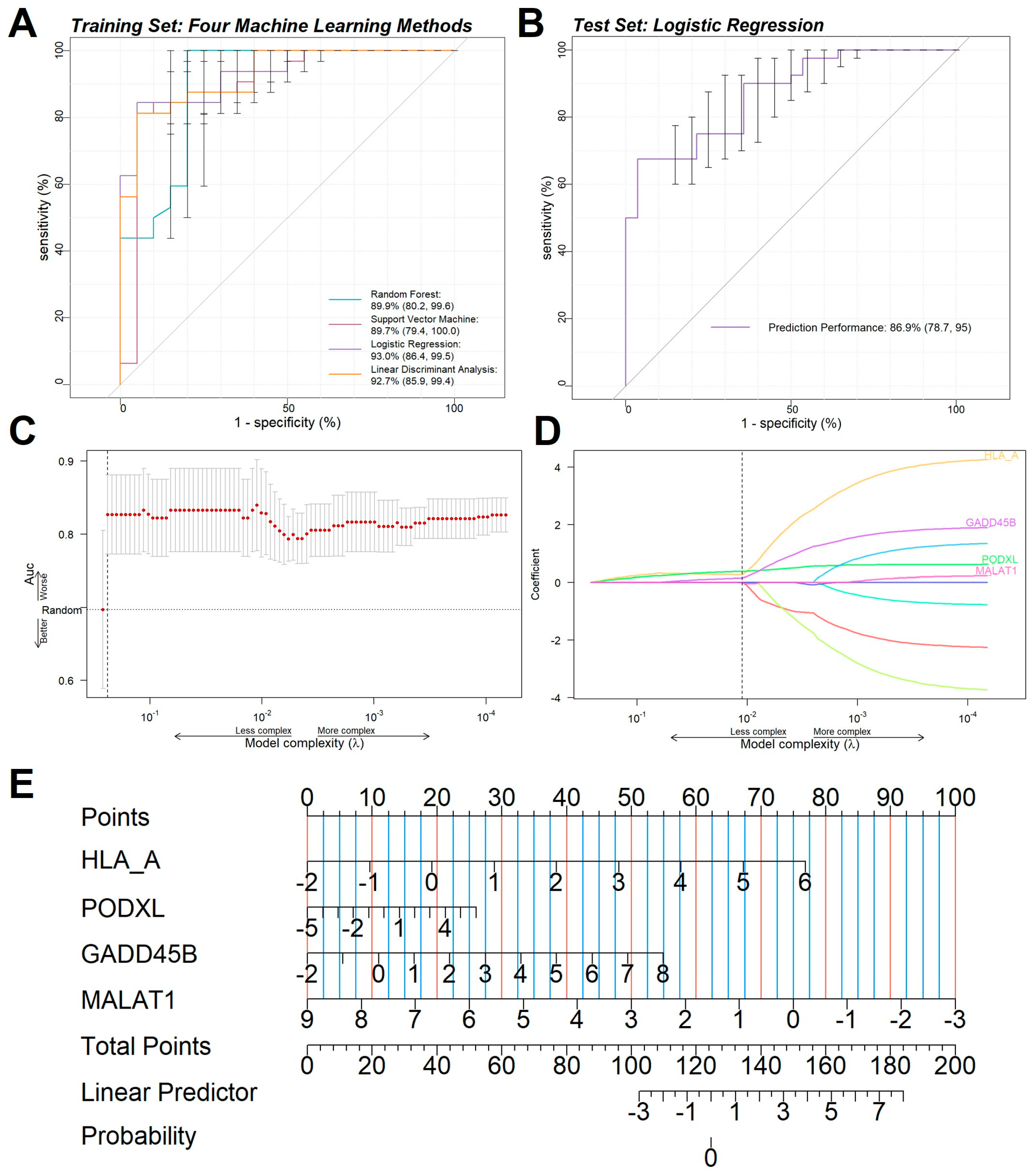
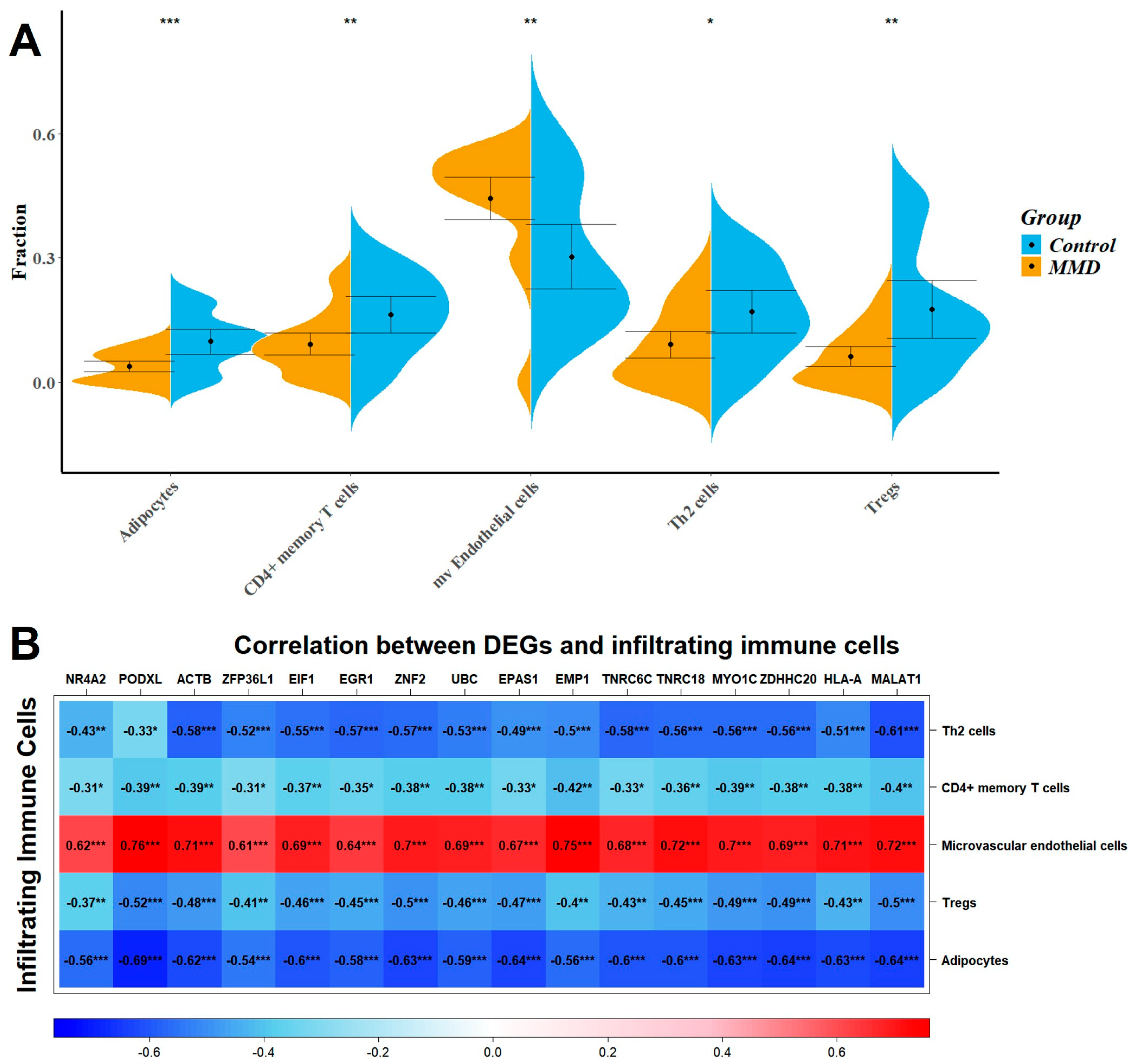
2.5. Immune Infiltration Analysis
3. Discussion
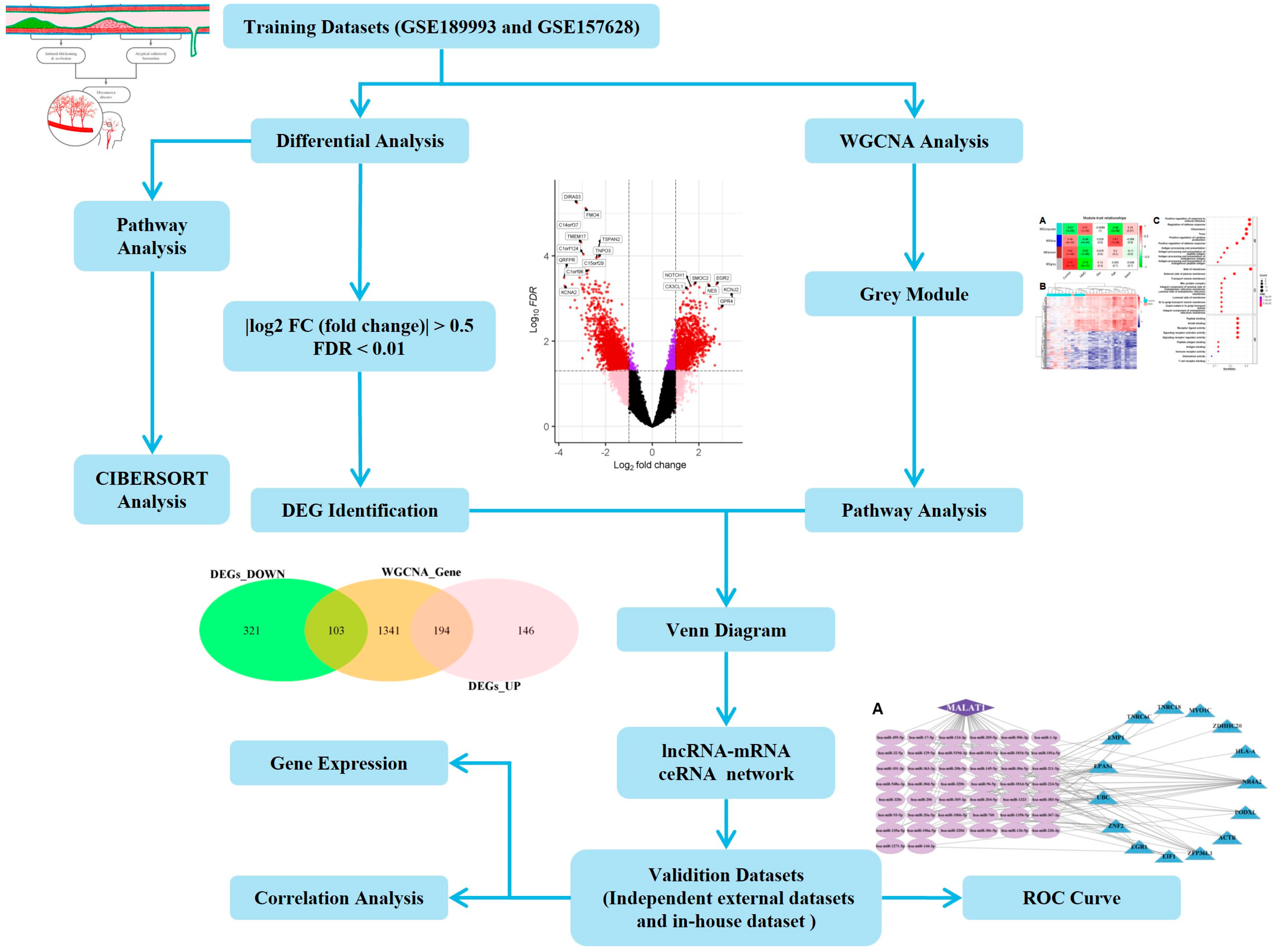
4. Materials and Methods
4.1. Selection and Preparation of Datasets
4.2. Weighted Gene Co-Expression Network Analysis—WGCNA
4.3. Identification of Differentially Expressed Genes and Venn Diagram Analysis
4.4. Biological Function and Pathway Enrichment Analyses
4.5. CeRNA Regulatory Network
4.6. Machine Learning and Feature Selection
4.7. Evaluation of Tissue-Infiltrating Immune Cells
4.8. Extracellular Vesicle Isolation and RNA Extraction
4.9. MiRNA Sequencing and Bioinformatic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Scott, R.M.; Smith, E.R. Moyamoya disease and moyamoya syndrome. N. Engl. J. Med. 2009, 360, 1226–1237. [Google Scholar] [CrossRef]
- Kuroda, S.; Houkin, K. Moyamoya disease: Current concepts and future perspectives. Lancet Neurol. 2008, 7, 1056–1066. [Google Scholar] [CrossRef]
- Suzuki, J.; Kodama, N. Moyamoya disease—A review. Stroke 1983, 14, 104–109. [Google Scholar] [CrossRef]
- Asselman, C.; Hemelsoet, D.; Eggermont, D.; Dermaut, B.; Impens, F. Moyamoya disease emerging as an immune-related angiopathy. Trends Mol. Med. 2022, 28, 939–950. [Google Scholar] [CrossRef]
- Bower, R.S.; Mallory, G.W.; Nwojo, M.; Kudva, Y.C.; Flemming, K.D.; Meyer, F.B. Moyamoya disease in a primarily white, midwestern US population: Increased prevalence of autoimmune disease. Stroke 2013, 44, 1997–1999. [Google Scholar] [CrossRef]
- Ahmad, A.S.; Tahir, R.A.; Mitsias, P.D. Moyamoya vasculopathy with anti-SCL-70 antibodies: A case report and review of the literature. J. Clin. Neurosci. 2018, 56, 177–179. [Google Scholar] [CrossRef]
- Hiruma, M.; Watanabe, N.; Mitsumatsu, T.; Suzuki, N.; Fukushita, M.; Matsumoto, M.; Yoshihara, A.; Yoshimura Noh, J.; Sugino, K.; Ito, K. Clinical features of moyamoya disease with Graves’ disease: A retrospective study of 394,422 patients with thyroid disease. Endocr. J. 2023, 70, 141–148. [Google Scholar] [CrossRef]
- Bolem, N.; Nga, V.D.W.; Chou, N.; Yeo, T.T.; Lin, J.; Sharma, V.K. Coexisting Moyamoya Syndrome and Type 1 Diabetes Mellitus: A Case Report and Review of the Literature. Asian J. Neurosurg. 2020, 15, 194–197. [Google Scholar] [CrossRef]
- Ni, J.; Zhou, L.X.; Wei, Y.P.; Li, M.L.; Xu, W.H.; Gao, S.; Cui, L.Y. Moyamoya syndrome associated with Graves’ disease: A case series study. Ann. Transl. Med. 2014, 2, 77. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, X.; Liu, K.; Hamblin, M.H.; Yin, K.J. Long Noncoding RNA Malat1 Regulates Cerebrovascular Pathologies in Ischemic Stroke. J. Neurosci. 2017, 37, 1797–1806. [Google Scholar] [CrossRef]
- Dorschel, K.B.; Wanebo, J.E. Physiological and pathophysiological mechanisms of the molecular and cellular biology of angiogenesis and inflammation in moyamoya angiopathy and related vascular diseases. Front. Neurol. 2023, 14, 661611. [Google Scholar] [CrossRef]
- Ng, S.Y.; Lin, L.; Soh, B.S.; Stanton, L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013, 29, 461–468. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zörnig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef]
- Dhamija, S.; Diederichs, S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer 2016, 139, 269–280. [Google Scholar] [CrossRef]
- Kölling, M.; Genschel, C.; Kaucsar, T.; Hübner, A.; Rong, S.; Schmitt, R.; Sörensen-Zender, I.; Haddad, G.; Kistler, A.; Seeger, H.; et al. Hypoxia-induced long non-coding RNA Malat1 is dispensable for renal ischemia/reperfusion-injury. Sci. Rep. 2018, 8, 3438. [Google Scholar] [CrossRef]
- Wang, S.; Ren, L.; Shen, G.; Liu, M.; Luo, J. The knockdown of MALAT1 inhibits the proliferation, invasion and migration of hemangioma endothelial cells by regulating MiR-206/VEGFA axis. Mol. Cell. Probes 2020, 51, 101540. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.W.; Liu, M.M.; Duan, R.; Tao, Y.F.; Zhou, J.S.; Fang, W.R.; Zhu, J.R.; Niu, L.; Sun, J.G. The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol. Sin. 2020, 41, 22–33. [Google Scholar] [CrossRef]
- Santoro, J.D.; Lee, S.; Wang, A.C.; Ho, E.; Nagesh, D.; Khoshnood, M.; Tanna, R.; Durazo-Arvizu, R.A.; Manning, M.A.; Skotko, B.G.; et al. Increased Autoimmunity in Individuals With Down Syndrome and Moyamoya Disease. Front. Neurol. 2021, 12, 724969. [Google Scholar] [CrossRef]
- Chen, J.B.; Liu, Y.; Zhou, L.X.; Sun, H.; He, M.; You, C. Prevalence of autoimmune disease in moyamoya disease patients in Western Chinese population. J. Neurol. Sci. 2015, 351, 184–186. [Google Scholar] [CrossRef]
- Gast, M.; Rauch, B.H.; Nakagawa, S.; Haghikia, A.; Jasina, A.; Haas, J.; Nath, N.; Jensen, L.; Stroux, A.; Böhm, A.; et al. Immune system-mediated atherosclerosis caused by deficiency of long non-coding RNA MALAT1 in ApoE-/-mice. Cardiovasc. Res. 2019, 115, 302–314. [Google Scholar] [CrossRef]
- Boon, R.A.; Jaé, N.; Holdt, L.; Dimmeler, S. Long Noncoding RNAs: From Clinical Genetics to Therapeutic Targets? J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar] [CrossRef]
- Chen, Y.; Li, S.; Zhang, Y.; Wang, M.; Li, X.; Liu, S.; Xu, D.; Bao, Y.; Jia, P.; Wu, N.; et al. The lncRNA Malat1 regulates microvascular function after myocardial infarction in mice via miR-26b-5p/Mfn1 axis-mediated mitochondrial dynamics. Redox Biol. 2021, 41, 101910. [Google Scholar] [CrossRef]
- Fantuzzi, G. Adipose tissue, adipokines, and inflammation. J. Allergy Clin. Immunol. 2005, 115, 911–919, quiz 920. [Google Scholar] [CrossRef]
- Pober, J.S.; Sessa, W.C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 2007, 7, 803–815. [Google Scholar] [CrossRef]
- Wensveen, F.M.; Valentić, S.; Šestan, M.; Wensveen, T.T.; Polić, B. Interactions between adipose tissue and the immune system in health and malnutrition. Semin. Immunol. 2015, 27, 322–333. [Google Scholar] [CrossRef]
- Kleinewietfeld, M.; Hafler, D.A. The plasticity of human Treg and Th17 cells and its role in autoimmunity. Semin. Immunol. 2013, 25, 305–312. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Zhang, T.; Su, L.; Liu, B.; Zhu, Z.; Li, C. LncRNA MALAT1 promotes gastric cancer progression via inhibiting autophagic flux and inducing fibroblast activation. Cell Death Dis. 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Dong, N.; Xu, B.; Shi, H. Long noncoding RNA MALAT1 acts as a competing endogenous RNA to regulate Amadori-glycated albumin-induced MCP-1 expression in retinal microglia by a microRNA-124-dependent mechanism. Inflamm. Res. 2018, 67, 913–925. [Google Scholar] [CrossRef]
- Liu, H.; Shi, C.; Deng, Y. MALAT1 affects hypoxia-induced vascular endothelial cell injury and autophagy by regulating miR-19b-3p/HIF-1alpha axis. Mol. Cell. Biochem. 2020, 466, 25–34. [Google Scholar] [CrossRef]
- Zhao, R.; Zhu, Y.; Yang, X.; Duan, T.; Yang, J.; Chen, B.; Yang, X.; Ni, G.; Li, S.; He, Y. Inorganic Arsenic-Induced Up-Regulation of MALAT1 Affects Cell Apoptosis via Disrupting the Binding Between IKBalpha and IKKbeta, P65. J. Biochem. Mol. Toxicol. 2025, 39, e70374. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.T.; Xu, D.Y.; Wang, G.Y.; Sun, J.L.; Huang, Y.; Wang, S.Z. IL-6 induced lncRNA MALAT1 enhances TNF-alpha expression in LPS-induced septic cardiomyocytes via activation of SAA3. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 302–309. [Google Scholar]
- Tian, H.; Wu, M.; Zhou, P.; Huang, C.; Ye, C.; Wang, L. The long non-coding RNA MALAT1 is increased in renal ischemia-reperfusion injury and inhibits hypoxia-induced inflammation. Ren. Fail. 2018, 40, 527–533. [Google Scholar] [CrossRef]
- Rectenwald, J.E.; Moldawer, L.L.; Huber, T.S.; Seeger, J.M.; Ozaki, C.K. Direct evidence for cytokine involvement in neointimal hyperplasia. Circulation 2000, 102, 1697–1702. [Google Scholar] [CrossRef] [PubMed]
- Reid, A.J.; Bhattacharjee, M.B.; Regalado, E.S.; Milewicz, A.L.; El-Hakam, L.M.; Dauser, R.C.; Milewicz, D.M. Diffuse and uncontrolled vascular smooth muscle cell proliferation in rapidly progressing pediatric moyamoya disease. J. Neurosurg. Pediatr. 2010, 6, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Pang, Q.; Shi, Y.; Chen, X.; Tu, F. Long noncoding RNA MALAT1 promotes angiogenesis through the caveolin-1/VEGF pathway after cerebral ischemic injury. Neuroreport 2025, 36, 350–363. [Google Scholar] [CrossRef]
- Berry, J.A.; Cortez, V.; Toor, H.; Saini, H.; Siddiqi, J. Moyamoya: An Update and Review. Cureus 2020, 12, e10994. [Google Scholar] [CrossRef]
- Fang, Y.C.; Wei, L.F.; Hu, C.J.; Tu, Y.K. Pathological Circulating Factors in Moyamoya Disease. Int. J. Mol. Sci. 2021, 22, 1696. [Google Scholar] [CrossRef]
- Kanamori, F.; Yokoyama, K.; Ota, A.; Yoshikawa, K.; Karnan, S.; Maruwaka, M.; Shimizu, K.; Ota, S.; Uda, K.; Araki, Y.; et al. Transcriptome-wide analysis of intracranial artery in patients with moyamoya disease showing upregulation of immune response, and downregulation of oxidative phosphorylation and DNA repair. Neurosurg. Focus. 2021, 51, E3. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Arikawa, E.; Sun, Y.; Wang, J.; Zhou, Q.; Ning, B.; Dial, S.L.; Guo, L.; Yang, J. Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study. BMC Genom. 2008, 9, 328. [Google Scholar] [CrossRef]
- López-Romero, P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genom. 2011, 12, 64. [Google Scholar] [CrossRef]
- Zahurak, M.; Parmigiani, G.; Yu, W.; Scharpf, R.B.; Berman, D.; Schaeffer, E.; Shabbeer, S.; Cope, L. Pre-processing Agilent microarray data. BMC Bioinform. 2007, 8, 142. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Sturm, G.; Finotello, F.; List, M. Immunedeconv: An R Package for Unified Access to Computational Methods for Estimating Immune Cell Fractions from Bulk RNA-Sequencing Data. Methods Mol. Biol. 2020, 2120, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, F.; Coates, B.; Zhang, Y.; Zhou, X.; Cheng, D. Comparative profiling of microRNAs in the winged and wingless English grain aphid, Sitobion avenae (F.) (Homoptera: Aphididae). Sci. Rep. 2016, 6, 35668. [Google Scholar] [CrossRef] [PubMed]
| Disease | GEO Accession | Tissue Sources | Data | DEG Count | GEO GPL | Assay Type | ||
|---|---|---|---|---|---|---|---|---|
| Case | Control | Up | Down | |||||
| MDD Train_dataset | GSE189993 | MCA | 21 | 11 | 424 | 340 | GPL16699 | Array |
| MDD Train_dataset | GSE157628 | MCA | 11 | 9 | GPL16699 | Array | ||
| MDD Val_dataset | GSE141022 | MCA | 4 | 4 | 567 | 165 | GPL16699 | Array |
| MDD Val_dataset | GSE141024 | MCA | 4 | 4 | 1381 | 661 | GPL16699 | Array |
| Total Sample | MMD, N = 32 | Control, N = 20 | Statistics/df | p Value | |
|---|---|---|---|---|---|
| Gender (% female) | 38 (73.08%) | 25 (78.13%) | 13 (65.00%) | 0.51377/1 | 0.4735 |
| Age, y, mean ± SD | 47.4 ± 19.5 | 43.5 ± 13.7 | 53.8 ± 25.4 | −1.6327/32 | 0.1124 |
| Atherosclerosis risk factor, mmol/L | |||||
| Glucose | 105.7 ± 23.3 | 105.9 ± 23.3 | 105.3 ± 24.5 | −0.11593/37 | 0.9083 |
| Triglyceride | 96.8 ± 63.1 | 107.8 ± 73.4 | 75.8 ± 28.9 | 1.8641/47 | 0.06863 |
| TC | 193.7 ± 39.1 | 200.0 ± 41.6 | 181.7 ± 32.2 | 1.4093/45 | 0.1744 |
| LDL | 118.2 ± 32.7 | 122.0 ± 36.7 | 110.9 ± 23.1 | 0.90712/50 | 0.3687 |
| HDL | 56.5 ± 12.5 | 58.4 ± 11.6 | 53.0 ± 13.8 | 1.7623/44 | 0.08499 |
| Total Sample | MMD, N = 10 | Control, N = 10 | Statistics/df | p Value | |
|---|---|---|---|---|---|
| Gender (% female) | 12 (60.00%) | 6 (60.00%) | 6 (60.00%) | 0/1 | 1 |
| Age, y, mean ± SD | 38.7 ± 9.1 | 38.9 ± 9.6 | 38.4 ± 9.1 | 0.11936/18 | 0.9063 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Fu, H.; Fang, S.; Ni, J.; Peng, B. Construction and Analysis of Immune Infiltration and Competing Endogenous RNA Network in Moyamoya Disease. Int. J. Mol. Sci. 2025, 26, 7957. https://doi.org/10.3390/ijms26167957
Liu W, Fu H, Fang S, Ni J, Peng B. Construction and Analysis of Immune Infiltration and Competing Endogenous RNA Network in Moyamoya Disease. International Journal of Molecular Sciences. 2025; 26(16):7957. https://doi.org/10.3390/ijms26167957
Chicago/Turabian StyleLiu, Wenhao, Hanhui Fu, Shiyuan Fang, Jun Ni, and Bin Peng. 2025. "Construction and Analysis of Immune Infiltration and Competing Endogenous RNA Network in Moyamoya Disease" International Journal of Molecular Sciences 26, no. 16: 7957. https://doi.org/10.3390/ijms26167957
APA StyleLiu, W., Fu, H., Fang, S., Ni, J., & Peng, B. (2025). Construction and Analysis of Immune Infiltration and Competing Endogenous RNA Network in Moyamoya Disease. International Journal of Molecular Sciences, 26(16), 7957. https://doi.org/10.3390/ijms26167957





