BRAF V600E Mutation Has Variable Tumor-Specific Effects on Expression of MAPK Pathway Genes That Could Affect Patient Outcome
Abstract
1. Introduction
2. Results
3. Discussion
4. Methods and Materials
4.1. DNA Next-Generation Sequencing (NGS)
4.2. Whole Transcriptomic Sequencing
4.3. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404, Erratum in: Cancer Discov. 2012, 2, 960. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Roa, P.; Bremer, N.V.; Foglizzo, V.; Cocco, E. Mutations in the Serine/Threonine Kinase BRAF: Oncogenic Drivers in Solid Tumors. Cancers 2024, 16, 1215. [Google Scholar] [CrossRef]
- Savoia, P.; Fava, P.; Casoni, F.; Cremona, O. Targeting the ERK Signaling Pathway in Melanoma. Int. J. Mol. Sci. 2019, 20, 1483. [Google Scholar] [CrossRef]
- Priantti, J.N.; Vilbert, M.; Madeira, T.; Moraes, F.C.A.; Hein, E.C.K.; Saeed, A.; Cavalcante, L. Efficacy and Safety of Rechallenge with BRAF/MEK Inhibitors in Advanced Melanoma Patients: A Systematic Review and Meta-Analysis. Cancers 2023, 15, 3754. [Google Scholar] [CrossRef]
- Klein, O.; Clements, A.; Menzies, A.M.; O’tOole, S.; Kefford, R.F.; Long, G.V. BRAF inhibitor activity in V600R metastatic melanoma. Eur. J. Cancer 2013, 49, 1073–1079. [Google Scholar] [CrossRef]
- Al-Masri, M.; Al-Shobaki, T.; Al-Najjar, H.; Iskanderian, R.; Younis, E.; Abdallah, N.; Tbakhi, A.; Haddad, H.; Al-Masri, M.; Obeid, Z.; et al. BRAF V600E mutation in papillary thyroid carcinoma: It’s relation to clinical features and oncologic outcomes in a single cancer centre experience. Endocr. Connect. 2021, 10, 1531–1537. [Google Scholar] [CrossRef]
- Podolski, A.; Castellucci, E.; Halmos, B. Precision medicine: BRAF mutations in thyroid cancer. Precis. Cancer Med. 2019, 2, 29. [Google Scholar] [CrossRef]
- Falchook, G.S.; Millward, M.; Hong, D.; Naing, A.; Piha-Paul, S.; Waguespack, S.G.; Cabanillas, M.E.; Sherman, S.I.; Ma, B.; Curtis, M.; et al. BRAF inhibitor dabrafenib in patients with metastatic BRAF-mutant thyroid cancer. Thyroid 2015, 25, 71–77. [Google Scholar] [CrossRef]
- Brose, M.S.; E Cabanillas, M.; Cohen, E.E.W.; Wirth, L.J.; Riehl, T.; Yue, H.; I Sherman, S.; Sherman, E.J. Vemurafenib in patients with BRAF(V600E)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: A non-randomised, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1272–1282. [Google Scholar] [CrossRef]
- Wagle, M.-C.; Kirouac, D.; Klijn, C.; Liu, B.; Mahajan, S.; Junttila, M.; Moffat, J.; Merchant, M.; Huw, L.; Wongchenko, M.; et al. A transcriptional MAPK Pathway Activity Score (MPAS) is a clinically relevant biomarker in multiple cancer types. NPJ Precis. Oncol. 2018, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Huijberts, S.C.; van Geel, R.M.; Bernards, R.; Beijnen, J.H.; Steeghs, N. Encorafenib, binimetinib and cetuximab combined therapy for patients with BRAFV600E mutant metastatic colorectal cancer. Future Oncol. 2020, 16, 161–173. [Google Scholar] [CrossRef] [PubMed]
- da Silva, I.P.; Wang, K.Y.; Wilmott, J.S.; Holst, J.; Carlino, M.S.; Park, J.J.; Quek, C.; Wongchenko, M.; Yan, Y.; Mann, G.; et al. Distinct Molecular Profiles and Immunotherapy Treatment Outcomes of V600E and V600K BRAF-Mutant Melanoma. Clin. Cancer Res. 2019, 25, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Ascierto, P.A.; Kirkwood, J.M.; Grob, J.-J.; Simeone, E.; Grimaldi, A.M.; Maio, M.; Palmieri, G.; Testori, A.; Marincola, F.M.; Mozzillo, N. The role of BRAF V600 mutation in melanoma. J. Transl. Med. 2012, 10, 85. [Google Scholar] [CrossRef]
- Kebebew, E.; Weng, J.B.; Bauer, J.; Ranvier, G.; Clark, O.H.; Duh, Q.-Y.; Shibru, D.; Bastian, B.; Griffin, A. The prevalence and prognostic value of BRAF mutation in thyroid cancer. Ann. Surg. 2007, 246, 466–470. [Google Scholar] [CrossRef]
- Dankner, M.; Wang, Y.; Fazelzad, R.; Johnson, B.; Nebhan, C.A.; Dagogo-Jack, I.; Myall, N.J.; Richtig, G.; Bracht, J.W.; Gerlinger, M.; et al. Clinical Activity of Mitogen-Activated Protein Kinase–Targeted Therapies in Patients With Non–V600 BRAF-Mutant Tumors. JCO Precis. Oncol. 2022, 6, e2200107. [Google Scholar] [CrossRef]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. FDA Grants Accelerated Approval to Dabrafenib in Combination with Trametinib for Unresectable or Metastatic Solid Tumors with BRAF V600E Mutation; US Food and Drug Administration: Silver Spring, MD, USA, 2022.
- Stenger, M. Dabrafenib in Combination with Trametinib for Unresectable or Metastatic Solid Tumors with BRAF V600E Mutation; The ASCO Post: Huntington, NY, USA, 2022. [Google Scholar]
- Gouda, M.A.; Subbiah, V. Expanding the Benefit: Dabrafenib/Trametinib as Tissue-Agnostic Therapy for BRAF V600E–Positive Adult and Pediatric Solid Tumors. Am. Soc. Clin. Oncol. Educ. Book 2023, 43, e404770. [Google Scholar] [CrossRef]
- Girotti, M.R.; Marais, R. Déjà Vu: EGF receptors drive resistance to BRAF inhibitors. Cancer Discov. 2013, 3, 487–490. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Shi, Q.; Smyrk, T.C.; Thibodeau, S.N.; Dienstmann, R.; Guinney, J.; Bot, B.M.; Tejpar, S.; Delorenzi, M.; Goldberg, R.M.; et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 2015, 148, 88–99. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Ebi, H.; Turke, A.B.; Coffee, E.M.; Nishino, M.; Cogdill, A.P.; Brown, R.D.; Della Pelle, P.; Dias-Santagata, D.; Hung, K.E.; et al. EGFR-mediated re activation of MAPK signaling contributes to insensitivity of BRAF-mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012, 2, 227–235. [Google Scholar] [CrossRef]
- Xing, M. BRAF mutation in thyroid cancer. Endocr. Relat. Cancer 2005, 12, 245–262. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef]
- Rusinek, D.; Swierniak, M.; Chmielik, E.; Kowal, M.; Kowalska, M.; Cyplinska, R.; Czarniecka, A.; Piglowski, W.; Korfanty, J.; Chekan, M.; et al. BRAFV600E-Associated Gene Expression Profile: Early Changes in the Transcriptome, Based on a Transgenic Mouse Model of Papillary Thyroid Carcinoma. PLoS ONE 2015, 10, e0143688. [Google Scholar] [CrossRef] [PubMed]
- Byron, A.; Bernhardt, S.; Ouine, B.; Cartier, A.; Macleod, K.G.; Carragher, N.O.; Sibut, V.; Korf, U.; Serrels, B.; de Koning, L. Integrative analysis of multi-platform reverse-phase protein array data for the pharmacodynamic assessment of response to targeted therapies. Sci. Rep. 2020, 10, 21985. [Google Scholar] [CrossRef] [PubMed]
- Gouda, M.A.; Ong, E.; Huang, H.J.; McPhaul, L.W.; Yoon, S.; Janku, F.; Gianoukakis, A.G. Ultrasensitive detection of BRAF V600E mutations in circulating tumor DNA of patients with metastatic thyroid cancer. Endocrine 2022, 76, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.M.; Yu, H.; Edwards, R.; Chen, L.; Kazianis, S.; Brafford, P.; Acs, G.; Herlyn, M.; Xu, X. Mutant V600E BRAF increases hypoxia inducible factor-1alpha expression in melanoma. Cancer Res. 2007, 67, 3177–3184. [Google Scholar] [CrossRef]
- Wang, J.; Huang, S.K.; Marzese, D.M.; Hsu, S.C.; Kawas, N.P.; Chong, K.K.; Long, G.V.; Menzies, A.M.; Scolyer, R.A.; Izraely, S.; et al. Epigenetic changes of EGFR have an important role in BRAF inhibitor–resistant cutaneous melanomas. J. Investig. Dermatol. 2015, 135, 532–541. [Google Scholar] [CrossRef]
- Podium Presentation. In Proceedings of the Western Surgical Association 132nd Scientific Session, Colorado Springs, CO, USA, 2–5 November 2024.
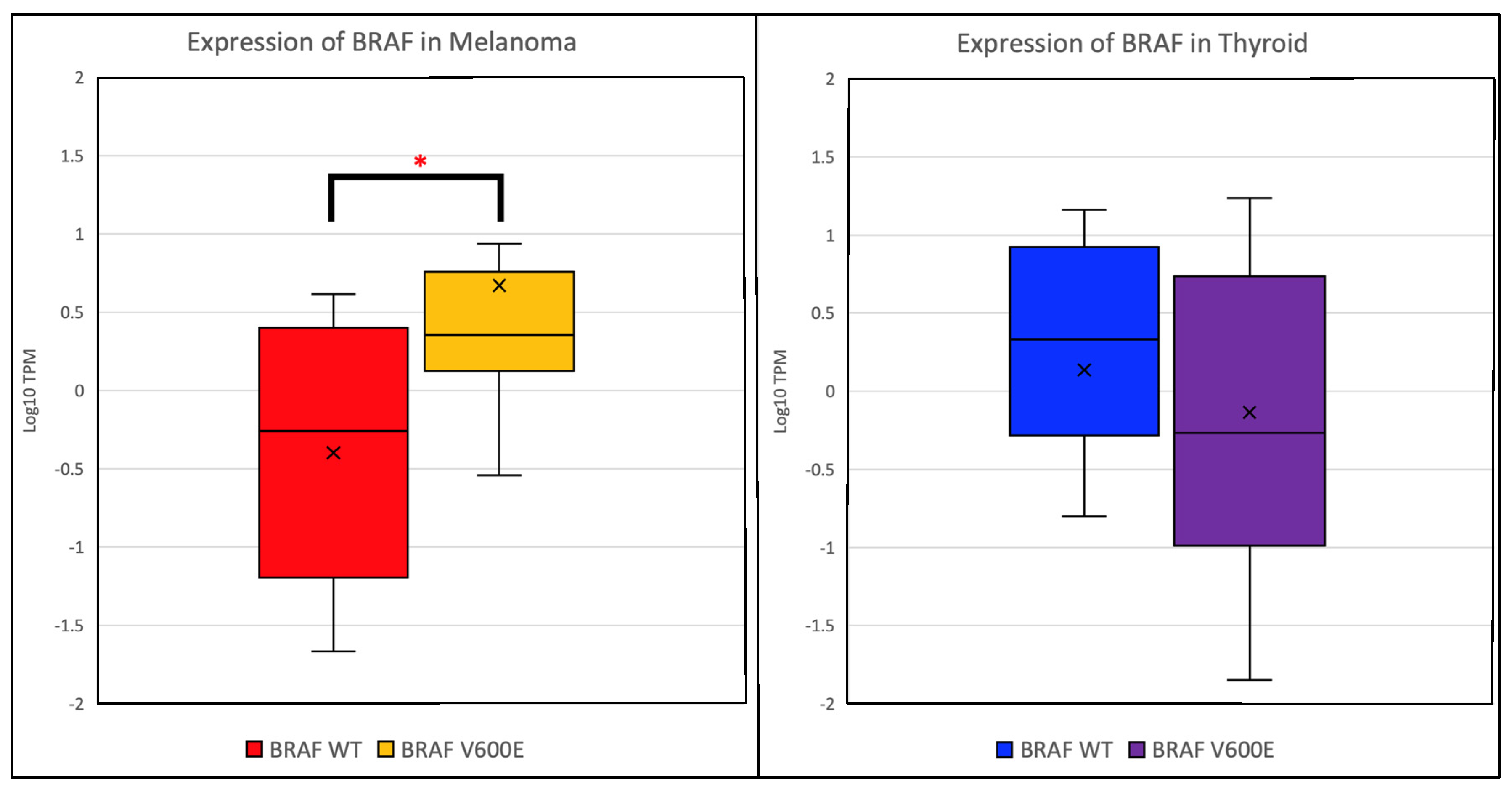
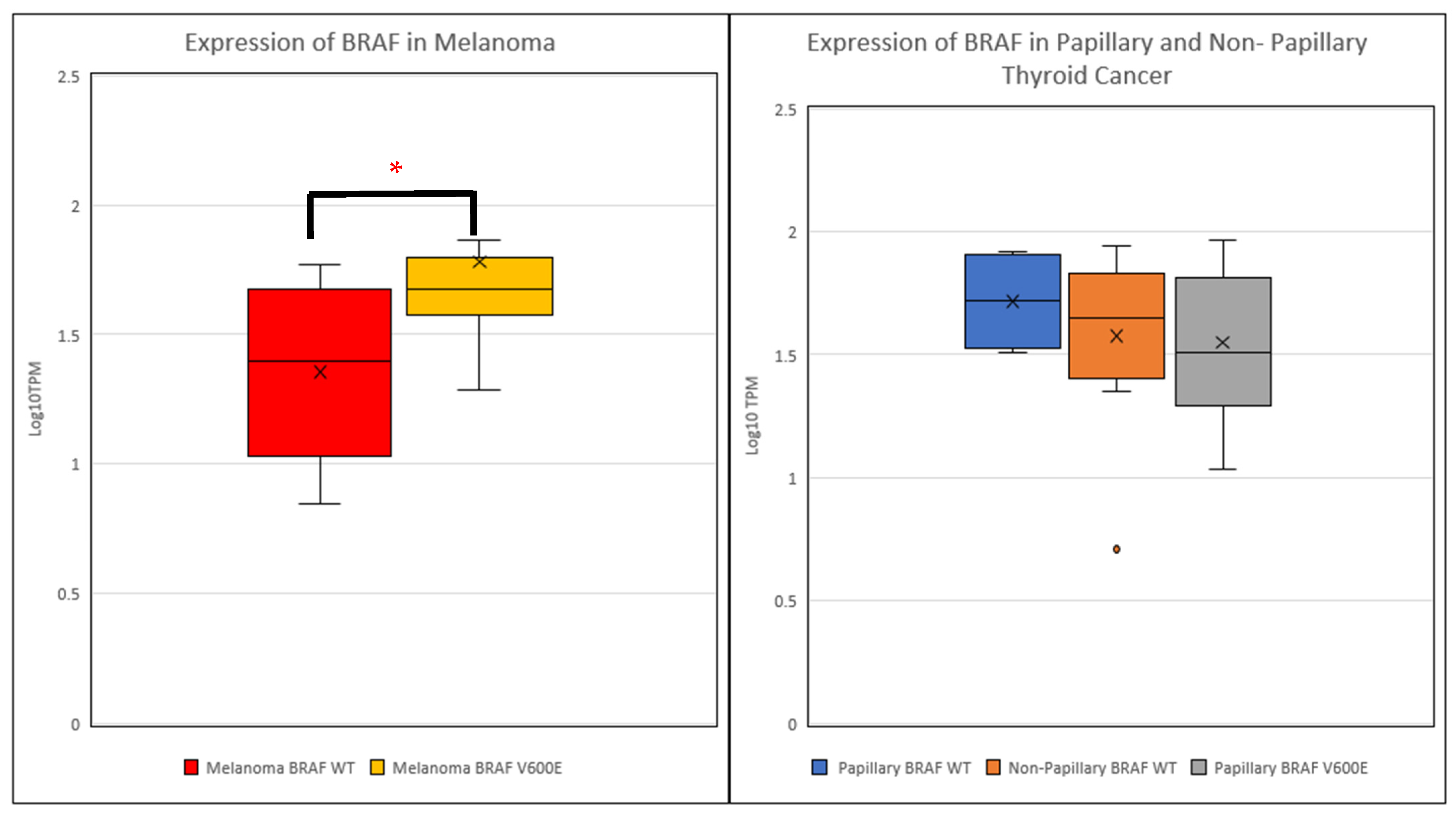

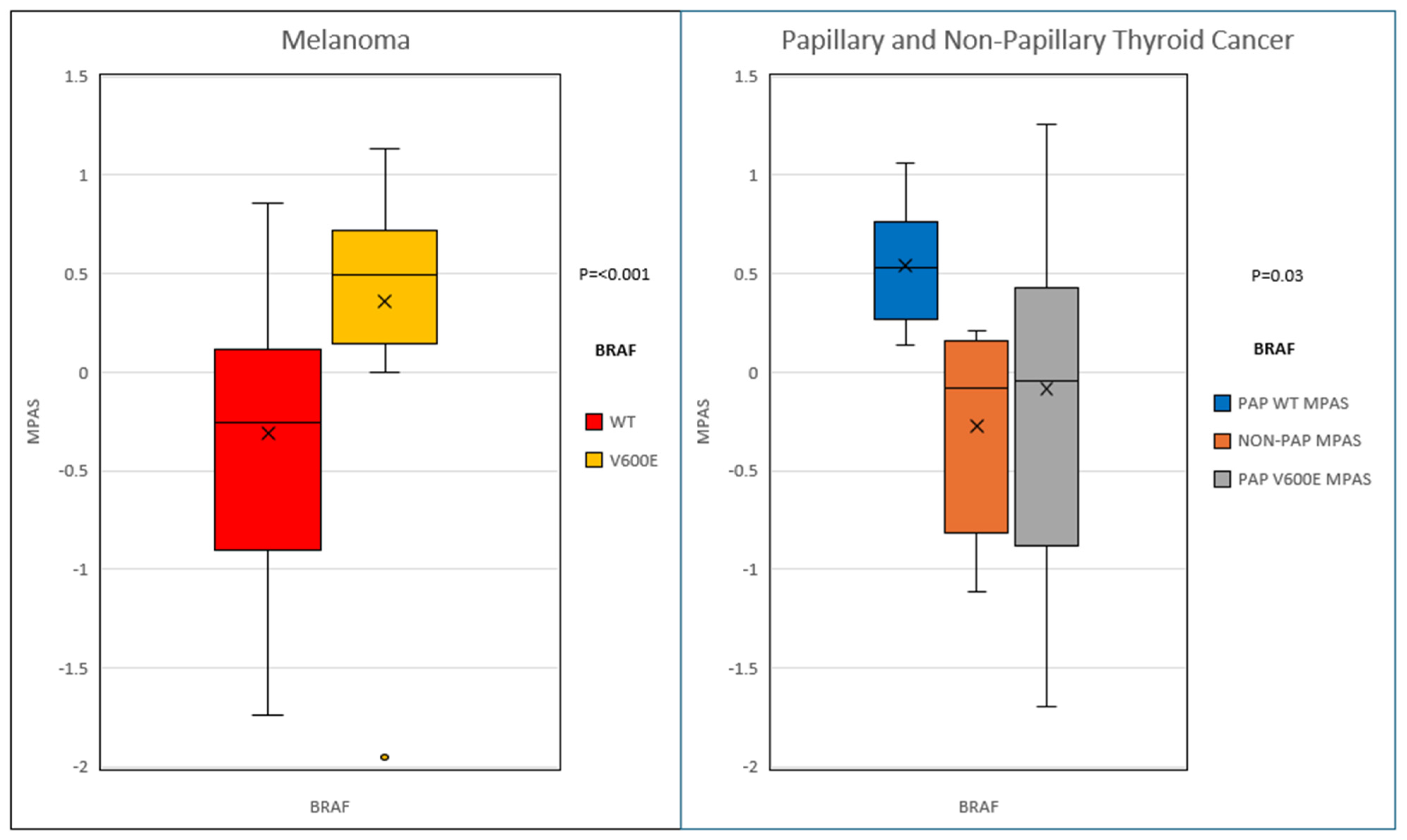
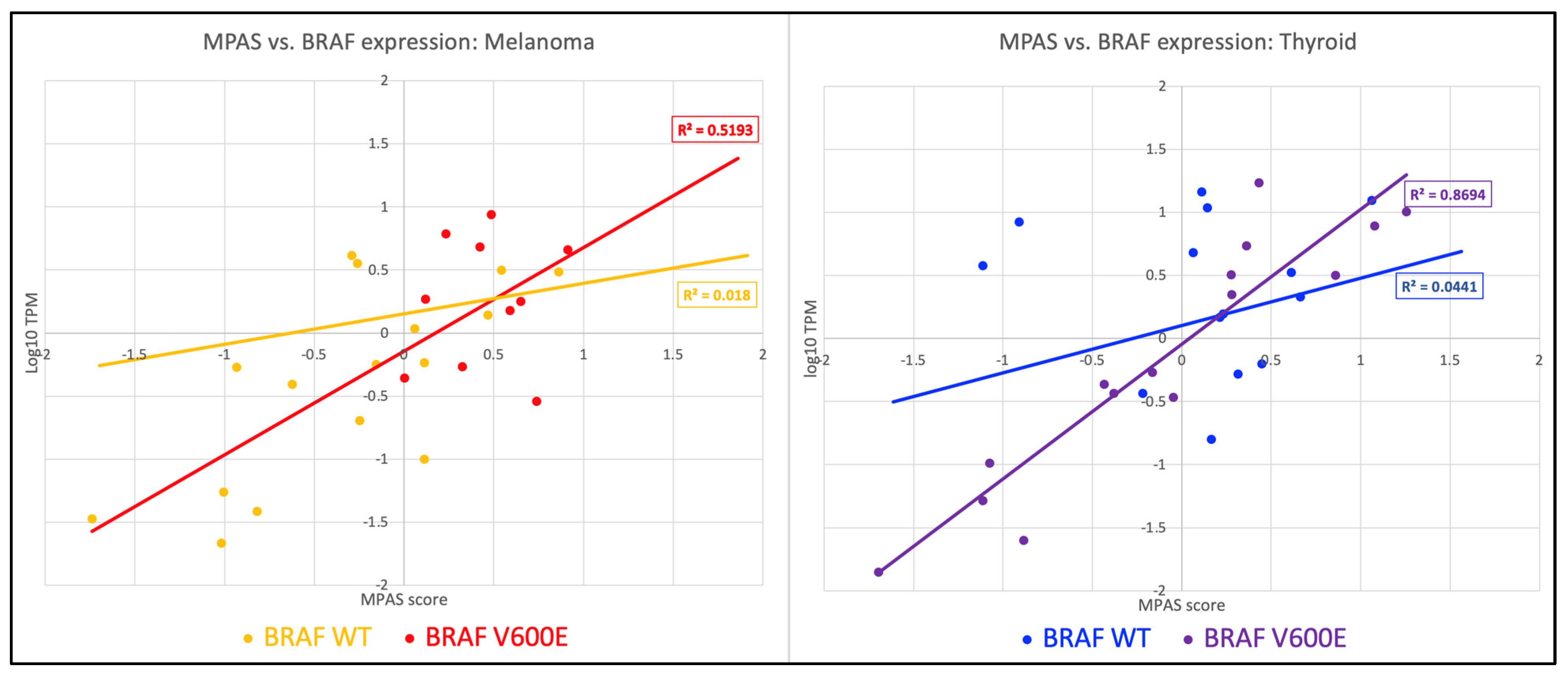
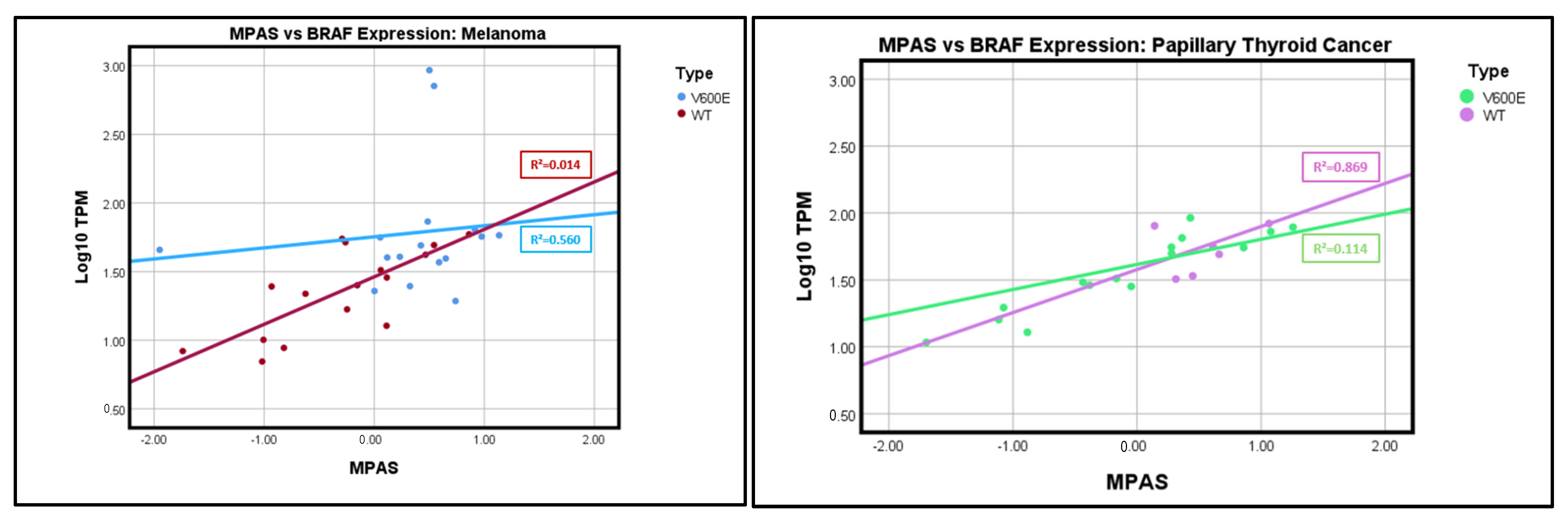
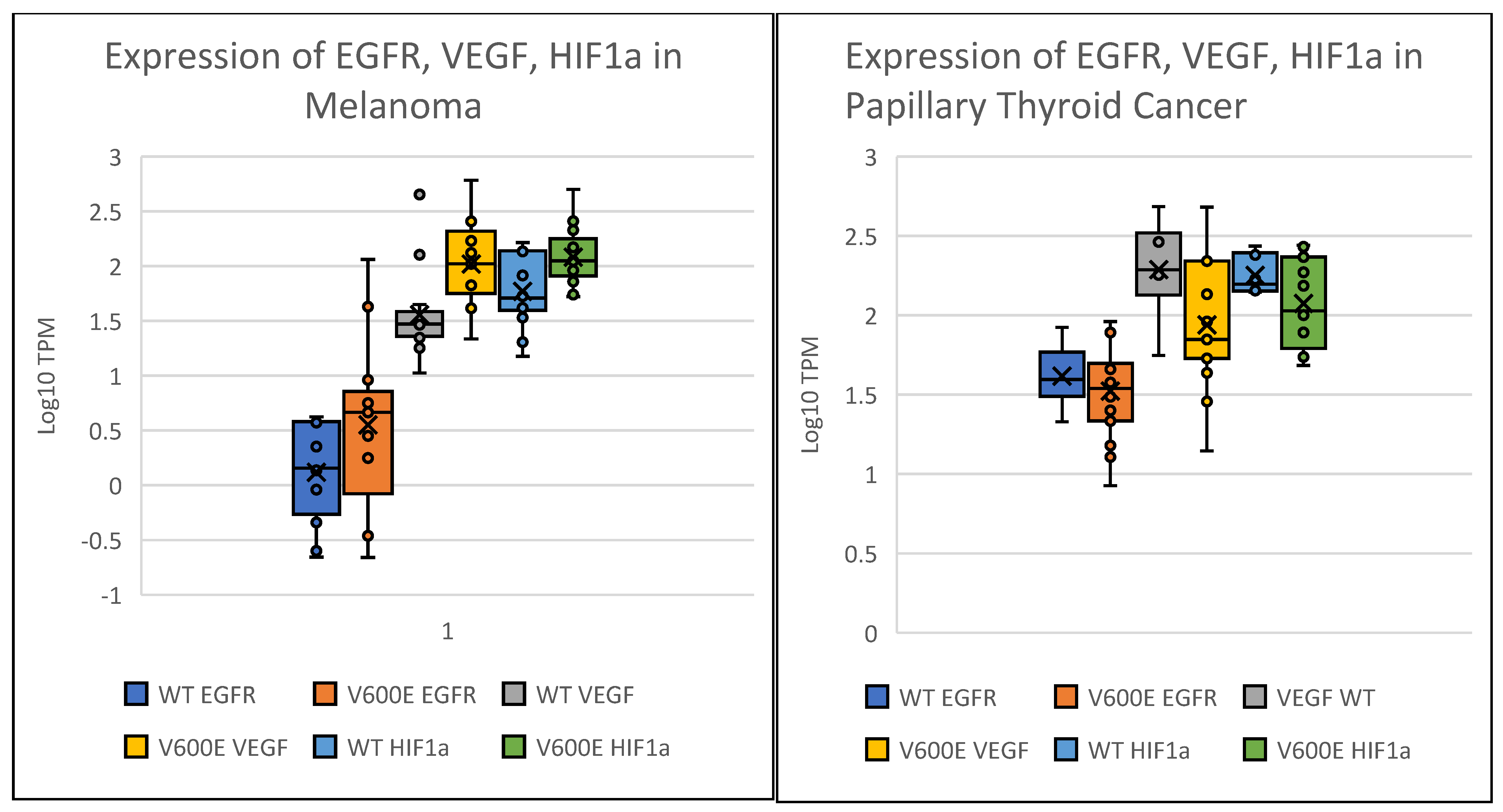
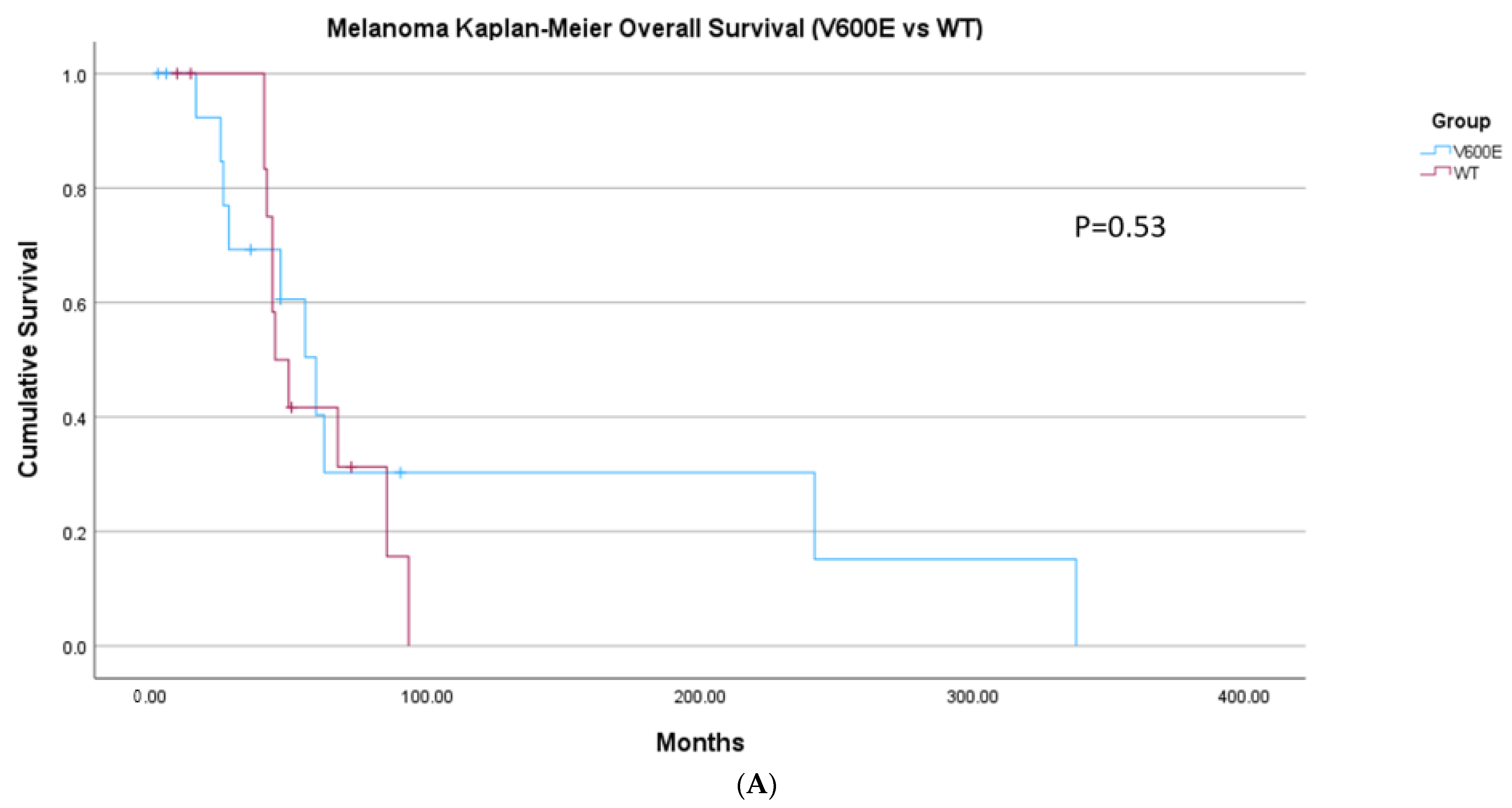
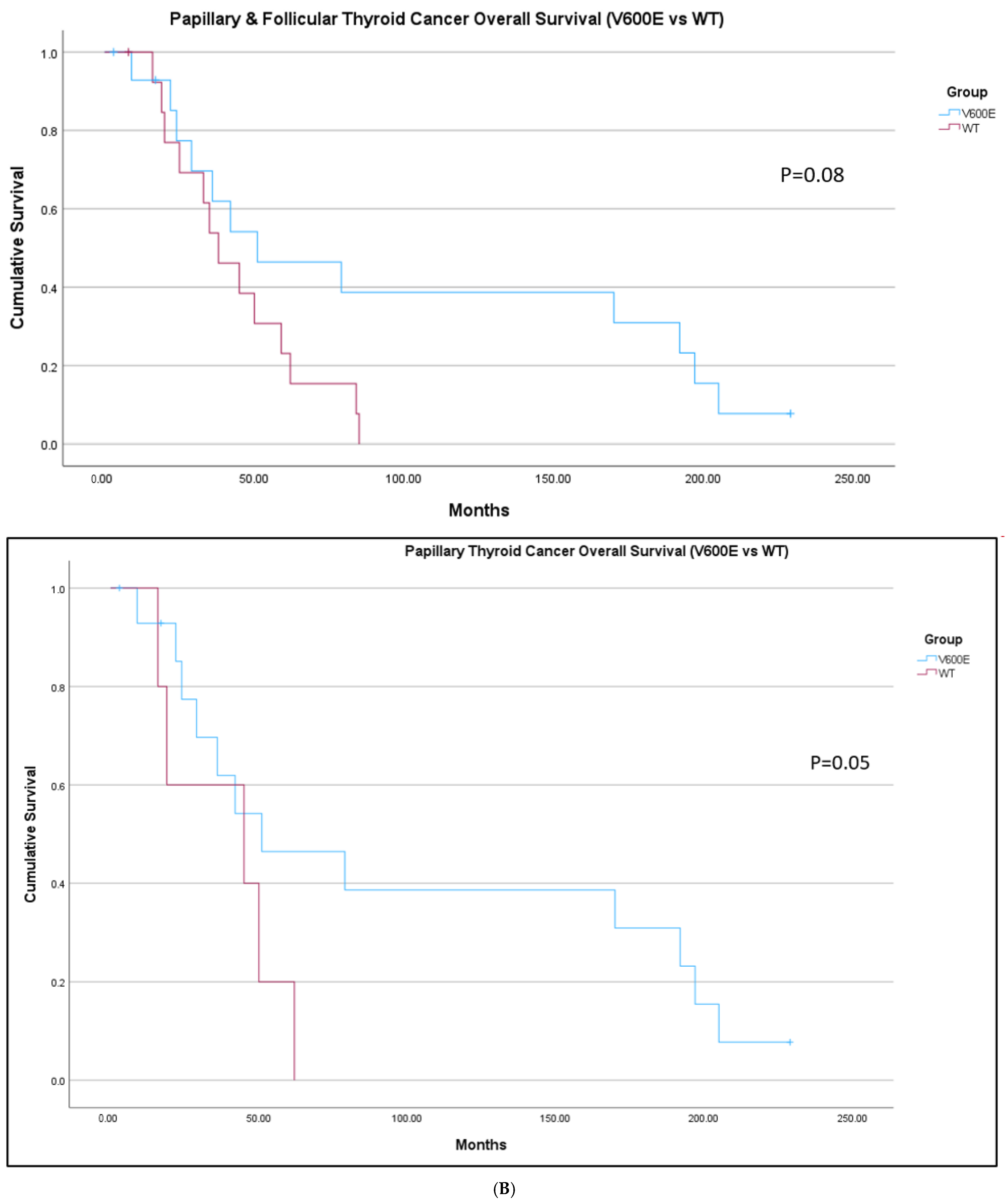
| Group | Melanoma | Thyroid Cancer |
|---|---|---|
| BRAF WT | 134 | 29 |
| BRAF V600E Mutation | 33 | 19 |
| Other BRAF V600 Mutations | 14 | 0 |
| Miscellaneous BRAF mutations | 19 | 1 |
| Patient | Age at Diagnosis | Gender | Diagnosis | Stage | BRAF V600E Y/N |
|---|---|---|---|---|---|
| 1 | 60 | Male | Medullary thyroid carcinoma | 3 | N |
| 2 | 36 | Female | Papillary thyroid carcinoma | 1 | Y |
| 3 | 68 | Female | Papillary thyroid carcinoma | 2 | Y |
| 4 | 36 | Female | Papillary thyroid carcinoma | 1 | N |
| 5 | 69 | Female | Follicular thyroid carcinoma | 1 | N |
| 6 | 51 | Male | Papillary thyroid carcinoma | 1 | Y |
| 7 | 43 | Male | Medullary thyroid carcinoma | 4A | N |
| 8 | 37 | Female | Papillary thyroid carcinoma | 1 | N |
| 9 | 62 | Male | Papillary thyroid carcinoma | 2 | Y |
| 10 | 50 | Male | Papillary thyroid carcinoma | 1 | Y |
| 11 | 35 | Female | Papillary thyroid carcinoma | 1 | Y |
| 12 | 51 | Male | Papillary thyroid carcinoma | 4 | N |
| 13 | 44 | Male | Papillary thyroid carcinoma | 4 | N |
| 14 | 46 | Male | Papillary thyroid carcinoma | 2 | Y |
| 15 | 60 | Female | Medullary thyroid carcinoma | 2A | Y |
| 16 | 62 | Female | Papillary thyroid carcinoma | 2 | Y |
| 17 | 62 | Female | Follicular thyroid carcinoma | 1 | N |
| 18 | 55 | Male | Papillary thyroid carcinoma | 4B | Y |
| 19 | 52 | Male | Papillary thyroid carcinoma | 1 | Y |
| 20 | 59 | Male | Papillary thyroid carcinoma | 2 | N |
| 21 | 47 | Male | Follicular thyroid carcinoma | 1 | N |
| 22 | 49 | Male | Papillary thyroid carcinoma | 2 | Y |
| 23 | 41 | Male | Papillary thyroid carcinoma | 1 | N |
| 24 | 55 | Female | Papillary thyroid carcinoma | 2 | N |
| 25 | 21 | Female | Papillary thyroid carcinoma | 1 | Y |
| 26 | 40 | Female | Follicular thyroid carcinoma | 1 | N |
| 27 | 23 | Female | Papillary thyroid carcinoma | 2 | Y |
| 28 | 81 | Male | Anaplastic thyroid carcinoma | 4B | N |
| 29 | 79 | Male | Anaplastic thyroid carcinoma | 4A | N |
| Patient | Age at Diagnosis | Gender | Stage | BRAF V600E Y/N |
|---|---|---|---|---|
| 1 | 48 | Female | 4 | Y |
| 2 | 43 | Female | 4 | Y |
| 3 | 59 | Male | 4 | N |
| 4 | 21 | Male | 1B | Y |
| 5 | 79 | Female | 4 | N |
| 6 | 57 | Male | 4 | Y |
| 7 | 54 | Male | 4 | N |
| 8 | 50 | Female | 4 | N |
| 9 | 58 | Female | 4 | N |
| 10 | 37 | Female | 4 | Y |
| 11 | 67 | Male | 4 | N |
| 12 | 64 | Male | 3B | N |
| 13 | 77 | Male | 3C | N |
| 14 | 65 | Male | 3C | Y |
| 15 | 53 | Female | 3B | Y |
| 16 | 72 | Female | 4 | N |
| 17 | 83 | Male | 4 | N |
| 18 | 47 | Male | 3 | N |
| 19 | 71 | Male | 4 | N |
| 20 | 88 | Female | 3C | N |
| 21 | 34 | Female | 4 | Y |
| 22 | 63 | Male | 4 | Y |
| 23 | 49 | Male | 4 | Y |
| 24 | 39 | Female | 3C | Y |
| 25 | 64 | Male | 3C | Y |
| 26 | 55 | Female | 4 | Y |
| 27 | 66 | Male | 3 | Y |
| 28 | 78 | Male | 4 | N |
| 29 | 83 | Male | 3C | Y |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darabi, S.; Stafford, P.; Braxton, D.R.; Zuazo, C.E.; Brodie, T.J.; Demeure, M.J. BRAF V600E Mutation Has Variable Tumor-Specific Effects on Expression of MAPK Pathway Genes That Could Affect Patient Outcome. Int. J. Mol. Sci. 2025, 26, 7910. https://doi.org/10.3390/ijms26167910
Darabi S, Stafford P, Braxton DR, Zuazo CE, Brodie TJ, Demeure MJ. BRAF V600E Mutation Has Variable Tumor-Specific Effects on Expression of MAPK Pathway Genes That Could Affect Patient Outcome. International Journal of Molecular Sciences. 2025; 26(16):7910. https://doi.org/10.3390/ijms26167910
Chicago/Turabian StyleDarabi, Sourat, Phillip Stafford, David R. Braxton, Carlos E. Zuazo, Taylor J. Brodie, and Michael J. Demeure. 2025. "BRAF V600E Mutation Has Variable Tumor-Specific Effects on Expression of MAPK Pathway Genes That Could Affect Patient Outcome" International Journal of Molecular Sciences 26, no. 16: 7910. https://doi.org/10.3390/ijms26167910
APA StyleDarabi, S., Stafford, P., Braxton, D. R., Zuazo, C. E., Brodie, T. J., & Demeure, M. J. (2025). BRAF V600E Mutation Has Variable Tumor-Specific Effects on Expression of MAPK Pathway Genes That Could Affect Patient Outcome. International Journal of Molecular Sciences, 26(16), 7910. https://doi.org/10.3390/ijms26167910







