Associations Between Genetic Variants in MCT2 (rs3763980, rs995343, rs3763979) and MCT4 (rs11323780) with Blood Lactate Kinetics Before and After Supramaximal Exercise
Abstract
1. Introduction
2. Results
2.1. MCT2 rs3763980
2.2. MCT2 rs995343
2.3. MCT2 rs3763979
2.4. MCT4 rs11323780
2.5. Correlation Analysis
3. Discussion
Limitations
4. Materials and Methods
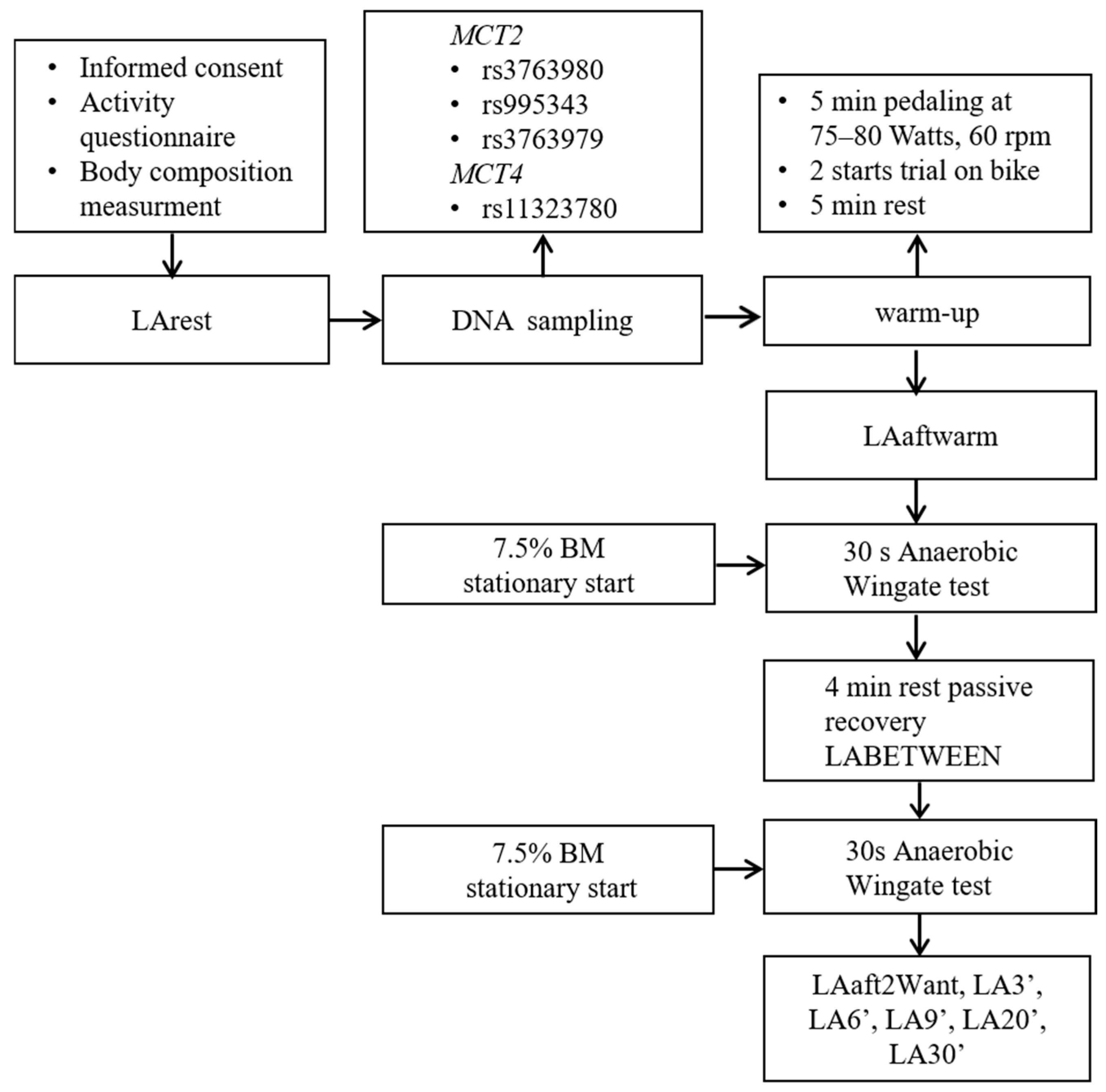
4.1. Study Design
4.2. Participants
4.3. Intermittent All-Out Wingate Tests
4.4. Blood LA Measurement
4.5. DNA Sampling and Isolation
4.6. Genotyping Analyses
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ginevičienė, V.; Utkus, A.; Pranckevičienė, E.; Semenova, E.A.; Hall, E.C.R.; Ahmetov, I.I. Perspectives in Sports Genomics. Biomedicines 2022, 10, 298. [Google Scholar] [CrossRef]
- Semenova, E.A.; Hall, E.C.R.; Ahmetov, I.I. Genes and Athletic Performance: The 2023 Update. Genes 2023, 14, 1235. [Google Scholar] [CrossRef]
- Mandadzhiev, N. The contemporary role of lactate in exercise physiology and exercise prescription—A review of the literature. Folia Med. 2025, 67, e144693. [Google Scholar] [CrossRef]
- Spriet, L.L.; Howlett, R.A.; Heigenhauser, G.J.F. An enzymatic approach to lactate production in human skeletal muscle during exercise. Med. Sci. Sports Exerc. 2000, 32, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.; Morris, M. Monocarboxylate Transporters: Therapeutic Targets and Prognostic. Clin. Pharmacol. Ther. 2016, 100, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Halestrap, A.P. The SLC16 gene family-Structure, role and regulation in health and disease. Mol. Asp. Med. 2013, 34, 337–349. [Google Scholar] [CrossRef]
- Juel, C.; Halestrap, A.P. Lactate transport in skeletal muscle—Role and regulation of the monocarboxylate transporter. J. Physiol. 1999, 517, 633–642. [Google Scholar] [CrossRef]
- Grollman, E.F.; Philp, N.J.; McPhie, P.; Ward, R.D.; Sauer, B. Determination of transport kinetics of chick MCT3 monocarboxylate transporter from retinal pigment epithelium by expression in genetically modified yeast. Biochemistry 2000, 39, 9351–9357. [Google Scholar] [CrossRef]
- Talbot, J.; Mavez, L. Skeletal muscle fiber type: Using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 518–534. [Google Scholar] [CrossRef]
- Plotkin, D.L.; Roberts, M.D.; Haun, C.T.; Schoenfeld, B.J. Muscle fiber type transitions with exercise training: Shifting perspectives. Sport 2021, 9, 127. [Google Scholar] [CrossRef]
- Van Hall, G. Lactate kinetics in human tissues at rest and during exercise. Acta Physiol. 2010, 199, 499–508. [Google Scholar] [CrossRef]
- Van Hall, G.; Jensen-Urstad, M.; Rosdahl, H.; Holmberg, H.C.; Saltin, B.; Calbet, J.A.L. Leg and arm lactate and substrate kinetics during exercise. Am. J. Physiol.-Endocrinol. Metab. 2003, 284, 193–205. [Google Scholar] [CrossRef]
- Yoshida, Y.; Holloway, G.P.; Ljubicic, V.; Hatta, H.; Spriet, L.L.; Hood, D.A.; Bonen, A. Negligible direct lactate oxidation in subsarcolemmal and intermyofibrillar mitochondria obtained from red and white rat skeletal muscle. J. Physiol. 2007, 582, 1317–1335. [Google Scholar] [CrossRef]
- Gladden, L.B. Lactate metabolism during exercise. In Principles of Exercise Biochemistry; Poortmans, J.R., Ed.; Karger: Basel, Switzerland, 2003; pp. 152–196. [Google Scholar]
- Brooks, G.A. Bioenergetics of exercising humans. Compr. Physiol. 2012, 2, 537–562. [Google Scholar] [CrossRef]
- Billat, L.V. Use of blood lactate measurements for prediction of exercise performance and for control of training. Recommendations for long-distance running. Sports Med. 1996, 22, 157–175. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, M.L.; Harris, J.E.; Hernández, A.; Gladden, L.B. Blood lactate measurements and analysis during exercise: A guide for clinicians. J. Diabetes Sci. Technol. 2007, 1, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Beneke, R.; Leithäuser, R.M.; Ochentel, O. Blood lactate diagnostics in exercise testing and training. Int. J. Sports Physiol. Perform. 2011, 6, 8–24. [Google Scholar] [CrossRef]
- Gurovich, A.N.; Heiser, B.; Hayes, C.; Marshall, E.; Roath, S.; Kabous, N.G. Clinical Markers of Exercise Intensity as a Surrogate for Blood Lactate Levels Only During Low-Intensity Exercise in Patients With Coronary Artery Disease. Cardiopulm. Phys. Ther. J. 2018, 29, 144–151. [Google Scholar] [CrossRef]
- Lacour, J.R.; Bouvat, E.; Barthélémy, J.C. Post-competition blood lactate concentrations as indicators of anaerobic energy expenditure during 400-m and 800-m races. Eur. J. Appl. Physiol. Occup. Physiol. 1990, 61, 172–176. [Google Scholar] [CrossRef]
- Maculewicz, E.; Mastalerz, A.; Garbacz, A.; Mróz, A.; Stastny, P.; Petr, M.; Kolinger, D.; Vostatková, P.; Bojarczuk, A. The interactions between monocarboxylate transporter genes MCT1, MCT2, and MCT4 and the kinetics of blood lactate production and removal after high-intensity efforts in elite males: A cross-sectional study. BMC Genom. 2025, 26, 133. [Google Scholar] [CrossRef]
- Gene [Internet]. National Library of Medicine (US) NC for BI: Bethesda, MD, USA. 2004. Available online: https://www.ncbi.nlm.nih.gov/gene/9123 (accessed on 10 November 2024).
- Guo, X.; Chen, C.; Liu, B.; Wu, Y.; Chen, Y.; Zhou, X.; Huang, X.; Li, X.; Yang, H.; Chen, Z.; et al. Genetic variations in monocarboxylate transporter genes as predictors of clinical outcomes in non-small cell lung cancer. Tumor Biol. 2015, 36, 3931–3939. [Google Scholar] [CrossRef] [PubMed]
- Fei, F.; Guo, X.; Chen, Y.; Liu, X.; Tu, J.; Xing, J.; Chen, Z.; Ji, J.; He, X. Polymorphisms of monocarboxylate transporter genes are associated with clinical outcomes in patients with colorectal cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Van Dijkhuizen, P.; Wulffraat, N. Prediction of methotrexate efficacy and adverse events in patients with juvenile idiopathic arthritis: A systematic literature review. Pediatr. Rheumatol. 2014, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Moncrieffe, H.; Hinks, A.; Ursu, S.; Kassoumeri, L.; Etheridge, A.; Hubank, M.; Martin, P.; Weiler, T.; Glass, D.N.; Thompson, S.D.; et al. Generation of novel pharmacogenomic candidates in response to methotrexate in juvenile idiopathic arthritis: Correlation between gene expression and genotype. Pharmacogenet. Genom. 2010, 20, 665–676. [Google Scholar] [CrossRef]
- Jorfeldt, L.; Juhlin-Dannfelt, A.; Karlsson, J. Lactate release in relation to tissue lactate in human skeletal muscle during exercise. J. Appl. Physiol. Respir. Env. Exerc. Physiol. 1978, 44, 350–352. [Google Scholar] [CrossRef]
- Thomas, C.; Perrey, S.; Lambert, K.; Hugon, G.; Mornet, D.; Mercier, J. Monocarboxylate transporters, blood lactate removal after supramaximal exercise, and fatigue indexes in humans. J. Appl. Physiol. 2005, 98, 804–809. [Google Scholar] [CrossRef]
- Maciejewski, H.; Bourdin, M.; Féasson, L.; Dubouchaud, H.; Denis, C.; Freund, H.; Messonnier, L.A. Muscle MCT4 content is correlated with the lactate removal ability during recovery following all-out supramaximal exercise in highly-trained rowers. Front. Physiol. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Gladden, L.B. Lactate metabolism: A new paradigm for the third millennium. J. Physiol. 2004, 558, 5–30. [Google Scholar] [CrossRef]
- Menzies, P.; Menzies, C.; McIntyre, L.; Paterson, P.; Wilson, J.; Kemi, O.J. Blood lactate clearance during active recovery after an intense running bout depends on the intensity of the active recovery. J. Sports Sci. 2010, 28, 975–982. [Google Scholar] [CrossRef]
- Bangsbo, J.; Hellsten, Y. Muscle blood flow and oxygen uptake in recovery from exercise. Acta Physiol. Scand. 1998, 162, 305–312. [Google Scholar] [CrossRef]
- Merezhinskaya, N.; Fishbein, W.N. Monocarboxylate transporters: Past, present, and future. Histol. Histopathol. 2009, 24, 243–264. [Google Scholar] [CrossRef] [PubMed]
- Lund, J.; Aas, V.; Tingstad, R.H.; Van Hees, A.; Nikolić, N. Utilization of lactic acid in human myotubes and interplay with glucose and fatty acid metabolism. Sci. Rep. 2018, 8, 9814. [Google Scholar] [CrossRef] [PubMed]
- Fishbein, W.N.; Merezhinskaya, N.; Foellmer, J.W. Relative distribution of three major lactate transporters in frozen human tissues and their localization in unfixed skeletal muscle. Muscle Nerve 2002, 26, 101–112. [Google Scholar] [CrossRef]
- Hashimoto, T.; Masuda, S.; Taguchi, S.; Brooks, G.A. Immunohistochemical analysis of MCT1, MCT2 and MCT4 expression in rat plantaris muscle. J. Physiol. 2005, 567, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef]
- Brooks, G.A.; Curl, C.C.; Leija, R.G.; Osmond, A.D.; Duong, J.J.; Arevalo, J.A. Tracing the lactate shuttle to the mitochondrial reticulum. Exp. Mol. Med. 2022, 54, 1332–1347. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Wilson, M.C. The monocarboxylate transporter family-Role and regulation. IUBMB Life 2012, 64, 109–119. [Google Scholar] [CrossRef]
- Petr, M.; Štastný, P.; Pecha, O.; Šteffl, M.; Šeda, O.; Kohlíková, E. PPARA intron polymorphism associated with power performance in 30-s anaerobic Wingate Test. PLoS ONE 2014, 10, e0134424. [Google Scholar] [CrossRef]
- MacIntosh, B.R.; Rishaug, P.; Svedahl, K. Assessment of peak power and short-term work capacity. Eur. J. Appl. Physiol. 2003, 88, 572–579. [Google Scholar] [CrossRef]
- Lopez, E.I.D.; Smoliga, J.M.; Zavorsky, G.S. The effect of passive versus active recovery on power output over six repeated Wingate sprints. Res. Q. Exerc. Sport 2014, 85, 519–526. [Google Scholar] [CrossRef]
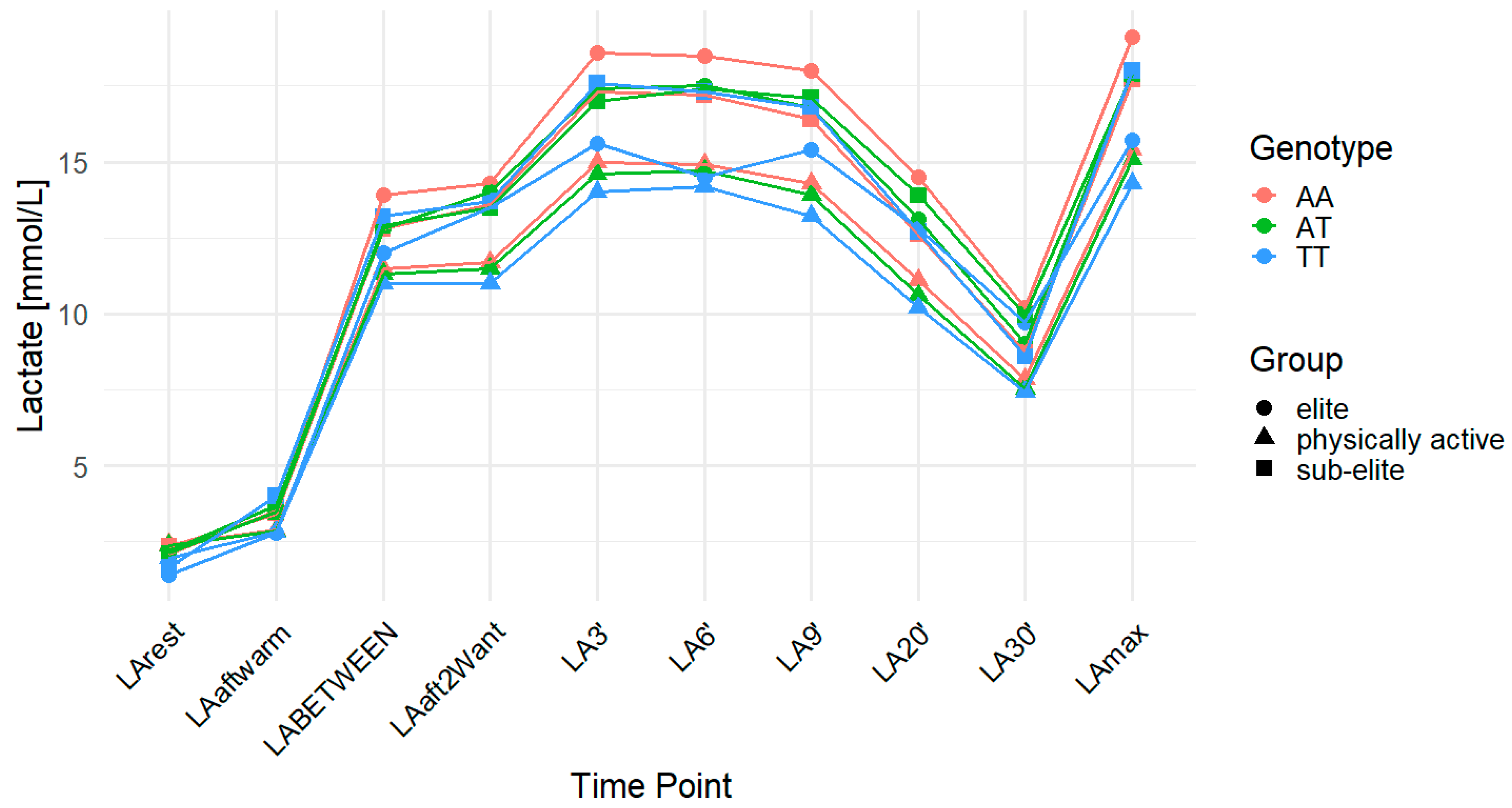

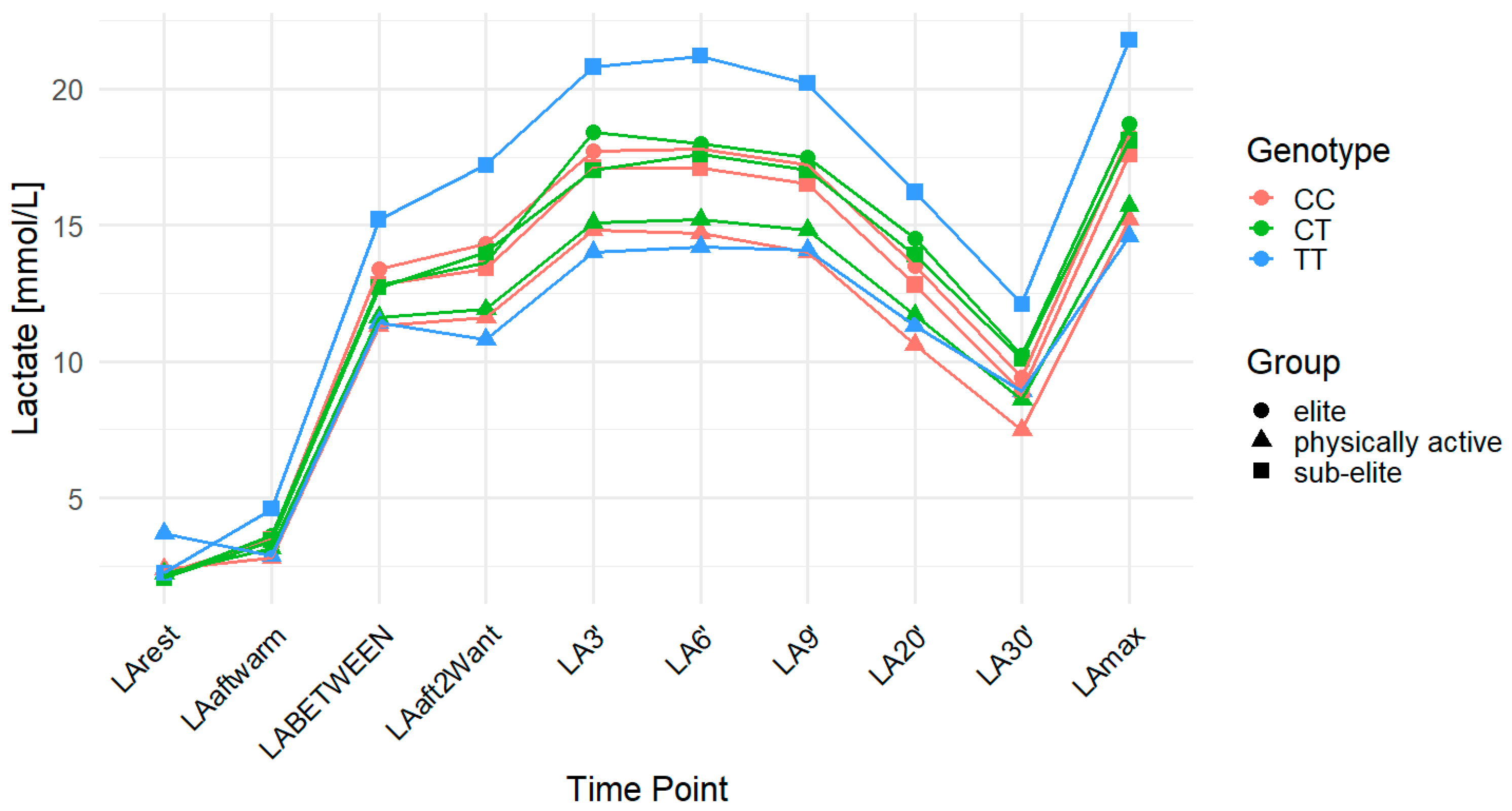
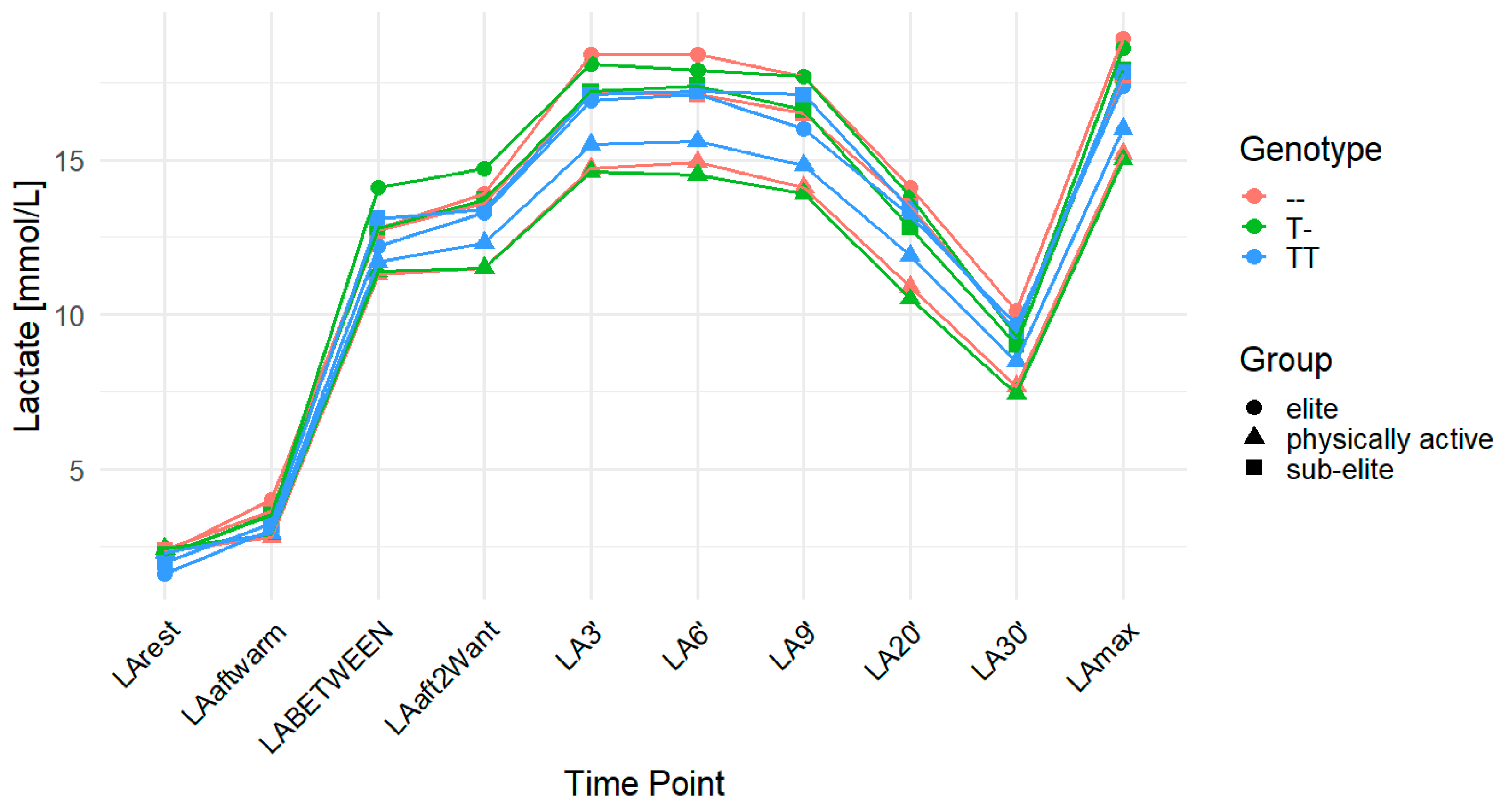
| Group | n | Age (Years) | Weight (kg) | Height (cm) |
|---|---|---|---|---|
| Elite | 42 | 20.48 ± 2.80 | 73.65 ± 14.72 | 182.87 ± 6.42 |
| Sub-elite | 145 | 19.79 ± 2.80 | 72.92 ± 9.24 | 182.67 ± 5.67 |
| Physically active | 192 | 20.94 ± 1.90 | 77.44 ± 9.61 | 180.73 ± 6.50 |
| MAF (%) | HWE p-Value All | HWE p-Value Elite | HWE p-Value Sub-Elite | HWE p-Value Physically Active | |
|---|---|---|---|---|---|
| MCT2 (rs3763980) | allele T (24.18) | 0.38 | 0.45 | 0.62 | 0.40 |
| MCT2 (rs995343) | allele G (43.77) | 0.45 | 0.52 | 0.22 | 0.88 |
| MCT2 (rs3763979) | allele T (12.46) | 0.22 | 1.00 | 0.66 | 0.19 |
| MCT4 (rs11323780) | allele T (47.33) | 1.00 | 0.76 | 0.69 | 0.66 |
| Genotype | Models | Effect (η2, d); (95% CI); Post Hoc Power | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AT | TT | Codominant | Dominant | Recessive | Codominant | Dominant | Recessive | ||
| AA vs. AT vs. TT | AA vs. AT + TT | AA + AT vs. TT | AA vs. AT vs. TT | AA vs. AT + TT | AA + AT vs. TT | |||||
| LArest | All | 2.35 | 2.26 | 1.76 | 0.03/0.04 * | 0.07 | 0.02/0.04 * | 0.02; (0.00–0.10) | 0.16; (−0.05–0.38) | 0.63; (0.11–1.15) |
| Elite | 2.08 | 2.21 | 1.4 | 0.31 | 0.78 | 0.16 | 0.06; (0.00–0.36) | −0.09; (−0.69–0.52) | 1.05; (−0.40–2.48) | |
| Sub-elite | 2.33 | 2.11 | 1.65 | 0.16 | 0.13 | 0.13 | 0.04; (0.00–0.22) | 0.30; (−0.09–0.69) | 0.64; (−0.19–1.47) | |
| Physically active | 2.41 | 2.37 | 1.96 | 0.43 | 0.54 | 0.21 | 0.009; (0.00–0.09) | 0.09; (−0.20–0.38) | 0.48; (−0.27–1.24) | |
| LA30′ | All | 8.34 | 8.56 | 8.2 | 0.99 | 0.88 | 1 | 0.002; (0.00–0.03) | −0.06; (−0.28–0.15) | 0.08; (−0.44–0.60) |
| Elite | 10.21 | 9.01 | 9.72 | 0.34 | 0.15 | 0.95 | 0.05; (0.00–0.35) | 0.45; (−0.17–1.06) | −0.04; (−1.46–1.38) | |
| Sub-elite | 8.72 | 9.96 | 8.6 | 0.13 | 0.08 | 0.6 | 0.04; (0.00–0.23) | −0.35; (−0.73–0.05) | 0.22; (−0.61–1.05) | |
| Physically active | 7.84 | 7.53 | 7.43 | 0.71 | 0.41 | 0.77 | 0.004; (0.00–0.06) | 0.12; (−0.17–0.41) | 0.11; (−0.64–0.87) | |
| LAmax | All | 16.47 | 16.42 | 15.94 | 0.68 | 0.46 | 0.51 | 0.002; (0.00–0.03) | 0.03; (−0.18–0.25) | 0.19; (−0.33–0.70) |
| Elite | 19.1 | 17.99 | 15.69 | 0.06 | 0.06 | 0.08 | 0.14; (0.00–0.49) | 0.61; (−0.01–1.23) | 1.31; (−0.15–2.75) | |
| Sub-elite | 17.73 | 17.85 | 17.97 | 0.97 | 0.81 | 0.87 | 0.001; (0.00–0.01) | −0.05; (−0.43–0.34) | −0.07 (−0.89–0.76) | |
| Physically active | 15.44 | 15.06 | 14.27 | 0.26 | 0.17 | 0.23 | 0.01; (0.00–0.11) | 0.20; (−0.09–0.49) | 0.46; (−0.29–1.22) | |
| ACC | All | 14.12 | 14.16 | 14.18 | 0.98 | 0.87 | 0.92 | 0; (0.00–0.00) | −0.02; (−0.23–0.20) | −0.02; (−0.53–0.50) |
| Elite | 17.02 | 15.78 | 14.29 | 0.04/0.06 * | 0.02/0.06 * | 0.14 | 0.2; (0.00–0.52); 0.8 | 0.8; (0.12–1.38); 0.7 | 1.10; (−0.34–2.54) | |
| Sub-elite | 15.4 | 15.73 | 16.32 | 0.66 | 0.45 | 0.5 | 0.008; (0.00–0.11) | −0.15; (−0.54–0.24) | −0.29; (−1.11–0.54) | |
| Physically active | 13.03 | 12.7 | 12.31 | 0.48 | 0.26 | 0.49 | 0.008; (0.00–0.08) | 0.17; (−0.12–0.46) | 0.27; (−0.49–1.02) | |
| DCC | All | 8.13 | 7.87 | 7.74 | 0.51 | 0.3 | 0.45 | 0.004; (0.00–0.05) | 0.12; (−0.09–0.34) | 0.13; (−0.39–0.64) |
| Elite | 8.89 | 8.98 | 5.97 | 0.24 | 0.81 | 0.09 | 0.07 (0.00–0.39) | 0.07; (−0.53–0.68) | 1.26; (−0.20–2.69) | |
| Sub-elite | 9 | 7.88 | 9.38 | 0.15 | 0.06 | 0.34 | 0.07; (0.00–0.30) | 0.44; (0.05–0.83) | −0.40; (−1.23–0.42) | |
| Physically active | 7.61 | 7.53 | 6.84 | 0.66 | 0.67 | 0.38 | 0.004; (0.00–0.06) | 0.06; (−0.23–0.35) | 0.34; (−0.42–1.09) | |
| Genotype | Models | Effect (η2, d); (95% CI); Post Hoc Power | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | Codominant | Dominant | Recessive | Codominant | Dominant | Recessive | ||
| AA vs. AG vs. GG | AA vs. AG + GG | AA + AG vs. GG | AA vs. AG vs. GG | AA vs. AG + GG | AA + AG vs. GG | |||||
| LArest | All | 2.10 | 2.40 | 2.30 | 0.03/0.04 * | 0.01/0.03 * | 0.94 | 0.02; (0.00–0.11) | −0.31; (−0.54–−0.08) | −0.01; (−0.29–0.27) |
| Elite | 1.98 | 2.18 | 2.12 | 0.72 | 0.43 | 0.98 | 0.02; (0.00–0.23) | −0.26; (−0.91–0.38) | −0.01; (−0.95–0.92) | |
| Sub-elite | 1.93 | 2.29 | 2.46 | 0.1 | 0.04/0.12 * | 0.25 | 0.05; (0.00–0.24) | −0.44; (−0.87–−0.02) | −0.33; (−0.90–0.23) | |
| Physically active | 2.23 | 2.51 | 2.27 | 0.12 | 0.15 | 0.37 | 0.02; (0.00–0.14) | −0.23; (−0.55–0.08) | 0.16; (−0.19–0.51) | |
| LA30′ | All | 8.42 | 8.54 | 8.05 | 0.53 | 0.99 | 0.28 | 0.004; (0.00–0.04) | 0.00; (−0.23–0.24) | 0.15; (−0.13–0.44) |
| Elite | 9.56 | 9.44 | 10.57 | 0.68 | 0.92 | 0.38 | 0.02; (0.00–0.24) | −0.03; (−0.67–0.61) | −0.42; (−1.36–0.52) | |
| Sub-elite | 9.05 | 9.6 | 8.25 | 0.32 | 0.68 | 0.2 | 0.02; (0.00–0.17) | −0.09; (−0.51–0.33) | 0.37; (−0.20–0.94) | |
| Physically active | 7.77 | 7.7 | 7.67 | 0.98 | 0.86 | 0.91 | 0; (0.00–0.00) | 0.03; (−0.28–0.34) | 0.02; (−0.33–0.37) | |
| LAmax | All | 16.17 | 16.49 | 16.68 | 0.47 | 0.26 | 0.43 | 0.004; (0.00–0.05) | −0.13; (−0.37–0.10) | −0.11; (−0.39–0.17) |
| Elite | 18.41 | 18.27 | 19.04 | 0.79 | 0.99 | 0.51 | 0.01; (0.00–0.20) | 0.004; (−0.64–0.65) | −0.32; (−1.26–0.62) | |
| Sub-elite | 17.6 | 17.88 | 17.86 | 0.89 | 0.64 | 0.92 | 0.002; (0.00–0.06) | −0.10; (−0.52–0.32) | −0.03; (−0.59–0.54) | |
| Physically active | 14.77 | 15.25 | 15.98 | 0.03/0.04 * | 0.05 | 0.02/0.04 * | 0.04; (0.00–0.18) | −0.32; (−0.63–−0.00) | −0.41; (−0.76–−0.06) | |
| ACC | All | 14.07 | 14.09 | 14.38 | 0.75 | 0.78 | 0.45 | 0.002; (0.00–0.03) | −0.03; (−0.27–0.20) | −0.11; (−0.39–0.17) |
| Elite | 16.43 | 16.08 | 16.93 | 0.66 | 0.76 | 0.45 | 0.02; (0.00–0.25) | 0.10; (−0.54–0.74) | −0.36; (−1.30–0.58) | |
| Sub-elite | 15.68 | 15.59 | 15 | 0.95 | 0.83 | 0.77 | 0.001 (0.00–0.03) | 0.05 (−0.37–0.46) | 0.08; (−0.48–0.65) | |
| Physically active | 12.54 | 12.74 | 13.71 | 0.02/0.03 * | 0.17 | 0.01/0.03 * | 0.04; (0.00–0.18) | −0.22; (−0.53–0.09) | −0.48; (−0.84–−0.13) | |
| DCC | All | 7.75 | 7.95 | 8.63 | 0.05 | 0.17 | 0.02/0.06 * | 0.02; (0.00–0.10) | −0.16; (−0.40–0.07) | −0.33; (−0.62–−0.05) |
| Elite | 8.86 | 8.83 | 8.47 | 0.95 | 0.91 | 0.76 | 0.002; (0.00–0.06) | 0.04; (−0.60–0.68) | 0.15; (−0.79–1.08) | |
| Sub-elite | 8.55 | 8.28 | 9.61 | 0.12 | 0.99 | 0.05 | 0.04 (0.00–0.23) | 0.003; (−0.42–0.42) | −0.57; (−1.14–−0.002) | |
| Physically active | 7 | 7.55 | 8.31 | 0.01/0.02 * | 0.03/0.03 * | 0.01/0.02 * | 0.04; (0.003–0.19) | −0.36; (−0.67–−0.04) | −0.44; (−0.80–−0.09) | |
| Genotype | Models | Effect (η2, d); (95% CI); Post Hoc Power | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | Codominant | Dominant | Recessive | Codominant | Dominant | Recessive | ||
| CC vs. CT vs. TT | CC vs. CT + TT | CC + CT vs. TT | CC vs. CT vs. TT | CC vs. CT + TT | CC + CT vs. TT | |||||
| LArest | All | 2.3 | 2.16 | 3.28 | 0.67 | 0.76 | 0.003/0.009 * | 0.03; (0.01–0.13) | 0.04; (−0.22–0.29) | −1.14; (−1.89–−0.38) |
| Elite | 2.09 | 2.11 | - | - | - | - | - | - | - | |
| Sub-elite | 2.23 | 2.07 | 2.25 | 0.75 | 0.47 | 0.94 | 0.006; (0.00–0.10) | 0.17; (−0.29–0.62) | −0.06; (−1.46–1.34) | |
| Physically active | 2.37 | 2.24 | 3.69 | 0.003/0.005 * | 0.8 | 0.001/0.003 * | 0.06; (0.01–0.22) | −0.05; (−0.39–0.30) | −1.51; (−2.41–−0.60) | |
| LA30′ | All | 8.13 | 9.33 | 9.84 | 0.67 | 0.001/0.003 * | 0.19 | 0.03; (0.01; 0.14) | −0.44; (−0.70–−0.18) | −0.51; (−1.26–0.24) |
| Elite | 10.22 | 9.4 | - | - | - | - | - | - | - | |
| Sub-elite | 8.92 | 10.15 | 12.13 | 0.11 | 0.06 | 0.19 | 0.04; (0.00–0.24) | −0.45; (−0.91–0.01) | −0.94; (−2.35–0.47) | |
| Physically active | 7.46 | 8.59 | 8.92 | 0.03/0.04 * | 0.01/0.03 * | 0.29 | 0.04; (0.00–0.18) | −0.46; (−0.81–−0.12) | −0.48; (−1.37–0.41) | |
| LAmax | All | 16.3 | 16.87 | 16.68 | 0.67 | 0.12 | 0.81 | 0.007; (0.00–0.06) | −0.20; (−0.45–0.06) | −0.09; (−0.84–0.66) |
| Elite | 18.73 | 18.3 | - | - | - | - | - | - | - | |
| Sub-elite | 17.64 | 17.97 | 21.78 | 0.1 | 0.31 | 0.04/0.12 * | 0.05; (0.00–0.24) | −0.24; (−0.70–0.22) | −1.52; (−2.93–−0.10) | |
| Physically active | 15.19 | 15.65 | 14.64 | 0.43 | 0.37 | 0.53 | 0.009; (0.00–0.09) | −0.16; (−0.50–0.19) | 0.29; (−0.60–1.18) | |
| ACC | All | 14 | 14.7 | 13.4 | 0.67 | 0.1 | 0.48 | 0.01; (0.00–0.08) | −0.21; (−0.47–0.04) | 0.27; (−0.48–1.02) |
| Elite | 16.63 | 16.18 | - | - | - | - | - | - | - | |
| Sub-elite | 15.41 | 15.91 | 19.53 | 0.08 | 0.2 | 0.04/0.12 * | 0.05; (0.00–0.25) | −0.30; (−0.76–0.16) | −1.52; (−2.93–−0.10) | |
| Physically active | 12.82 | 13.41 | 10.96 | 0.05 | 0.43 | 0.05 | 0.03; (0.00–0.17) | −0.14; (−0.48–0.20) | 0.90; (0.01–1.80) | |
| DCC | All | 8.17 | 7.53 | 6.84 | 0.67 | 0.02/0.06 * | 0.17 | 0.02; (0.00–0.10) | 0.31; (0.05–0.57) | 0.53; (−0.23–1.28) |
| Elite | 8.5 | 8.9 | - | - | - | - | - | - | - | |
| Sub-elite | 8.72 | 7.83 | 9.65 | 0.19 | 0.15 | 0.48 | 0.03; (0.00–0.21) | 0.34; (−0.12–0.80) | −0.51; (−1.91–0.89) | |
| Physically active | 7.73 | 7.07 | 5.72 | 0.04/0.06 * | 0.03/0.06 * | 0.06 | 0.03; (0.00–0.17); 0.8 | 0.38; (0.04–0.73) | 0.87; (−0.03–1.76) | |
| Genotype | Models | Effect (η2, d); (95% CI); Post Hoc Power | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| -- | T- | TT | Codominant | Dominant | Recessive | Codominant | Dominant | Recessive | ||
| -- vs. T- vs. TT | -- vs. T- + TT | -- + T- vs. TT | -- vs. T- vs. TT | -- vs. T- + TT | -- + T- vs. TT | |||||
| LArest | All | 2.35 | 2.35 | 2.09 | 0.07 | 0.49 | 0.02/0.06 * | 0.02; (0.00–0.09) | 0.08; (−0.15–0.32) | 0.30; (0.04–0.55) |
| Elite | 2.31 | 2.33 | 1.62 | 0.04/0.06 * | 0.26 | 0.01/0.03 * | 0.2; (0.00–0.51); 0.8 | 0.39; (−0.28–1.07) | 0.96; (0.21–1.69); 0.8 | |
| Sub-elite | 2.38 | 2.22 | 1.98 | 0.24 | 0.27 | 0.14 | 0.03; (0.00–0.19) | 0.27; (−0.18–0.72) | 0.33; (−0.11–0.76) | |
| Physically active | 2.28 | 2.34 | 2.44 | 0.61 | 0.67 | 0.47 | 0.005; (0.00–0.07) | −0.07; (−0.38–0.24) | 0.13; (−0.23–0.49) | |
| LA30′ | All | 8.47 | 8.12 | 9 | 0.09 | 0.49 | 0.05 | 0.01; (0.00–0.09) | 0.03; (−0.21–0.27) | −0.26; (−0.52–−0.00) |
| Elite | 10.11 | 9.29 | 9.66 | 0.69 | 0.43 | 0.95 | 0.02; (0.00–0.24) | 0.27; (−0.40–0.94) | −0.02; (−0.73–0.69) | |
| Sub-elite | 9.43 | 9.03 | 9.44 | 0.8 | 0.73 | 0.69 | 0.004; (0.00–0.08) | 0.08; (−0.37–0.52) | −0.09; (−0.52–0.35) | |
| Physically active | 8.49 | 7.68 | 7.43 | 0.1 | 0.9 | 0.04/0.12 * | 0.02; (0.00–0.14) | −0.02; (−0.33–0.29) | −0.38; (−0.74–−0.02) | |
| LAmax | All | 16.36 | 16.27 | 16.86 | 0.29 | 0.49 | 0.12 | 0.007; (0.00–0.06) | −0.04; (−0.28–0.20) | −0.20; (−0.46–0.05) |
| Elite | 18.94 | 18.6 | 17.39 | 0.24 | 0.34 | 0.1 | 0.07; (0.00–0.39) | 0.33; (−0.35–1.00) | 0.61; (−0.12–1.33) | |
| Sub-elite | 17.61 | 17.86 | 17.83 | 0.93 | 0.7 | 0.94 | 0.001; (0.00–0.04) | −0.09; (−0.53–0.36) | −0.02; (−0.45–0.42) | |
| Physically active | 16 | 15.22 | 15 | 0.06 | 0.85 | 0.02/0.06 * | 0.03; (0.00–0.16) | −0.03; (−0.34–0.28) | −0.42; (−0.78–−0.06) | |
| ACC | All | 14.01 | 13.92 | 14.77 | 0.07 | 0.49 | 0.02/0.06 * | 0.02; (0.00–0.09) | −0.06; (−0.30–0.17) | −0.30; (−0.56–−0.04) |
| Elite | 16.63 | 16.37 | 15.77 | 0.59 | 0.5 | 0.33 | 0.03; (0.00–0.27) | 0.23; (−0.44–0.90) | 0.36; (−0.36–1.07) | |
| Sub-elite | 15.24 | 15.64 | 15.85 | 0.7 | 0.43 | 0.56 | 0.007 (0.00–0.11) | −0.18; (−0.63–0.27) | −0.13 (−0.56–0.31) | |
| Physically active | 13.72 | 12.88 | 12.56 | 0.02/0.03 * | 0.99 | 0.01/0.03 * | 0.04; (0.00–0.19) | −0.00; (−0.31–0.31) | −0.48; (−0.84–−0.12) | |
| DCC | All | 7.88 | 8.15 | 7.86 | 0.54 | 0.49 | 0.52 | 0.004; (0.00–0.04) | −0.08; (−0.32–0.16) | 0.08; (−0.17–0.34) |
| Elite | 8.83 | 9.31 | 7.73 | 0.25 | 0.96 | 0.11 | 0.07; (0.00–0.38) | 0.02; (−0.65–0.69) | 0.59; (−0.14–1.31) | |
| Sub-elite | 8.18 | 8.83 | 8.38 | 0.43 | 0.33 | 0.64 | 0.02; (0.00–0.15) | −2.22; (−0.67–0.23) | 0.10; (−0.33–0.54) | |
| Physically active | 7.52 | 7.54 | 7.57 | 0.99 | 0.97 | 0.92 | 0; (0.00–0.00) | −0.01; (−0.32–0.30) | 0.02; (−0.34–0.37) | |
| LArest | LAaftwarm | LABETWEEN | LAaft2WAnt | LA3′ | LA6′ | LA9′ | LA20′ | LA30′ | LAmax | ACC | DCC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LArest | 1.00 | 0.28 | 0.16 | 0.10 | 0.16 | 0.15 | 0.06 | 0.06 | 0.08 | 0.16 | 0.08 | |
| LAaftwarm | 0.28 | 1.00 | 0.38 | 0.33 | 0.33 | 0.32 | 0.26 | 0.26 | 0.26 | 0.33 | 0.24 | 0.07 |
| LABETWEEN | 0.16 | 0.38 | 1.00 | 0.78 | 0.80 | 0.75 | 0.72 | 0.61 | 0.50 | 0.77 | 0.72 | 0.30 |
| LAaft2WAnt | 0.10 | 0.33 | 0.78 | 1.00 | 0.75 | 0.71 | 0.67 | 0.53 | 0.43 | 0.72 | 0.69 | 0.32 |
| LA3′ | 0.16 | 0.33 | 0.80 | 0.75 | 1.00 | 0.92 | 0.86 | 0.73 | 0.60 | 0.95 | 0.90 | 0.39 |
| LA6′ | 0.15 | 0.32 | 0.75 | 0.71 | 0.92 | 1.00 | 0.93 | 0.81 | 0.69 | 0.97 | 0.92 | 0.30 |
| LA9′ | 0.06 | 0.26 | 0.72 | 0.67 | 0.86 | 0.93 | 1.00 | 0.85 | 0.75 | 0.93 | 0.91 | 0.17 |
| LA20′ | 0.06 | 0.26 | 0.61 | 0.53 | 0.73 | 0.81 | 0.85 | 1.00 | 0.93 | 0.79 | 0.77 | −0.22 |
| LA30′ | 0.08 | 0.26 | 0.50 | 0.43 | 0.60 | 0.69 | 0.75 | 0.93 | 1.00 | 0.67 | 0.64 | |
| LAmax | 0.16 | 0.33 | 0.77 | 0.72 | 0.95 | 0.97 | 0.93 | 0.79 | 0.67 | 1.00 | ||
| ACC | 0.24 | 0.72 | 0.69 | 0.90 | 0.92 | 0.91 | 0.77 | 0.64 | 1.00 | 0.33 | ||
| DCC | 0.08 | 0.07 | 0.30 | 0.32 | 0.39 | 0.30 | 0.17 | −0.22 | 0.33 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maculewicz, E.; Bojarczuk, A.; Mastalerz, A.; Johne, M.; Mróz, A.; Garbacz, A.; Stastny, P. Associations Between Genetic Variants in MCT2 (rs3763980, rs995343, rs3763979) and MCT4 (rs11323780) with Blood Lactate Kinetics Before and After Supramaximal Exercise. Int. J. Mol. Sci. 2025, 26, 7865. https://doi.org/10.3390/ijms26167865
Maculewicz E, Bojarczuk A, Mastalerz A, Johne M, Mróz A, Garbacz A, Stastny P. Associations Between Genetic Variants in MCT2 (rs3763980, rs995343, rs3763979) and MCT4 (rs11323780) with Blood Lactate Kinetics Before and After Supramaximal Exercise. International Journal of Molecular Sciences. 2025; 26(16):7865. https://doi.org/10.3390/ijms26167865
Chicago/Turabian StyleMaculewicz, Ewelina, Aleksandra Bojarczuk, Andrzej Mastalerz, Monika Johne, Anna Mróz, Aleksandra Garbacz, and Petr Stastny. 2025. "Associations Between Genetic Variants in MCT2 (rs3763980, rs995343, rs3763979) and MCT4 (rs11323780) with Blood Lactate Kinetics Before and After Supramaximal Exercise" International Journal of Molecular Sciences 26, no. 16: 7865. https://doi.org/10.3390/ijms26167865
APA StyleMaculewicz, E., Bojarczuk, A., Mastalerz, A., Johne, M., Mróz, A., Garbacz, A., & Stastny, P. (2025). Associations Between Genetic Variants in MCT2 (rs3763980, rs995343, rs3763979) and MCT4 (rs11323780) with Blood Lactate Kinetics Before and After Supramaximal Exercise. International Journal of Molecular Sciences, 26(16), 7865. https://doi.org/10.3390/ijms26167865









