Full-Length Transcriptome Analysis of Alternative Splicing and Polyadenylation in the Molecular Regulation of Labor Division in Apis cerana cerana
Abstract
1. Introduction
2. Results
2.1. AS Functional Annotation
2.2. Differential AS Events
2.3. Key DASEs Related to Development
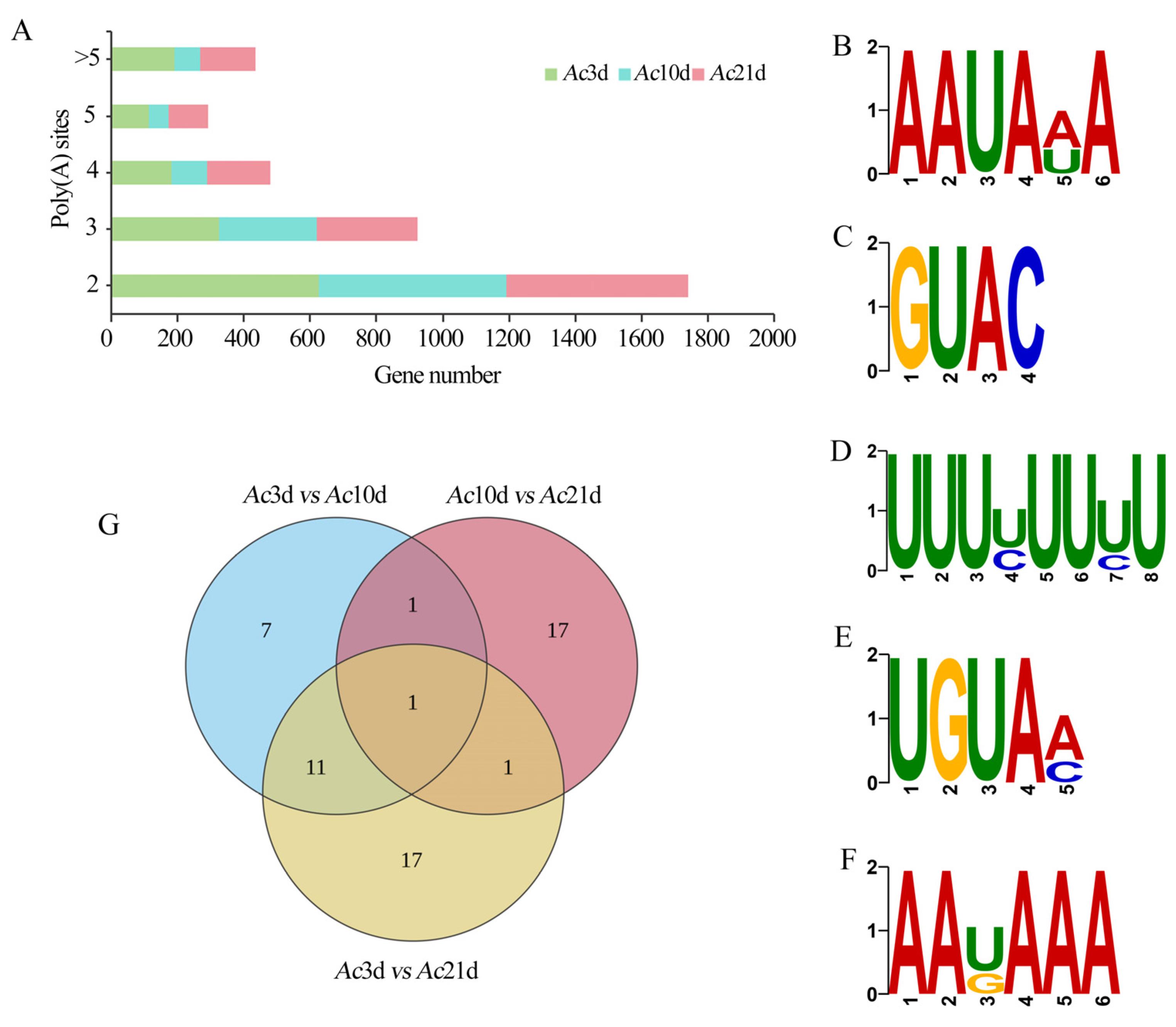
2.4. Profiling of Alternative Polyadenylation Sites
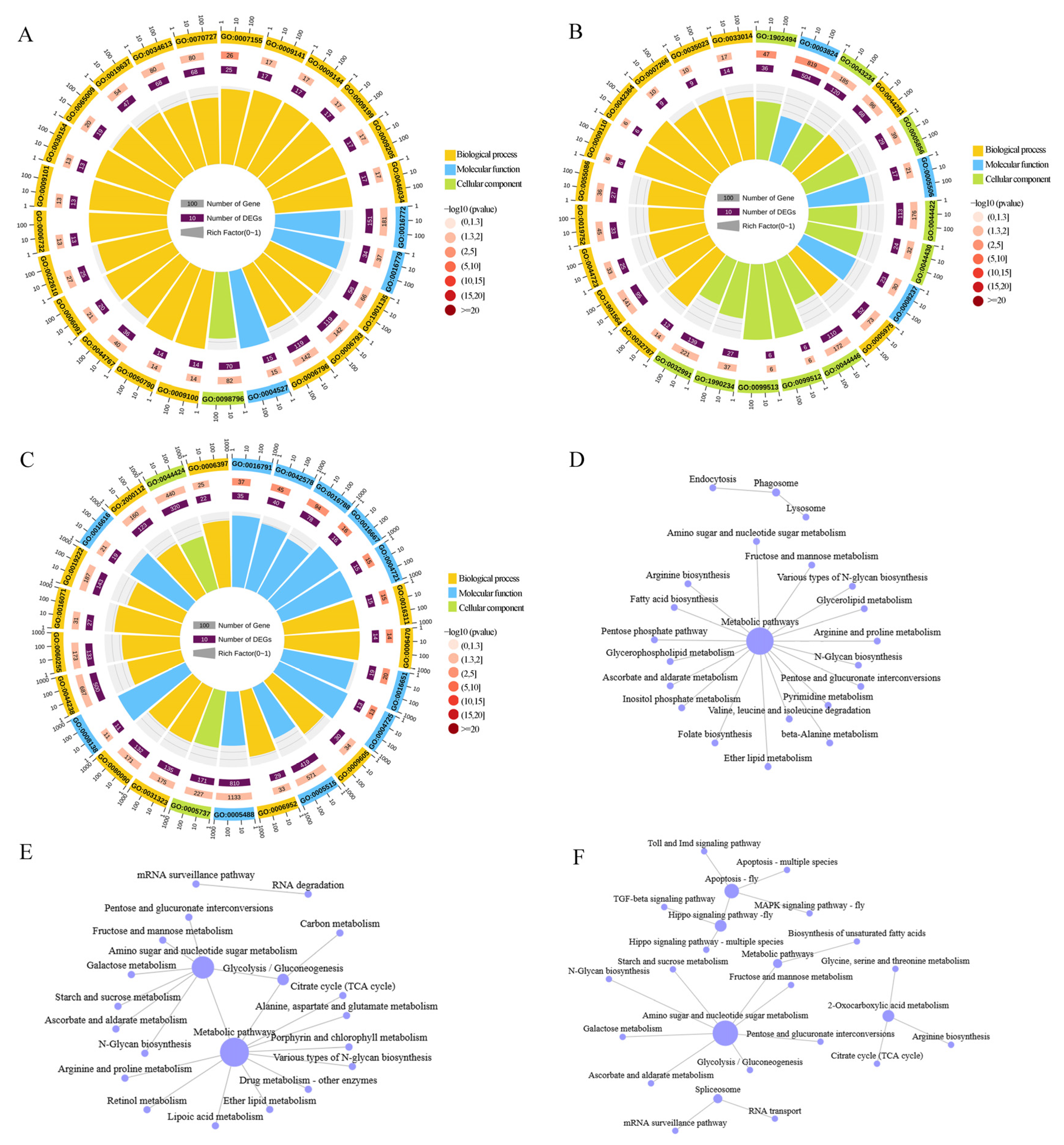
2.5. APA Functional Annotation
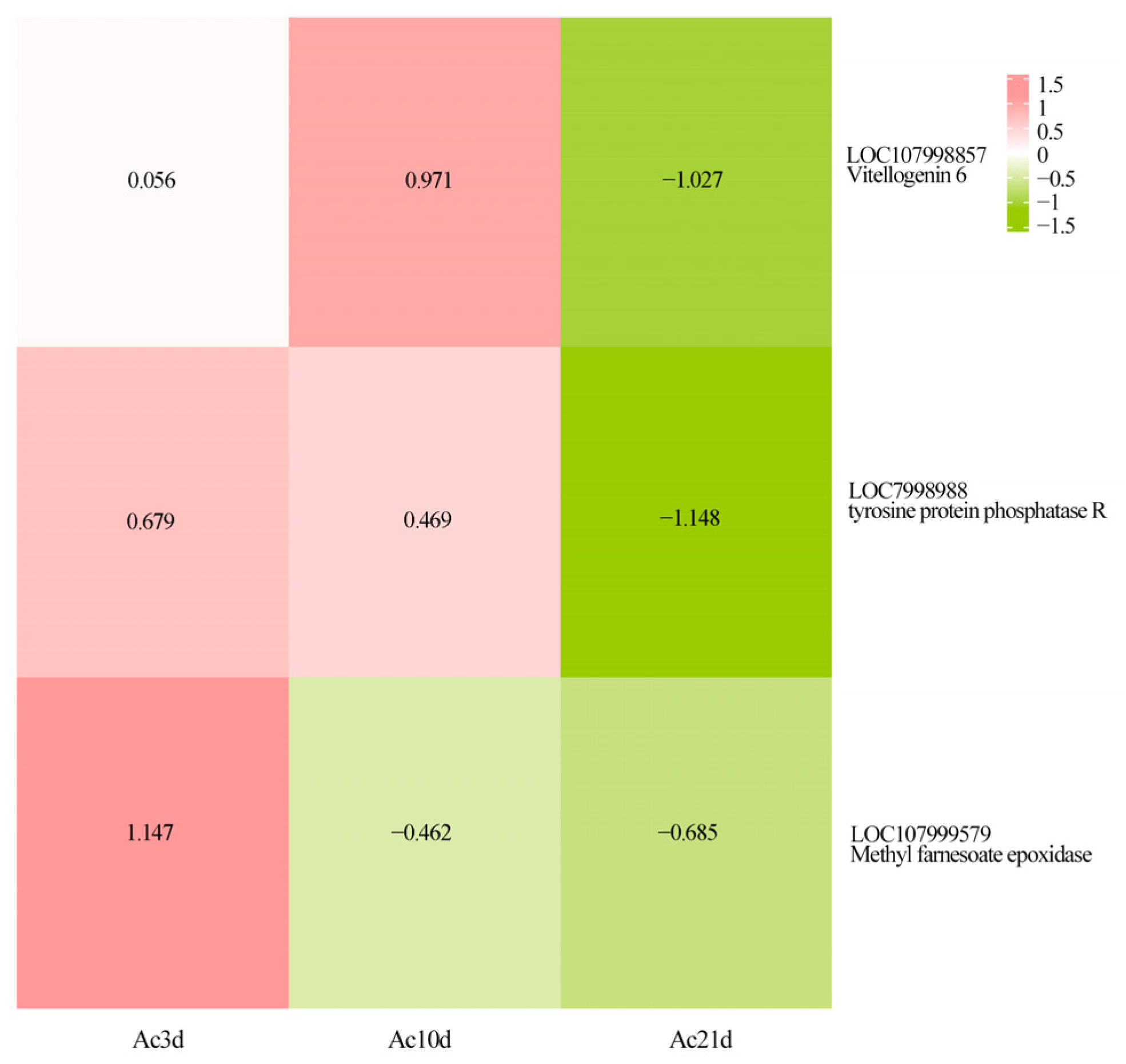
2.6. Key DAPAGs Related to Development
3. Discussion
4. Materials and Methods
4.1. Honey Bees
4.2. Full-Length Transcriptome Data Source
4.3. Alternative Splicing Isoform Analysis
4.4. Bee Poly(A) Analysis
4.5. GO Enrichment and KEGG Annotation Analysis
4.6. RNA Extraction and RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, B.R. Division of labor in honeybees: Form, function, and proximate mechanisms. Behav. Ecol. Sociobiol. 2010, 64, 305–316. [Google Scholar] [CrossRef]
- Robinson, G.E.; Grozinger, C.M.; Whitfield, C.W. Sociogenomics: Social life in molecular terms. Nat. Rev. Genet. 2005, 6, 257–270. [Google Scholar] [CrossRef]
- Schilcher, F.; Scheiner, R. New insight into molecular mechanisms underlying division of labor in honeybees. Curr. Opin. Insect Sci. 2023, 59, 101080. [Google Scholar] [CrossRef]
- Ma, R.; Rangel, J.; Grozinger, C.M. Honey bee (Apis mellifera) larval pheromones may regulate gene expression related to foraging task specialization. BMC Genom. 2019, 20, 592. [Google Scholar] [CrossRef]
- Whitfield, C.W.; Cziko, A.M.; Robinson, G.E. Gene expression profiles in the brain predict behavior in individual honey bees. Science 2003, 302, 296–299. [Google Scholar] [CrossRef]
- Mu, X.; Zhang, Z.; Liu, Q.; Ma, J.; Qin, Y.; Lang, H.; Zhang, Y.; Zhang, N.; Guo, Q.; Zhang, P.; et al. Single-nucleus and spatial transcriptomics identify brain landscape of gene regulatory networks associated with behavioral maturation in honeybees. Nat. Commun 2025, 16, 3343. [Google Scholar] [CrossRef]
- O’Connell, D.P.; Baker, B.M.; Atauri, D.; Jones, J.C. Increasing temperature and time in glasshouses increases honey bee activity and affects internal brood conditions. J. Insect Physiol. 2024, 155, 104635. [Google Scholar] [CrossRef] [PubMed]
- Winston, M.L.; Mitchell, S.R.; Punnett, E.N. Feasibility of package honey bee (Hymenoptera: Apidae) production in Southwestern British Columbia, Canada. J. Econ. Entomol. 1985, 5, 1037–1041. [Google Scholar] [CrossRef]
- Robinson, G.E.; Huang, Z.Y. Colony integration in honey bees: Genetic, endocrine and social control of division of labor. Apidologie 1998, 29, 159–170. [Google Scholar] [CrossRef]
- Schulz, D.J.; Huang, Z.Y.; Robinson, G.E. Effects of colony food shortage on behavioral development in honey bees. Behav. Ecol. Sociobiol. 1998, 42, 295–303. [Google Scholar] [CrossRef]
- Traynor, K.S.; Conte, Y.L.; Page, R.E. Age matters: Pheromone profiles of larvae differentially influence foraging behaviour in the honeybee, Apis mellifera. Anim. Behav. 2015, 99, 1–8. [Google Scholar] [CrossRef]
- Traynor, K.S.; Wang, Y.; Brent, C.S.; Amdam, G.; Page, J.R.E. Young and old honeybee (Apis mellifera) larvae differentially prime the developmental maturation of their caregivers. Anim. Behav. 2017, 124, 193–202. [Google Scholar] [CrossRef]
- Withers, G.S.; Fahrbach, S.E.; Robinson, G.E. Selective neuroanatomical plasticity and division of labour in the honeybee. Nature 1993, 364, 238–240. [Google Scholar] [CrossRef]
- Sullivan, J.P.; Fahrbach, S.E.; Harrison, J.F.; Capaldi, E.A.; Fewell, J.H.; Robinson, G.E. Juvenile hormone and division of labor in honey bee colonies: Effects of allatectomy on flight behavior and metabolism. J. Exp. Biol. 2003, 206, 2287–2296. [Google Scholar] [CrossRef]
- Harris, J.W.; Woodring, J. Effects of stress, age, season, and source colony on levels of octopamine, dopamine and serotonin in the honey bee (Apis mellifera L.) brain. J. Insect Physiol. 1992, 38, 29–35. [Google Scholar] [CrossRef]
- Wagener-Hulme, C.; Kuehn, J.C.; Schulz, D.J.; Robinson, G.E. Biogenic amines and division of labor in honey bee colonies. J. Comp. Physiol. A 1999, 184, 471–479. [Google Scholar] [CrossRef]
- Liu, F.; Shi, T.F.; Qi, L. Progress in the regulating factors of behavioral transition between nurses and foragers in honeybees. Acta Entomol. Sin. 2018, 61, 1350–1355. [Google Scholar] [CrossRef]
- Kuang, B.Y.; Kuang, H.O. Bee Biology; Yunnan Science and Technology Press: Kunming, China, 2002; pp. 47–48. [Google Scholar]
- Hu, D.B.; Xiao, S.; Wang, Y.; Hua, H.X. Notch is an alternative splicing gene in brown planthopper, Nilaparvata lugens. Arch. Insect Biochem. Physiol. 2022, 110, e21894. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, X.; Liu, C.; Wang, Z.; Lei, W.; Li, Q.; Zhao, Y.; Wang, X. Dynamic Alternative Polyadenylation during Litopenaeus Vannamei Metamorphosis Development. Genes 2024, 15, 837. [Google Scholar] [CrossRef] [PubMed]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Nygard, A.B.; Cirera, S.; Gilchrist, M.J.; Gorodkin, J.; Jørgensen, C.B.; Fredholm, M. A study of alternative splicing in the pig. BMC Res. Notes 2010, 3, 123. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef]
- Chen, H.Z.; Fan, X.X.; Fan, Y.C.; Wang, J.; Zhu, Z.W. Analysis of alternative splicing and alternative polyadenylation of Nosema ceranae genes. Mycosystema 2021, 40, 161–173. [Google Scholar] [CrossRef]
- Sun, M.H.; Liu, J.M.; Wang, S.Y.; Zhu, L.R.; Wang, Z.X.; Ye, Y.; Qian, J.; Gu, X.; Xu, X.; Chen, D.; et al. Unraveling the complexity of transcriptome in Ascosphaera apis spore: Based on third-generation and next-generation sequencing. Acta Microbiol. Sin. 2022, 62, 2981–2994. [Google Scholar]
- Fan, X.X.; Wang, S.Y.; Zhu, L.R.; Jing, X.; Guo, S.J.; Niu, Q.; Jiang, H.; Xu, X.; Chen, D.; Fu, Z.; et al. Identification of endocytosis-associated genes, and full-length transcripts, in Apis mellifera ligustica. J. Appl. Entomol. 2023, 60, 825–834. [Google Scholar]
- Hu, X.F.; Jin, M.J.; Gong, Z.X.; Lin, Z.Z.; Zhang, L.Z.; Zeng, Z.J.; Wang, Z.L. Full-Length Transcriptome Profile of Apis cerana Revealed by Nanopore Sequencing. Int. J. Mol. Sci. 2024, 25, 10833. [Google Scholar] [CrossRef]
- Elkon, R.; Ugalde, A.P.; Agami, R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat. Rev. Genet. 2013, 14, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Smibert, P.; Miura, P.; Westholm, J.O.; Shenker, S.; May, G.; Duff, M.O.; ZHANG, D.; Eads, B.D.; Carlson, J.; Brown, J.B.; et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012, 1, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Kasowitz, S.D.; Ma, J.; Anderson, S.J.; Leu, N.A.; Xu, Y.; Gregory, B.D.; Schultz, R.M.; Wang, P.J.; Yan, W. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018, 14, e1007412. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; An, B.; Liang, M.; Duan, X.; Du, L.; Cai, W.; Zhu, B.; Gao, X.; Chen, Y.; Xu, L.; et al. PacBio Single-Molecule Long-Read Sequencing Provides New Light on the Complexity of Full-Length Transcripts in Cattle. Front. Genet. 2021, 12, 664974. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Hamilton, M.; Jacobi, J.L.; Ngam, P.; Devitt, N.; Schilkey, F.; Ben-Hur, A.; Reddy, A.S.N. A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 2016, 7, 11706. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xiao, Y.; Li, Y.; Wang, X.; Qi, S.; Wang, Y.; Zhao, L.; Wang, K.; Peng, W.; Luo, G.-Z.; et al. RNA m6A Modification Functions in Larval Development and Caste Differentiation in Honeybee (Apis mellifera). Cell Rep. 2021, 34, 108580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, L.; Zhao, Y.; Wang, Y.; Chen, C.; Hu, Y.; Zhu, Y.; Sun, H.; Cheng, Y.; Sun, Q.; et al. Single-cell transcriptomic analysis of honeybee brains identifies vitellogenin as caste differentiation-related factor. iScience 2022, 25, 104643. [Google Scholar] [CrossRef]
- Harwood, G.; Amdam, G. Vitellogenin in the honey bee midgut. Apidologie 2021, 52, 837–847. [Google Scholar] [CrossRef]
- Guo, Q.; Zu, M.; Liu, D.; Yan, Y.; Yang, W.; Xu, K. Roles of Vitellogenin and Its Receptor Genes in Female Reproduction of the Cigarette Beetle, Lasioderma serricorne. Insects 2025, 16, 175. [Google Scholar] [CrossRef] [PubMed]
- Salmela, H.; Amdam, G.V.; Freitak, D. Transfer of immunity from mother to offspring is mediated via egg-yolk protein vitellogenin. PLoS Pathog. 2015, 11, e1005015. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Zhou, W.C.; Zhan, H.P.; Yu, Y.L.; He, X.J.; Wan, W.; Wei, X.P. Construction of the full-length transcriptome of the Chinese honey bee (Apis cerana cerana) workers under low temperature stress based on PacBio Iso-Seq. Acta Entomol. Sin. 2024, 67, 611–621. [Google Scholar] [CrossRef]
- Li, J.; Tracy, J.W.; Christensen, B.M. Relationship of hemolymph phenol oxidase and mosquito age in Aedes aegypti. J. Invertebr. Pathol. 1992, 60, 188–191. [Google Scholar] [CrossRef]
- Xue, C.B.; Chen, Q.X.; Wang, Q.; Ke, L.N.; Luo, W.K. Comparison on properties of polyphenoloxidase from Pieris rapae (L.) in different stages and instars. Acta Entomol. Sin. 2004, 47, 305–309. [Google Scholar] [CrossRef]
- Li, X.Z.; Liu, Y.H. Characterization of polyphenol oxidase from the fruit fly Bactrocera tau (Walker) (Diptera: Tephritidae). Acta Entomol. Sin. 2011, 54, 982–988. [Google Scholar] [CrossRef]
- Kanost, M.R.; Jiang, H.; Yu, X.Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef]
- Theopold, U.; Schmidt, O.; Söderhäll, K.; Dushay, M.S. Coagulation in arthropods: Defence, wound closure and healing. Trends Immunol. 2004, 25, 289–294. [Google Scholar] [CrossRef]
- Dudzic, J.P.; Kondo, S.; Ueda, R.; Bergman, C.M.; Lemaitre, B. Drosophila innate immunity: Regional and functional specialization of prophenoloxidases. BMC Biol. 2015, 13, 81. [Google Scholar] [CrossRef]
- Shin, M.; Chang, E.; Lee, D.; Kim, N.; Cho, B.; Cha, N.; Koranteng, F.; Song, J.-J.; Shim, J. Drosophila immune cells transport oxygen through PPO2 protein phase transition. Nature 2024, 631, 350–359. [Google Scholar] [CrossRef]
- Calderone, N.W. Proximate mechanisms of age polyethism in the honey bee, Apis mellifera L. Apidologie 1998, 29, 127–158. Apidologie 1998, 29, 127–158. [Google Scholar] [CrossRef]
- Johnson, B.R. Global information sampling in the honey bee. Naturwissenschaften 2008, 95, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.B. Within-nest temporal polyethism in the honey bee. Behav. Ecol. Sociobiol. 2008, 62, 777–784. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef] [PubMed]
- Alamancos, G.P.; Pagès, A.; Trincado, J.L.; Bellora, N.; Eyras, E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 2015, 21, 1521–1531. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, D.; Fan, Y.; Zhou, W.; Zhan, H.; Yu, Y.; Wei, X. Full-Length Transcriptome Analysis of Alternative Splicing and Polyadenylation in the Molecular Regulation of Labor Division in Apis cerana cerana. Int. J. Mol. Sci. 2025, 26, 7859. https://doi.org/10.3390/ijms26167859
Yao D, Fan Y, Zhou W, Zhan H, Yu Y, Wei X. Full-Length Transcriptome Analysis of Alternative Splicing and Polyadenylation in the Molecular Regulation of Labor Division in Apis cerana cerana. International Journal of Molecular Sciences. 2025; 26(16):7859. https://doi.org/10.3390/ijms26167859
Chicago/Turabian StyleYao, Dan, Yuanchan Fan, Wencai Zhou, Hongpin Zhan, Yinglong Yu, and Xiaoping Wei. 2025. "Full-Length Transcriptome Analysis of Alternative Splicing and Polyadenylation in the Molecular Regulation of Labor Division in Apis cerana cerana" International Journal of Molecular Sciences 26, no. 16: 7859. https://doi.org/10.3390/ijms26167859
APA StyleYao, D., Fan, Y., Zhou, W., Zhan, H., Yu, Y., & Wei, X. (2025). Full-Length Transcriptome Analysis of Alternative Splicing and Polyadenylation in the Molecular Regulation of Labor Division in Apis cerana cerana. International Journal of Molecular Sciences, 26(16), 7859. https://doi.org/10.3390/ijms26167859





