Association of PAX3 Gene Polymorphism with Three-Dimensional Nasal Root Morphology
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. Genotyping
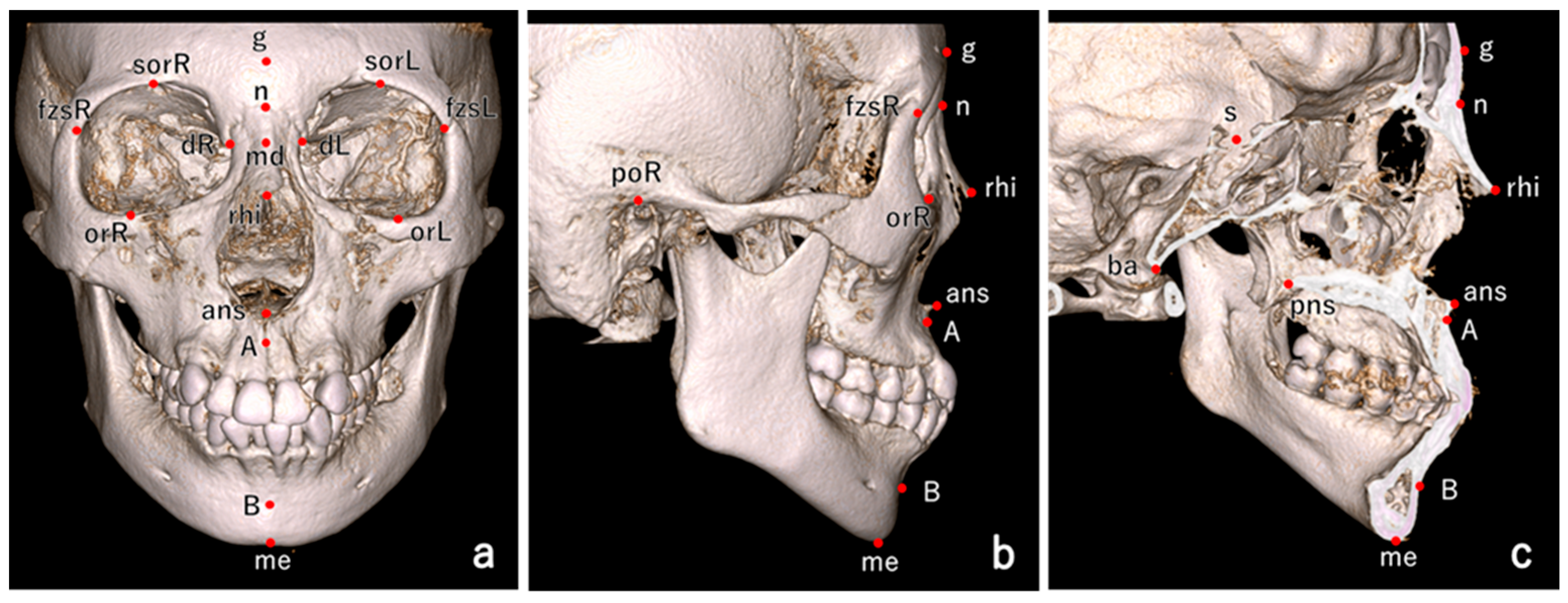
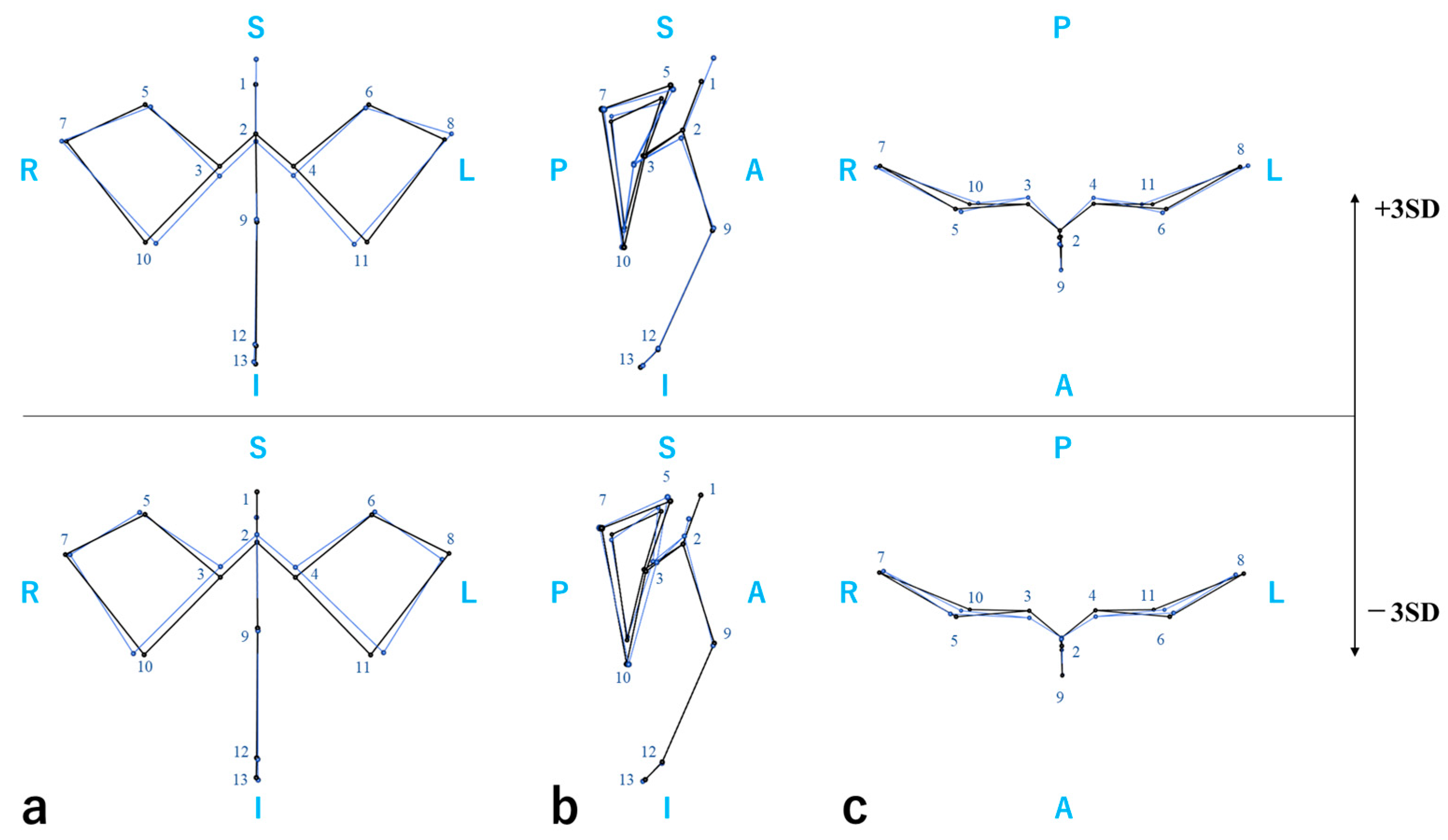
4.3. Craniofacial Measurements
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PAX3 | Paired box gene 3 |
| WS | Waardenburg syndrome |
| CDHS | Craniofacial-deafness-hand syndrome |
| GWAS | Genome-Wide Association Study |
| SNP | Single-nucleotide polymorphism |
| CBCT | Cone-beam computed tomography |
| MDCT | Multi-detector computed tomography |
| LD | Linkage disequilibrium |
| PCA | Principal component analysis |
| PC | Principal component |
| GPA | Generalized Procrustes analysis |
References
- Martínez-Abadías, N.; Esparza, M.; Sjøvold, T.; González-José, R.; Santos, M.; Hernández, M. Heritability of human cranial dimensions: Comparing the evolvability of different cranial regions. J. Anat. 2009, 214, 19–35. [Google Scholar] [CrossRef]
- van Wezel, N.A.; Bos, A.; Prahl, C. Expectations of treatment and satisfaction with dentofacial appearance in patients applying for orthodontic treatment. Am. J. Orthod. Dentofac. Orthop. 2015, 147, 698–703. [Google Scholar] [CrossRef]
- Harvati, K.; Weaver, T.D. Human cranial anatomy and the differential preservation of population history and climate signatures. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2006, 288, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Al-Obaidi, S.; Papageorgiou, S.N.; Saade, M.; Caradonna, K.M.; Kantarci, A.; Will, L.; Motro, M. Influence of genetic and environmental factors on transverse growth. Eur. J. Orthod. 2025, 47, cjaf003. [Google Scholar] [CrossRef] [PubMed]
- Petrides, G.; Clark, J.R.; Low, H.; Lovell, N.; Eviston, T.J. Three-dimensional scanners for soft-tissue facial assessment in clinical practice. J. Plast. Reconstr. Aesthetic Surg. 2021, 74, 605–614. [Google Scholar] [CrossRef]
- White, J.D.; Indencleef, K.; Naqvi, S.; Eller, R.J.; Hoskens, H.; Roosenboom, J.; Lee, M.K.; Li, J.; Mohammed, J.; Richmond, S.; et al. Insights into the genetic architecture of the human face. Nat. Genet. 2021, 53, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Cordero, D.R.; Brugmann, S.; Chu, Y.; Bajpai, R.; Jame, M.; Helms, J.A. Cranial neural crest cells on the move: Their roles in craniofacial development. Am. J. Med. Genet. A 2011, 155A, 270–279. [Google Scholar] [CrossRef]
- Nie, X.; Luukko, K.; Kettunen, P. FGF signalling in craniofacial development and developmental disorders. Oral Dis. 2006, 12, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Graf, D.; Malik, Z.; Hayano, S.; Mishina, Y. Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev. 2016, 27, 129–139. [Google Scholar] [CrossRef]
- Dworkin, S.; Boglev, Y.; Owens, H.; Goldie, S.J. The Role of Sonic Hedgehog in Craniofacial Patterning, Morphogenesis and Cranial Neural Crest Survival. J. Dev. Biol. 2016, 4, 24. [Google Scholar] [CrossRef]
- Reynolds, K.; Kumari, P.; Sepulveda, R.L.; Gu, R.; Ji, Y.; Kumar, S.; Zhou, C.J. Wnt signaling in orofacial clefts: Crosstalk, pathogenesis and models. Dis. Model. Mech. 2019, 12, dmm037051. [Google Scholar] [CrossRef]
- Barber, T.D.; Barber, M.C.; Cloutier, T.E.; Friedman, T.B. PAX3 gene structure, alternative splicing and evolution. Gene 1999, 237, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, C.T.; Hoth, C.F.; Amos, J.A.; da-Silva, E.O.; Milunsky, A. An exonic mutation in the HuP2 paired domain gene causes Waardenburg’s syndrome. Nature 1992, 355, 637–638. [Google Scholar] [CrossRef] [PubMed]
- Zlotogora, J.; Lerer, I.; Bar-David, S.; Ergaz, Z.; Abeliovich, D. Homozygosity for Waardenburg syndrome. Am. J. Hum. Genet. 1995, 56, 1173–1178. [Google Scholar]
- Gad, A.; Laurino, M.; Maravilla, K.R.; Matsushita, M.; Raskind, W.H. Sensorineural deafness, distinctive facial features, and abnormal cranial bones: A new variant of Waardenburg syndrome? Am. J. Med. Genet. A 2008, 146A, 1880–1885. [Google Scholar] [CrossRef]
- Wu, M.; Li, J.; Engleka, K.A.; Zhou, B.; Lu, M.M.; Plotkin, J.B.; Epstein, J.A. Persistent expression of Pax3 in the neural crest causes cleft palate and defective osteogenesis in mice. J. Clin. Investig. 2008, 118, 2076–2087. [Google Scholar] [CrossRef]
- Paternoster, L.; Zhurov, A.I.; Toma, A.M.; Kemp, J.P.; St Pourcain, B.; Timpson, N.J.; McMahon, G.; McArdle, W.; Ring, S.M.; Smith, G.D.; et al. Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am. J. Hum. Genet. 2012, 90, 478–485. [Google Scholar] [CrossRef]
- Liu, F.; van der Lijn, F.; Schurmann, C.; Zhu, G.; Chakravarty, M.M.; Hysi, P.G.; Wollstein, A.; Lao, O.; de Bruijne, M.; Ikram, M.A.; et al. A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 2012, 8, e1002932. [Google Scholar] [CrossRef]
- Qian, W.; Zhang, M.; Wan, K.; Xie, Y.; Du, S.; Li, J.; Mu, X.; Qiu, J.; Xue, X.; Zhuang, X.; et al. Genetic evidence for facial variation being a composite phenotype of cranial variation and facial soft tissue thickness. J. Genet. Genom. 2022, 49, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Sved, J.A. Linkage disequilibrium and its expectation in human populations. Twin Res. Hum. Genet. 2009, 12, 35–43. [Google Scholar] [CrossRef]
- Adhikari, K.; Fuentes-Guajardo, M.; Quinto-Sánchez, M.; Mendoza-Revilla, J.; Camilo Chacón-Duque, J.; Acuña-Alonzo, V.; Jaramillo, C.; Arias, W.; Lozano, R.B.; Pérez, G.M.; et al. A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat. Commun. 2016, 7, 11616. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Berisa, T.; Liu, J.Z.; Ségurel, L.; Tung, J.Y.; Hinds, D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016, 48, 709–717, Erratum in: Nat. Genet. 2016, 48, 1296. [Google Scholar] [CrossRef]
- Claes, P.; Roosenboom, J.; White, J.D.; Swigut, T.; Sero, D.; Li, J.; Lee, M.K.; Zaidi, A.; Mattern, B.C.; Liebowitz, C.; et al. Genome-wide mapping of global-to-local genetic effects on human facial shape. Nat. Genet. 2018, 50, 414–423. [Google Scholar] [CrossRef]
- Xiong, Z.; Dankova, G.; Howe, L.J.; Lee, M.K.; Hysi, P.G.; de Jong, M.A.; Zhu, G.; Adhikari, K.; Li, D.; Li, Y.; et al. Novel genetic loci affecting facial shape variation in humans. Elife 2019, 8, e49898. [Google Scholar] [CrossRef]
- Bonfante, B.; Faux, P.; Navarro, N.; Mendoza-Revilla, J.; Dubied, M.; Montillot, C.; Wentworth, E.; Poloni, L.; Varón-González, C.; Jones, P.; et al. A GWAS in Latin Americans identifies novel face shape loci, implicating VPS13B and a Denisovan introgressed region in facial variation. Sci. Adv. 2021, 7, eabc6160. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, S.; Du, S.; Qian, W.; Chen, J.; Qiao, L.; Yang, Y.; Tan, J.; Yuan, Z.; Peng, Q.; et al. Genetic variants underlying differences in facial morphology in East Asian and European populations. Nat. Genet. 2022, 54, 403–411. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Faux, P.; Delgado, M.E.; Bonfante, B.; Fuentes-Guajardo, M.; Mendoza-Revilla, J.; Chacón-Duque, J.C.; Hurtado, M.; Villegas, V.; et al. Automatic landmarking identifies new loci associated with face morphology and implicates Neanderthal introgression in human nasal shape. Commun. Biol. 2023, 6, 481. [Google Scholar] [CrossRef] [PubMed]
- Macía, J.; Solé, R.V.; Elena, S.F. The causes of epistasis in genetic networks. Evolution 2012, 66, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The long-range interaction landscape of gene promoters. Nature 2012, 489, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; De Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-Wide Association Studies. Nat. Rev. Methods Primer 2021, 1, 59. [Google Scholar] [CrossRef]
- Movassaghi, K.; Altobelli, D.E.; Zhou, H. Frontonasal suture expansion in the rabbit using titanium screws. J. Oral Maxillofac. Surg. 1995, 53, 1033–1042. [Google Scholar] [CrossRef]
- Wang, M.M.; Haveles, C.S.; Zukotynski, B.K.; Reid, R.R.; Lee, J.C. The 27 Facial Sutures: Timing and Clinical Consequences of Closure. Plast. Reconstr. Surg. 2022, 149, 701–720. [Google Scholar] [CrossRef]
- Roth, D.M.; Souter, K.; Graf, D. Craniofacial sutures: Signaling centres integrating mechanosensation, cell signaling, and cell differentiation. Eur. J. Cell Biol. 2022, 101, 151258. [Google Scholar] [CrossRef]
- Domaracki, M.; Stephan, C.N. Facial soft tissue thicknesses in Australian adult cadavers. J. Forensic Sci. 2006, 51, 5–10. [Google Scholar] [CrossRef]
- Piombino, P.; Esposito, E.; Committeri, U.; Barone, S.; Arena, A.; Cataldo, R.; Carraturo, E.; Vaira, L.A.; De Riu, G.; Mariniello, D.; et al. Facial soft tissue thickness measurement method and relationship with BMI, age and sex. J. Stomatol. Oral Maxillofac. Surg. 2023, 124, 101420. [Google Scholar] [CrossRef]
- Rajderkar, S.S.; Paraiso, K.; Amaral, M.L.; Kosicki, M.; Cook, L.E.; Darbellay, F.; Spurrell, C.H.; Osterwalder, M.; Zhu, Y.; Wu, H.; et al. Dynamic enhancer landscapes in human craniofacial development. Nat. Commun. 2024, 15, 2030. [Google Scholar] [CrossRef]
- Saether, L.; Van Belle, W.; Laeng, B.; Brennen, T.; Øvervoll, M. Anchoring gaze when categorizing faces’ sex: Evidence from eye-tracking data. Vis. Res. 2009, 49, 2870–2880. [Google Scholar] [CrossRef]
- Hui, L.; DelMonte, T.; Ranade, K. Genotyping using the TaqMan assay. Curr. Protoc. Hum. Genet. 2008, 56, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Tolentino, E.S.; Yamashita, F.C.; de Albuquerque, S.; Walewski, L.A.; Iwaki, L.C.V.; Takeshita, W.M.; Silva, M.C. Reliability and accuracy of linear measurements in cone-beam computed tomography using different software programs and voxel sizes. J. Conserv. Dent. 2018, 21, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Ghamri, M.; Dritsas, K.; Probst, J.; Jäggi, M.; Psomiadis, S.; Schulze, R.; Verna, C.; Katsaros, C.; Halazonetis, D.; Gkantidis, N. Accuracy of facial skeletal surfaces segmented from CT and CBCT radiographs. Sci. Rep. 2023, 13, 21002. [Google Scholar] [CrossRef]
- Tsuboi, A.; Koizumi, S.; Takahashi, M.; Hikita, Y.; Yamaguchi, T. The Role of Mandibular Thickness in Determining Anteroposterior Skeletal Relationships. Dent. J. 2024, 13, 3. [Google Scholar] [CrossRef]
- Okumura, Y.; Koizumi, S.; Suginouchi, Y.; Hikita, Y.; Kim, Y.-I.; Adel, M.; Nadim, M.; Yamaguchi, T. Chin morphology in relation to the skeletal pattern, age, gender, and ethnicity. Appl. Sci. 2022, 12, 12717. [Google Scholar] [CrossRef]
- Caple, J.; Stephan, C.N. A standardized nomenclature for craniofacial and facial anthropometry. Int. J. Leg. Med. 2016, 130, 863–879. [Google Scholar] [CrossRef]
- Adams, G.L.; Gansky, S.A.; Miller, A.J.; Harrell, W.E., Jr.; Hatcher, D.C. Comparison between traditional 2-dimensional cephalometry and a 3-dimensional approach on human dry skulls. Am. J. Orthod. Dentofac. Orthop. 2004, 126, 397–409. [Google Scholar] [CrossRef]
- Shin, S.M.; Kim, Y.M.; Kim, N.R.; Choi, Y.S.; Park, S.B.; Kim, Y.I. Statistical shape analysis-based determination of optimal midsagittal reference plane for evaluation of facial asymmetry. Am. J. Orthod. Dentofac. Orthop. 2016, 150, 252–260. [Google Scholar] [CrossRef]
- Harris, E.F.; Smith, R.N. Accounting for measurement error: A critical but often overlooked process. Arch. Oral Biol. 2009, 54 (Suppl. S1), S107–S117. [Google Scholar] [CrossRef]
- van der Heijden, P.; Korsten-Meijer, A.G.; van der Laan, B.F.; Wit, H.P.; Goorhuis-Brouwer, S.M. Nasal growth and maturation age in adolescents: A systematic review. Arch. Otolaryngol. Head. Neck Surg. 2008, 134, 1288–1293. [Google Scholar] [CrossRef]
- Al-Taai, N.; Persson, M.; Ransjö, M.; Levring Jäghagen, E.; Fors, R.; Westerlund, A. Craniofacial changes from 13 to 62 years of age. Eur. J. Orthod. 2022, 44, 556–565. [Google Scholar] [CrossRef]
| Male | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category | Measurements | Japanese (n = 56) | Korean (n = 29) | Egyptian (n = 63) | Japanese vs. Korean | Japanese vs. Egyptian | Korean vs. Egyptian | |||
| Mean | SD | Mean | SD | Mean | SD | p-Value | p-Value | p-Value | ||
| Craniofacial Morphology | n-me (mm) | 126.03 | 5.92 | 128.73 | 5.96 | 122.15 | 6.48 | 0.14 | <0.01 | <0.01 |
| n-ans (mm) | 56.25 | 3.08 | 57.48 | 2.23 | 55.13 | 5.92 | 0.14 | 0.01 | <0.01 | |
| a-ba (mm) | 90.95 | 5.24 | 91.19 | 4.44 | 94.52 | 4.53 | 0.98 | <0.01 | 0.01 | |
| n-ba (mm) | 102.54 | 4.53 | 104.06 | 5.17 | 104.46 | 4.26 | 0.32 | 0.06 | 0.92 | |
| n-s (mm) | 66.47 | 2.79 | 68.37 | 3.29 | 69.64 | 2.82 | 0.01 | <0.01 | 0.13 | |
| poR-poL (mm) | 124.60 | 5.61 | 127.86 | 4.18 | 117.53 | 6.15 | 0.03 | <0.01 | <0.01 | |
| sorR-sorL (mm) | 59.54 | 3.29 | 62.51 | 2.72 | 56.98 | 3.98 | <0.01 | <0.01 | <0.01 | |
| fzsR-fzsL (mm) | 104.84 | 4.94 | 108.14 | 3.74 | 105.44 | 4.81 | 0.01 | 0.99 | 0.02 | |
| orR-orL (mm) | 60.44 | 3.76 | 63.21 | 3.00 | 57.06 | 4.20 | 0.02 | <0.01 | <0.01 | |
| dR-dL (mm) | 19.36 | 1.99 | 20.44 | 2.07 | 20.07 | 2.29 | 0.16 | 0.40 | 0.99 | |
| n-g (mm) | 11.94 | 2.13 | 11.72 | 2.39 | 10.12 | 2.21 | 0.90 | <0.01 | <0.01 | |
| n-rhi (mm) | 26.28 | 3.96 | 28.35 | 2.37 | 24.23 | 3.18 | 0.02 | <0.01 | <0.01 | |
| n-md (mm) | 11.53 | 1.98 | 12.04 | 1.76 | 12.75 | 1.63 | 0.43 | <0.01 | 0.18 | |
| Nasal Root Morphology | g-n-rhi (°) | 138.54 | 7.95 | 139.58 | 5.89 | 126.38 | 9.01 | 0.99 | <0.01 | <0.01 |
| md-n-g (°) | 150.69 | 8.36 | 150.70 | 7.70 | 142.39 | 7.20 | 0.99 | <0.01 | <0.01 | |
| s-n-g (°) | 113.43 | 5.83 | 112.76 | 5.11 | 116.47 | 6.27 | 0.87 | 0.02 | 0.02 | |
| s-n-rhi (°) | 107.95 | 6.66 | 107.58 | 5.48 | 117.05 | 6.57 | 0.97 | <0.01 | <0.01 | |
| s-n-md (°) | 37.45 | 6.75 | 38.42 | 6.57 | 26.13 | 6.16 | 0.79 | <0.01 | <0.01 | |
| dR-n-dL (°) | 80.71 | 10.88 | 81.06 | 8.60 | 76.55 | 7.11 | 0.98 | 0.03 | 0.07 | |
| dR-s-dL (°) | 18.97 | 2.07 | 19.47 | 1.90 | 19.44 | 2.33 | 0.57 | 0.46 | 0.99 | |
| n-md/dR-dL ratio | 0.60 | 0.11 | 0.59 | 0.09 | 0.64 | 0.08 | 0.93 | 0.09 | 0.09 | |
| Female | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category | Measurements | Japanese (n = 145) | Korean (n = 45) | Egyptian (n = 79) | Japanese vs. Korean | Japanese vs. Egyptian | Korean vs. Egyptian | |||
| Mean | SD | Mean | SD | Mean | SD | p-Value | p-Value | p-Value | ||
| Craniofacial Morphology | n-me (mm) | 118.63 | 5.64 | 119.83 | 5.35 | 114.70 | 5.65 | 0.42 | <0.01 | <0.01 |
| n-ans (mm) | 52.12 | 2.46 | 53.58 | 2.28 | 51.61 | 4.58 | 0.01 | 0.04 | <0.01 | |
| a-ba (mm) | 85.86 | 4.52 | 86.26 | 4.42 | 90.37 | 5.08 | 0.87 | <0.01 | <0.01 | |
| n-ba (mm) | 96.53 | 4.13 | 98.10 | 3.99 | 99.29 | 4.19 | 0.07 | <0.01 | 0.27 | |
| n-s (mm) | 62.64 | 2.74 | 64.04 | 2.93 | 66.14 | 2.95 | 0.01 | <0.01 | <0.01 | |
| poR-poL (mm) | 117.18 | 4.84 | 120.53 | 4.87 | 110.59 | 5.08 | <0.01 | <0.01 | <0.01 | |
| sorR-sorL (mm) | 57.69 | 2.80 | 59.13 | 2.76 | 55.23 | 3.36 | 0.01 | <0.01 | <0.01 | |
| fzsR-fzsL (mm) | 99.88 | 3.53 | 101.26 | 3.74 | 101.77 | 4.29 | 0.06 | <0.01 | 0.99 | |
| orR-orL (mm) | 57.91 | 3.59 | 59.19 | 2.31 | 54.68 | 3.84 | 0.15 | <0.01 | <0.01 | |
| dR-dL (mm) | 18.59 | 1.86 | 19.26 | 2.02 | 19.49 | 1.77 | 0.29 | 0.01 | 0.99 | |
| n-g (mm) | 13.96 | 3.14 | 13.23 | 2.42 | 11.80 | 3.26 | 0.90 | <0.01 | 0.02 | |
| n-rhi (mm) | 24.95 | 2.85 | 26.98 | 2.82 | 24.08 | 3.39 | <0.01 | 0.10 | <0.01 | |
| n-md (mm) | 10.43 | 1.48 | 10.68 | 1.62 | 11.63 | 1.66 | 0.61 | <0.01 | <0.01 | |
| Nasal Root Morphology | g-n-rhi (°) | 147.84 | 5.11 | 149.28 | 5.60 | 141.36 | 5.79 | 0.26 | <0.01 | <0.01 |
| md-n-g (°) | 147.00 | 6.20 | 147.60 | 7.71 | 136.07 | 8.63 | 0.88 | <0.01 | <0.01 | |
| s-n-g (°) | 106.16 | 4.40 | 104.93 | 5.83 | 105.24 | 5.46 | 0.32 | 0.38 | 0.94 | |
| s-n-rhi (°) | 105.93 | 5.19 | 105.73 | 5.70 | 113.26 | 5.16 | 0.97 | <0.01 | <0.01 | |
| s-n-md (°) | 41.02 | 5.29 | 43.03 | 5.49 | 31.03 | 6.90 | 0.11 | <0.01 | <0.01 | |
| dR-n-dL (°) | 83.75 | 8.13 | 84.43 | 8.33 | 80.38 | 8.08 | 0.88 | 0.01 | 0.02 | |
| dR-s-dL (°) | 19.09 | 1.94 | 19.26 | 2.29 | 19.52 | 1.90 | 0.99 | 0.71 | 0.74 | |
| n-md/dR-dL ratio | 0.56 | 0.08 | 0.56 | 0.08 | 0.60 | 0.09 | 0.89 | 0.01 | 0.02 | |
| rs Number | Chr: Position | Location | Alleles | Derived Allele Frequency | LD Coefficients (D’/r2) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ancestral | Derived | Japanese | Korean | Egyptian | Japanese | Korean | Egyptian | |||
| rs9288572 | chr2:222097644 | Intergenic | G | A | 0.572 | 0.561 | 0.197 | 0.181/0.019 | 0.019/0.028 | 0.063/0.001 |
| rs7559271 | chr2:222203567 | Intronic | A | G | 0.701 | 0.649 | 0.532 | |||
| Haplotype | Haplotype Frequency | ||
|---|---|---|---|
| Japanese | Korean | Egyptian | |
| hAG | 0.419 | 0.384 | 0.108 |
| hAA | 0.153 | 0.177 | 0.089 |
| hGG | 0.283 | 0.265 | 0.424 |
| hGA | 0.145 | 0.174 | 0.379 |
| Measurements | Japanese (n = 201) | Korean (n = 74) | Egyptian (n = 142) | Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SD | p-Value | B | SD | p-Value | B | SD | p-Value | B | SD | p-Value | |
| rs9288572 | ||||||||||||
| g-n-rhi | 1.090 | 0.606 | 7.5 × 10−2 | 0.426 | 0.894 | 6.4 × 10−1 | 0.950 | 1.12 | 4.0 × 10−1 | 0.892 | 0.458 | 5.1 × 10−2 |
| md-n-g | −1.621 | 0.684 | 1.9 × 10−2 | 0.616 | 1.197 | 6.1 × 10−1 | −0.470 | 1.216 | 7.0 × 10−1 | −0.955 | 0.534 | 7.4 × 10−2 |
| s-n-g | −0.784 | 0.485 | 1.1 × 10−1 | 0.833 | 0.859 | 3.4 × 10−1 | −0.010 | 0.884 | 9.9 × 10−1 | −0.322 | 0.381 | 4.0 × 10−1 |
| s-n-rhi | −0.260 | 0.571 | 6.5 × 10−1 | −1.270 | 0.859 | 1.4 × 10−1 | −0.992 | 0.875 | 2.6 × 10−1 | −0.666 | 0.418 | 1.1 × 10−1 |
| s-n-md | −0.799 | 0.576 | 1.7 × 10−1 | −0.242 | 0.923 | 7.9 × 10−1 | −0.365 | 0.997 | 7.2 × 10−1 | −0.589 | 0.439 | 1.8 × 10−1 |
| dR-n-dL | 0.468 | 0.908 | 6.1 × 10−1 | −1.265 | 1.304 | 3.4 × 10−1 | 1.560 | 1.15 | 1.8 × 10−1 | 0.389 | 0.625 | 5.3 × 10−1 |
| dR-s-dL | −0.111 | 0.199 | 5.8 × 10−1 | −0.469 | 0.329 | 1.6 × 10−1 | 0.288 | 0.318 | 3.7 × 10−1 | −0.097 | 0.150 | 5.2 × 10−1 |
| n-md/dR-dL | −0.003 | 0.009 | 7.3 × 10−1 | 0.014 | 0.013 | 3.0 × 10−1 | −0.017 | 0.013 | 1.8 × 10−1 | −0.002 | 0.006 | 7.2 × 10−1 |
| rs7559271 | ||||||||||||
| g-n-rhi | 0.258 | 0.675 | 7.0 × 10−1 | −0.440 | 0.994 | 6.6 × 10−1 | 0.720 | 0.815 | 3.8 × 10−1 | 0.256 | 0.461 | 5.8 × 10−1 |
| md-n-g | −1.878 | 0.758 | 1.4 × 10−2 | 1.660 | 1.328 | 2.2 × 10−1 | −2.469 | 0.863 | 5.0 × 10−3 | −1.546 | 0.523 | 3.1 × 10−3 |
| s-n-g | −1.343 | 0.533 | 1.3 × 10−2 | 1.930 | 0.941 | 4.4 × 10−2 | −1.941 | 0.624 | 2.0 × 10−3 | −1.044 | 0.372 | 5.0 × 10−3 |
| s-n-rhi | 1.090 | 0.629 | 8.4 × 10−2 | −1.469 | 0.963 | 1.3 × 10−1 | 1.214 | 0.636 | 5.8 × 10−2 | 0.686 | 0.406 | 9.1 × 10−2 |
| s-n-md | −0.519 | 0.641 | 4.2 × 10−1 | −0.354 | 1.032 | 7.3 × 10−1 | −0.504 | 0.727 | 4.9 × 10−1 | −0.484 | 0.436 | 2.7 × 10−1 |
| dR-n-dL | 1.836 | 0.997 | 6.7 × 10−2 | 0.601 | 1.467 | 6.8 × 10−1 | 0.270 | 0.848 | 7.5 × 10−1 | 0.874 | 0.591 | 1.4 × 10−1 |
| dR-s-dL | 0.114 | 0.221 | 6.1 × 10−1 | 0.557 | 0.368 | 1.3 × 10−1 | −0.217 | 0.232 | 3.5 × 10−1 | 0.052 | 0.147 | 7.2 × 10−1 |
| n-md/dR-dL | −0.019 | 0.010 | 6.4 × 10−2 | −0.005 | 0.015 | 7.6 × 10−1 | −0.002 | 0.009 | 8.3 × 10−1 | −0.009 | 0.006 | 1.5 × 10−1 |
| Measurements | Group | Genotype | p-Value | ||
|---|---|---|---|---|---|
| AA | AG | GG | |||
| rs7559271 | |||||
| md-n-g | Japanese | 151.44 ± 8.16 | 148.64 ± 7.41 | 146.90 ± 6.31 | 0.031 |
| Korean | 147.69 ± 3.86 | 147.75 ± 7.68 | 150.32 ± 8.69 | 0.378 | |
| Egyptian | 141.14 ± 8.03 | 139.70 ± 7.92 | 136.00 ± 9.25 | 0.015 | |
| s-n-g | Japanese | 111.88 ± 6.17 | 108.3 6± 5.73 | 107.42 ± 5.67 | 0.016 |
| Korean | 106.25 ± 5.15 | 106.94 ± 5.92 | 109.67 ± 7.72 | 0.188 | |
| Egyptian | 112.46 ± 8.70 | 110.50 ± 7.41 | 108.06 ± 8.00 | 0.044 | |
| Measurements | Explanatory Variable | Japanese (n = 201) | Korean (n = 74) | Egyptian (n = 142) | Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SD | p-Value | B | SD | p-Value | B | SD | p-Value | B | SD | p-Value | ||
| md-n-g | hAG | −3.296 | 0.968 | 8.1 × 10−4 | 1.127 | 2.137 | 6.0 × 10−1 | −2.974 | 1.553 | 5.8 × 10−2 | −2.648 | 0.767 | 5.5 × 10−4 |
| hAA | −2.643 | 1.291 | 4.2 × 10−2 | −0.537 | 2.380 | 8.2 × 10−1 | −0.104 | 1.782 | 9.5 × 10−1 | −1.570 | 0.957 | 1.0 × 10−1 | |
| hGG | −2.496 | 1.063 | 2.0 × 10−2 | 1.491 | 2.183 | 5.0 × 10−1 | −2.379 | 0.948 | 1.3 × 10−2 | −2.058 | 0.673 | 2.2 × 10−3 | |
| s-n-g | hAG | −1.702 | 0.686 | 1.4 × 10−2 | 1.984 | 1.493 | 1.9 × 10−1 | −2.180 | 1.121 | 5.4 × 10−2 | −1.324 | 0.545 | 1.5 × 10−2 |
| hAA | 0.282 | 0.914 | 7.6 × 10−1 | 1.147 | 1.663 | 4.9 × 10−1 | 0.765 | 1.287 | 5.5 × 10−1 | 0.561 | 0.680 | 4.1 × 10−1 | |
| hGG | −0.628 | 0.753 | 4.1 × 10−1 | 3.292 | 1.525 | 3.4 × 10−2 | −1.737 | 0.685 | 1.2 × 10−2 | −0.785 | 0.481 | 1.0 × 10−1 | |
| Measurements | Explanatory Variable | Japanese (n = 201) | Korean (n = 74) | Egyptian (n = 142) | Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SD | p-Value | B | SD | p-Value | B | SD | p-Value | B | SD | p-Value | ||
| md-n-g | rs9288572 | −2.922 | 1.631 | 7.5 × 10−2 | 0.087 | 2.914 | 9.8 × 10−1 | 0.787 | 2.005 | 7.0 × 10−1 | −1.202 | 1.161 | 3.0 × 10−1 |
| rs7559271 | −2.893 | 1.429 | 4.4 × 10−2 | 1.375 | 2.234 | 5.4 × 10−1 | −2.075 | 1.017 | 4.3 × 10−2 | −1.900 | 0.777 | 1.4 × 10−2 | |
| Interaction | 1.101 | 1.078 | 3.1 × 10−1 | 0.225 | 1.819 | 9.0 × 10−1 | −1.115 | 1.524 | 4.7 × 10−1 | 0.336 | 0.792 | 6.7 × 10−1 | |
| s-n-g | rs9288572 | 0.080 | 1.158 | 9.5 × 10−1 | −0.847 | 2.049 | 6.8 × 10−1 | 0.452 | 1.453 | 7.6 × 10−1 | 0.049 | 0.828 | 9.5 × 10−1 |
| rs7559271 | −0.669 | 1.014 | 5.1 × 10−1 | 0.866 | 1.571 | 5.8 × 10−1 | −1.809 | 0.736 | 1.5 × 10−2 | −1.129 | 0.557 | 4.3 × 10−2 | |
| Interaction | −0.513 | 0.765 | 5.0 × 10−1 | 0.994 | 1.279 | 4.4 × 10−1 | −0.380 | 1.104 | 7.3 × 10−1 | −0.185 | 0.564 | 7.4 × 10−1 | |
| Outcome Variable | Explanatory Variable | Japanese (n = 201) | Korean (n = 74) | Egyptian (n = 142) | Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SD | p-Value | B | SD | p-Value | B | SD | p-Value | B | SD | p-Value | ||
| PC1 | rs7559271 | 0.000 | 0.002 | 0.868 | 0.008 | 0.003 | 0.016 | −0.001 | 0.002 | 0.538 | 0.001 | 0.001 | 0.414 |
| PC5 | rs7559271 | −0.005 | 0.002 | 0.048 | 0.001 | 0.003 | 0.656 | 0.004 | 0.002 | 0.029 | −0.001 | 0.001 | 0.859 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueda, S.; Kimura, R.; Kim, Y.-I.; Adel, M.; Hikita, Y.; Hatanaka, R.; Takahashi, M.; Koizumi, S.; Yamaguchi, T. Association of PAX3 Gene Polymorphism with Three-Dimensional Nasal Root Morphology. Int. J. Mol. Sci. 2025, 26, 7842. https://doi.org/10.3390/ijms26167842
Ueda S, Kimura R, Kim Y-I, Adel M, Hikita Y, Hatanaka R, Takahashi M, Koizumi S, Yamaguchi T. Association of PAX3 Gene Polymorphism with Three-Dimensional Nasal Root Morphology. International Journal of Molecular Sciences. 2025; 26(16):7842. https://doi.org/10.3390/ijms26167842
Chicago/Turabian StyleUeda, Seishiro, Ryosuke Kimura, Yong-Il Kim, Mohamed Adel, Yu Hikita, Reina Hatanaka, Masahiro Takahashi, So Koizumi, and Tetsutaro Yamaguchi. 2025. "Association of PAX3 Gene Polymorphism with Three-Dimensional Nasal Root Morphology" International Journal of Molecular Sciences 26, no. 16: 7842. https://doi.org/10.3390/ijms26167842
APA StyleUeda, S., Kimura, R., Kim, Y.-I., Adel, M., Hikita, Y., Hatanaka, R., Takahashi, M., Koizumi, S., & Yamaguchi, T. (2025). Association of PAX3 Gene Polymorphism with Three-Dimensional Nasal Root Morphology. International Journal of Molecular Sciences, 26(16), 7842. https://doi.org/10.3390/ijms26167842






