Candidate Genes, Markers, Signatures of Selection, and Quantitative Trait Loci (QTLs) and Their Association with Economic Traits in Livestock: Genomic Insights and Selection
Abstract
1. Introduction
1.1. Historical Context of Livestock Domestication and Economic Significance
1.2. Genetic Diversity and Modern Breeding Challenges
1.3. Role of Genomics in Addressing Global Food Security
2. Scope and Literature Search Strategy
2.1. Objectives
- (a)
- Evaluate how advanced molecular tools enhance the identification of genetic markers (candidate genes, QTLs, SSs) associated with pivotal economic traits, such as productivity, product quality, and disease resistance.
- (b)
- Assess the efficacy of genomic selection (GS) and genomic mapping in improving the genetic diversity, production efficiency, disease resilience, and environmental sustainability, benchmarking these approaches against conventional breeding methods.
- (c)
- Analyse the role of adaptive selection and targeted breeding strategies in bolstering livestock resilience, economic sustainability, and climate adaptation.
- (d)
- Develop actionable strategies to conserve genetic diversity in endangered breeds while maximizing their economic value through integration into contemporary breeding programs.
- (e)
- Synthesize technological innovations and conservation insights to propose future pathways for personalized breeding and sustainable production systems, thereby creating a decision-making framework for stakeholders.
2.2. Literature Search Strategy
3. Advancements in Molecular Biology Technologies and Their Roles in Enhancing Genetics and Breeding in Livestock
3.1. High-Throughput Sequencing and Genomic Tools
3.2. Whole-Genome Sequence Data
3.2.1. The Role of Whole-Genome Sequence Data in Enhancing Genetics and Breeding in Livestock
3.2.2. Progress and Applications of WGS in Livestock GWASs
3.2.3. Methodological Constraints of GWASs in Livestock
3.2.4. Strategies for Enhanced Detection Power and Future Directions
| No. | Specific Animal | Breed/Details | Scientific Name | Genome Size (MB) | Year | Ref. |
|---|---|---|---|---|---|---|
| 1 | Chicken | Red Junglefowl ancestor | Gallus gallus | 1050 | 2004 | [60] |
| 2 | Sheep | Rambouillet ewe | Ovis aries | 2780 | 2008 | [61] |
| 3 | Pig | Duroc breed | Sus scrofa | 2200 | 2008 | [62] |
| 4 | Cattle | Hereford breed | Bos taurus | 2910 | 2009 | [63] |
| 5 | Horse | Thoroughbred (Twilight) | Equus caballus | 2470 | 2009 | [64] |
| 6 | Dromedary Camel | – | Camelus dromedarius | 2200 | 2011 | [65] |
| 7 | Goat | Yunnan Black (Female) | Capra hircus | 2660 | 2011/2012 | [45,66] |
| 8 | Mallard Duck | – | Anas platyrhynchos | 1070 | 2013 | [67] |
3.3. CRISPR-Based Editing: Applications, Challenges, and Global Regulatory
3.4. Bioinformatics and Multi-Omics Integration
4. Key Economic Traits in Livestock
4.1. Genetic Dissection of Production Traits
4.2. Disease Resistance and Climate Resilience
4.3. Reproductive Efficiency and Litter Size
5. Quantitative Trait Loci (QTLs) Mapping
5.1. QTL Discovery and Genome-Wide Association Studies (GWASs)
5.2. Functional Roles of QTLs in Livestock Improvement
5.3. Comparative QTL Analysis Across Different Species
5.4. Quantitative Traits
6. Signatures of Selection (SSs) and Adaptive Evolution
6.1. Genome-Wide Scans for SSs
6.2. Domestication History and Breed-Specific Adaptations
6.3. Case Studies: Performance, Disease Resistance, and Production Traits
6.4. The Contribution of Advanced Molecular Tools to the Detection of SSs in Livestock Populations
7. Candidate Genes Driving Economic Traits
7.1. Growth and Muscle Development
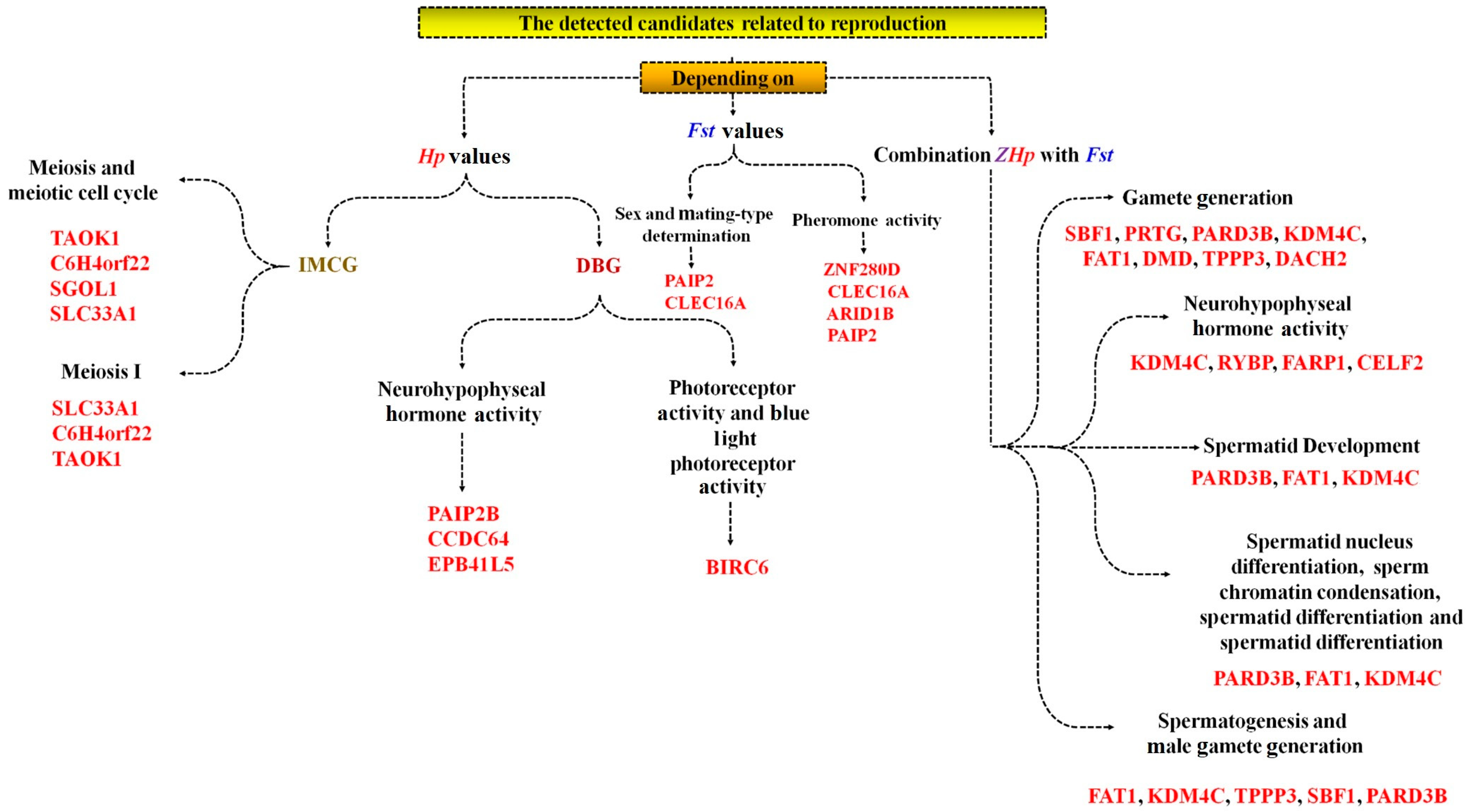
7.2. Reproduction and Fertility Traits
7.3. Milk Production and Composition
7.4. Fibre Production, Coat Colour, and Skin Sensitivity
7.5. Disease Resistance, Heat Tolerance, and Stress Response in Livestock
8. Challenges in GS
8.1. Applications of GS
8.2. Methodology of GS
8.3. Conceptual Evolution from MAS to GS
8.4. Challenges Faced by GS
9. Future Directions
9.1. Progress in Genomic Research for Ruminant Livestock
9.2. Advances in Gene Editing Technologies
9.3. Enhancing Genetic Diversity and Local Breed Resilience
9.4. Super-Pangenomes for Precision Breeding
9.5. Multi-Omics Integration for Trait Dissection
10. Ethical and Regulatory Imperatives
11. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ASF | African swine fever |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DArT | Diversity Arrays Technology |
| eQTLs | Expression quantitative trait loci |
| FAANG | Functional Annotation of Animal Genomes |
| FAO | Food and Agriculture Organization |
| GBLUP | Genomic best linear unbiased prediction |
| GBS | Genotyping by Sequencing |
| GEBV | Genomic-estimated breeding value |
| GS | Genomic selection |
| GWAS | Genome-wide association study |
| Indel | Insertion–Deletion |
| KASP | Kompetitive Allele-Specific PCR |
| LD | Linkage disequilibrium |
| MAS | Marker-assisted selection |
| MCMC | Monte Carlo Markov Chain |
| MHC | Major Histocompatibility Complex |
| mt-DNA | Mitochondrial DNA |
| NAGRP | National Animal Genome Research Program |
| NGS | Next-generation sequencing |
| QTLs | Quantitative trait loci |
| SDGs | Sustainable Development Goals |
| SNP | Single nucleotide polymorphism |
| SSs | Signatures of selection |
| SSRs | Simple Sequence Repeats |
| SVs | Structural variations |
| T2T | Telomere-to-telomere |
| TALEN | Transcription Activator-Like Effector Nuclease |
| WGS | Whole-genome sequencing |
| WGMs | Wide genetic markers |
| XP-CLR | Cross-Population Composite Likelihood Ratio |
| iHS | Integrated Haplotype Score |
| ZFN | Zinc finger nuclease |
References
- Legge, T. The beginning of caprine domestication in Southwest Asia. In The Origins and Spread of Agriculture and Pastoralism in Eurasia; Routledeg: Abingdon, UK, 1996; pp. 238–262. [Google Scholar]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, e4668. [Google Scholar] [CrossRef]
- Naderi, S.; Rezaei, H.-R.; Pompanon, F.; Blum, M.G.; Negrini, R.; Naghash, H.-R.; Balkız, Ö.; Mashkour, M.; Gaggiotti, O.E.; Ajmone-Marsan, P. The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals. Proc. Natl. Acad. Sci. USA 2008, 105, 17659–17664. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Servin, B.; Talenti, A.; Rochat, E.; Kim, E.S.; Oget, C.; Palhière, I.; Crisà, A.; Catillo, G.; Steri, R. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018, 50, 57. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.A.; Rashad, A.; Hassanine, N.N.A.M.; Sharaby, M.A. Modern Strategies to Enhance Goat Genetic Performance; Lambert Academic Publishing: Saarbrücken, Germany, 2019; ISBN 978-620-0-47020-1. [Google Scholar] [CrossRef]
- Clutton-Brock, J. A Natural History of Domesticated Mammals; Cambridge University Press: Cambridge, UK, 1999. [Google Scholar]
- Saleh, A.A.; Rashad, A.M.A.; Hassanine, N.N.A.M.; Sharaby, M.A.; Sallam, S.M.A. History of the Goat and Modern versus Old Strategies to Enhance the Genetic Performance. In Goat Science; Kukovics, S., Ed.; IntechOpen: Rijeka, Croatia, 2023. [Google Scholar] [CrossRef]
- MacHugh, D.E.; Bradley, D.G. Livestock genetic origins: Goats buck the trend. Proc. Natl. Acad. Sci. USA 2001, 98, 5382–5384. [Google Scholar] [CrossRef]
- Mohammadi, A.; Nassiry, M.; Mosafer, J.; Mohammadabadi, M.; Sulimova, G. Distribution of BoLA-DRB3 allelic frequencies and identification of a new allele in the Iranian cattle breed Sistani (Bos indicus). Russ. J. Genet. 2009, 45, 198–202. [Google Scholar] [CrossRef]
- Scherf, B.D.; Pilling, D. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; FAO: Rome, Italy, 2015. [Google Scholar]
- Shah, K.; Kaur, A.; Saxena, S.; Arora, S. CRISPR-Based Approach: A Way Forward to Sustainable Development Goals (SDGs). In Gene Editing in Plants: CRISPR-Cas and Its Applications; Springer: Singapore, 2024; pp. 709–733. [Google Scholar]
- Shah, A.A.; Zubi, R.; Thakur, S.; Chamoli, P. Modern Biotechnology Ventures to Accomplish the Targets of Zero Hunger and Adequate Nutrition. Int. J. Environ. Chem. 2025, 11, 1–14. [Google Scholar]
- Seilacher, A.; Bose, P.K.; Pflüger, F. Triploblastic animals more than 1 billion years ago: Trace fossil evidence from India. Science 1998, 282, 80–83. [Google Scholar] [CrossRef]
- Saleh, A.A.; Xue, L.; Zhao, Y. Screening Indels from the whole genome to identify the candidates and their association with economic traits in several goat breeds. Funct. Integr. Genom. 2023, 23, 58. [Google Scholar] [CrossRef]
- Camargo, L.S.A.; Saraiva, N.Z.; Oliveira, C.S.; Carmickle, A.; Lemos, D.R.; Siqueira, L.G.B.; Denicol, A.C. Perspectives of gene editing for cattle farming in tropical and subtropical regions. Anim. Reprod. 2023, 19, e20220108. [Google Scholar] [CrossRef]
- Husien, H.M.; Saleh, A.A.; Hassanine, N.; Rashad, A.M.A.; Sharaby, M.A.; Mohamed, A.Z.; Abdelhalim, H.; Hafez, E.E.; Essa, M.O.A.; Adam, S.Y.; et al. The Evolution and Role of Molecular Tools in Measuring Diversity and Genomic Selection in Livestock Populations (Traditional and Up-to-Date Insights): A Comprehensive Exploration. Vet. Sci. 2024, 11, 627. [Google Scholar] [CrossRef]
- Bilotto, F.; Christie-Whitehead, K.M.; Malcolm, B.; Barnes, N.; Cullen, B.; Ayre, M.; Harrison, M.T. Costs of transitioning the livestock sector to net-zero emissions under future climates. Nat. Commun. 2025, 16, 3810. [Google Scholar] [CrossRef]
- Hong, N.B.; Tuyet, T.T.; Linh, D.T.T. Consumers’ preferences for Vietnamese OCOP jerky beef: A market driving force for sustainable livestock development and poverty alleviation in the mountainous regions. Discov. Sustain. 2025, 6, 299. [Google Scholar] [CrossRef]
- Kasimanickam, R.; Ferreira, J.C.P.; Kastelic, J.; Kasimanickam, V. Application of Genomic Selection in Beef Cattle Disease Prevention. Animals 2025, 15, 277. [Google Scholar] [CrossRef]
- Bai, Y. Editorial: Enhancing Livestock Breeding Through Advanced Genetic Tools and Phenotyping Systems. Front. Genet. 2025, 16, 1617113. [Google Scholar] [CrossRef]
- Magoro, A.M.; Mtileni, B.; Hadebe, K.; Zwane, A. Assessment of genetic diversity and conservation in South African indigenous goat ecotypes: A review. Animals 2022, 12, 3353. [Google Scholar] [CrossRef]
- Jang, M.-J.; Lee, S.-H.; Kim, J.-M. Genome-Wide Association Studies: A Powerful Approach for Identifying Genomic Variants for Livestock Breeding and Disease Management. In Bioinformatics in Veterinary Science: Vetinformatics; Springer: Singapore, 2025; pp. 87–117. [Google Scholar]
- Gou, Y.; Jing, Y.; Wang, Y.; Li, X.; Yang, J.; Wang, K.; He, H.; Yang, Y.; Tang, Y.; Wang, C. AnimalGWASAtlas: Annotation and prioritization of GWAS loci and quantitative trait loci for animal complex traits. J. Biol. Chem. 2025, 301, 108267. [Google Scholar] [CrossRef]
- Van Laere, A.-S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, H.; Lei, A.; Zhou, J.; Zeng, W.; Zhu, H.; Dong, Z.; Niu, Y.; Shi, B.; Cai, B. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci. Rep. 2015, 5, 13878. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cai, B.; Zhou, S.; Zhu, H.; Qu, L.; Wang, X.; Chen, Y. RNA-seq reveals transcriptome changes in goats following myostatin gene knockout. PLoS ONE 2017, 12, e0187966. [Google Scholar] [CrossRef]
- Martin, P.; Palhière, I.; Maroteau, C.; Bardou, P.; Canale-Tabet, K.; Sarry, J.; Woloszyn, F.; Bertrand-Michel, J.; Racke, I.; Besir, H. A genome scan for milk production traits in dairy goats reveals two new mutations in Dgat1 reducing milk fat content. Sci. Rep. 2017, 7, 1872. [Google Scholar] [CrossRef]
- Gowane, G.; Akram, N.; Misra, S.; Prakash, V.; Kumar, A. Genetic diversity of Cahi DRB and DQB genes of caprine MHC class II in Sirohi goat. J. Genet. 2018, 97, 483–492. [Google Scholar] [CrossRef]
- Allais-Bonnet, A.; Richard, C.; André, M.; Gelin, V.; Deloche, M.-C.; Lamadon, A.; Morin, G.; Mandon-Pépin, B.; Canon, E.; Thépot, D. CRISPR/Cas9-editing of PRNP in Alpine goats. Vet. Res. 2025, 56, 11. [Google Scholar] [CrossRef]
- Wolf, M.; Yin, T.; Neumann, G.; Kokuć, P.; Brockmann, G.; König, S. Single-Step Breeding Value Estimations and Optimum Contribution Selection in Endangered Dual-Purpose German Black Pied Cattle (DSN) Using a Breed Specific SNP Chip. J. Anim. Breed. Genet. 2025, 106, 3345–3358. [Google Scholar] [CrossRef]
- Gaitri, N.; Sharma, A. Multi-omics: A Holistic View Towards Animal Health. Bio Vet. Innov. Mag. 2024, 1, 28–32. [Google Scholar]
- Clark, K.C.; Kwitek, A.E. Multi-Omic Approaches to Identify Genetic Factors in Metabolic Syndrome. Compr. Physiol. 2011, 12, 3045–3084. [Google Scholar] [CrossRef]
- Wróblewski, A.; Strycharz, J.; Świderska, E.; Drewniak, K.; Drzewoski, J.; Szemraj, J.; Kasznicki, J.; Śliwińska, A. Molecular insight into the interaction between epigenetics and leptin in metabolic disorders. Nutrients 2019, 11, 1872. [Google Scholar] [CrossRef]
- Tullo, E.; Finzi, A.; Guarino, M. Environmental impact of livestock farming and Precision Livestock Farming as a mitigation strategy. Sci. Total Environ. 2019, 650, 2751–2760. [Google Scholar] [CrossRef]
- Rupp, R.; Mucha, S.; Larroque, H.; McEwan, J.; Conington, J. Genomic application in sheep and goat breeding. Anim. Front. 2016, 6, 39–44. [Google Scholar] [CrossRef]
- Snyder, M.; Du, J.; Gerstein, M. Personal genome sequencing: Current approaches and challenges. Genes. Dev. 2010, 24, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.; Jamli, S. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef] [PubMed]
- CM Dekkers, J. Application of genomics tools to animal breeding. Curr. Genom. 2012, 13, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Talenti, A.; Palhière, I.; Tortereau, F.; Pagnacco, G.; Stella, A.; Nicolazzi, E.L.; Crepaldi, P.; Tosser-Klopp, G. Functional SNP panel for parentage assessment and assignment in worldwide goat breeds. Genet. Sel. Evol. 2018, 50, 55. [Google Scholar] [CrossRef]
- Burren, A.; Neuditschko, M.; Signer-Hasler, H.; Frischknecht, M.; Reber, I.; Menzi, F.; Drögemüller, C.; Flury, C. Genetic diversity analyses reveal first insights into breed-specific selection signatures within Swiss goat breeds. Anim. Genet. 2016, 47, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Bickhart, D.M.; Rosen, B.D.; Koren, S.; Sayre, B.L.; Hastie, A.R.; Chan, S.; Lee, J.; Lam, E.T.; Liachko, I.; Sullivan, S.T. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017, 49, 643. [Google Scholar] [CrossRef]
- Sharma, A.; Park, J.-E.; Chai, H.-H.; Jang, G.-W.; Lee, S.-H.; Lim, D. Next generation sequencing in livestock species-a review. J. Anim. Breed. Genom. 2017, 1, 23–30. [Google Scholar]
- Patel, S.M.; Koringa, P.G.; Nathani, N.M.; Patel, N.V.; Shah, T.M.; Joshi, C.G. Exploring genetic polymorphism in innate immune genes in Indian cattle (Bos indicus) and buffalo (Bubalus bubalis) using next generation sequencing technology. Meta Gene 2015, 3, 50–58. [Google Scholar] [CrossRef]
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 2014, 344, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2013, 31, 135. [Google Scholar] [CrossRef]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Sharma, A.; Lee, J.S.; Dang, C.G.; Sudrajad, P.; Kim, H.C.; Yeon, S.H.; Kang, H.S.; Lee, S.H. Stories and Challenges of Genome Wide Association Studies in Livestock—A Review. Asian-Australas. J. Anim. Sci. 2015, 28, 1371–1379. [Google Scholar] [CrossRef]
- Johnsson, M. Genomics in animal breeding from the perspectives of matrices and molecules. Hereditas 2023, 160, 20. [Google Scholar] [CrossRef]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef]
- Bolormaa, S.; Chamberlain, A.J.; Khansefid, M.; Stothard, P.; Swan, A.A.; Mason, B.; Prowse-Wilkins, C.P.; Duijvesteijn, N.; Moghaddar, N.; van der Werf, J.H.; et al. Accuracy of imputation to whole-genome sequence in sheep. Genet. Sel. Evol. 2019, 51, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, Y.; Zhang, Y.; Zhuang, Z.; Huang, J.; Luan, M.; Zhao, X.; Dong, L.; Ye, J.; Yang, M.; et al. A Comparative Study of Optimizing Genomic Prediction Accuracy in Commercial Pigs. Animals 2025, 15, 966. [Google Scholar] [CrossRef]
- Wang, B.; Li, P.; Hou, L.; Zhou, W.; Tao, W.; Liu, C.; Liu, K.; Niu, P.; Zhang, Z.; Li, Q. Genome-wide association study and genomic prediction for intramuscular fat content in Suhuai pigs using imputed whole-genome sequencing data. Evol. Appl. 2022, 15, 2054–2066. [Google Scholar] [CrossRef] [PubMed]
- Rubin, C.-J.; Megens, H.-J.; Barrio, A.M.; Maqbool, K.; Sayyab, S.; Schwochow, D.; Wang, C.; Carlborg, Ö.; Jern, P.; Jørgensen, C.B. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19529–19536. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, A.A.; Daetwyler, H.D.; Yudin, N.; Schnabel, R.D.; Vander Jagt, C.J.; Soloshenko, V.; Lhasaranov, B.; Popov, R.; Taylor, J.F.; Larkin, D.M. Scans for signatures of selection in Russian cattle breed genomes reveal new candidate genes for environmental adaptation and acclimation. Sci. Rep. 2018, 8, 12984. [Google Scholar] [CrossRef]
- Chen, L.; Pryce, J.E.; Hayes, B.J.; Daetwyler, H.D. Investigating the effect of imputed structural variants from whole-genome sequence on genome-wide association and genomic prediction in dairy cattle. Animals 2021, 11, 541. [Google Scholar] [CrossRef]
- van Den Berg, I.; Xiang, R.; Jenko, J.; Pausch, H.; Boussaha, M.; Schrooten, C.; Tribout, T.; Gjuvsland, A.B.; Boichard, D.; Nordbø, Ø. Meta-analysis for milk fat and protein percentage using imputed sequence variant genotypes in 94,321 cattle from eight cattle breeds. Genet. Sel. Evol. 2020, 52, 37. [Google Scholar] [CrossRef]
- Broekema, R.; Bakker, O.; Jonkers, I. A practical view of fine-mapping and gene prioritization in the post-genome-wide association era. Open Biol. 2020, 10, 190221. [Google Scholar] [CrossRef]
- Ros-Freixedes, R.; Johnsson, M.; Whalen, A.; Chen, C.-Y.; Valente, B.D.; Herring, W.O.; Gorjanc, G.; Hickey, J.M. Genomic prediction with whole-genome sequence data in intensely selected pig lines. Genet. Sel. Evol. 2022, 54, 65. [Google Scholar] [CrossRef]
- Ros-Freixedes, R. The contribution of whole-genome sequence data to genome-wide association studies in livestock: Outcomes and perspectives. Livest. Sci. 2024, 281, 105430. [Google Scholar] [CrossRef]
- Wallis, J.W.; Aerts, J.; Groenen, M.A.; Crooijmans, R.P.; Layman, D.; Graves, T.A.; Scheer, D.E.; Kremitzki, C.; Fedele, M.J.; Mudd, N.K. A physical map of the chicken genome. Nature 2004, 432, 761. [Google Scholar] [CrossRef]
- International Sheep Genomics Consortium; Archibald, A.L.; Cockett, N.E.; Dalrymple, B.P.; Faraut, T.; Kijas, J.W.; Maddox, J.F.; McEwan, J.C.; Oddy, V.H.; Raadsma, H.W.; et al. The sheep genome reference sequence: A work in progress. Anim. Genet. 2010, 41, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Groenen, M.A.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.J.; et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef]
- Bovine Genome Sequencing and Analysis Consortium; Elsik, C.G.; Tellam, R.L.; Worley, K.C.; Gibbs, R.A.; Muzny, D.M.; Weinstock, G.M.; Adelson, D.L.; Eichler, E.E.; Elnitski, L.; et al. The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science 2009, 324, 522–528. [Google Scholar] [CrossRef]
- Wade, C.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.; Adelson, D.; Bailey, E.; Bellone, R. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef]
- Ghosh, P.; Hsu, C.; Alyamani, E.J.; Shehata, M.M.; Al-Dubaib, M.A.; Al-Naeem, A.; Hashad, M.; Mahmoud, O.M.; Alharbi, K.B.; Al-Busadah, K. Genome-wide analysis of the emerging infection with Mycobacterium avium subspecies paratuberculosis in the Arabian camels (Camelus dromedarius). PLoS ONE 2012, 7, e31947. [Google Scholar] [CrossRef]
- Du, X.; Servin, B.; Womack, J.E.; Cao, J.; Yu, M.; Dong, Y.; Wang, W.; Zhao, S. An update of the goat genome assembly using dense radiation hybrid maps allows detailed analysis of evolutionary rearrangements in Bovidae. BMC Genom. 2014, 15, 625. [Google Scholar] [CrossRef]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.; Faircloth, B.C.; Nabholz, B.; Howard, J.T. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef]
- Urban, W.; Kropacz, M.; Łach, M.; Jankowska, A. CRISPR-Cas9 in the tailoring of genetically engineered animals. Curr. Issues Mol. Biol. 2025, 47, 330. [Google Scholar] [CrossRef]
- Shamshirgaran, Y.; Liu, J.; Sumer, H.; Verma, P.J.; Taheri-Ghahfarokhi, A. Tools for efficient genome editing; ZFN, TALEN, and CRISPR. Appl. Genome Modul. Ed. 2022, 2495, 29–46. [Google Scholar]
- Ain, Q.U.; Chung, J.Y.; Kim, Y.-H. Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN. J. Controll. Release 2015, 205, 120–127. [Google Scholar] [CrossRef]
- Gim, G.M.; Kwon, D.H.; Eom, K.H.; Moon, J.; Park, J.H.; Lee, W.W.; Jung, D.J.; Kim, D.H.; Yi, J.K.; Ha, J.J. Production of MSTN-mutated cattle without exogenous gene integration using CRISPR-Cas9. Biotechnol. J. 2022, 17, 2100198. [Google Scholar] [CrossRef]
- Gim, G.-M.; Uhm, K.-H.; Kwon, D.-H.; Kim, M.-J.; Jung, D.-J.; Kim, D.-H.; Yi, J.-K.; Ha, J.-J.; Yum, S.-Y.; Son, W.-J. Germline transmission of MSTN knockout cattle via CRISPR-Cas9. Theriogenology 2022, 192, 22–27. [Google Scholar] [CrossRef]
- Fajardo, C.; Macedo, M.; Buha, T.; De Donato, M.; Costas, B.; Mancera, J.M. Genetically Modified Animal-Derived Products: From Regulations to Applications. Animals 2025, 15, 1570. [Google Scholar] [CrossRef]
- Leone, L. Gene editing for the EU agrifood: Risks and promises in science regulation. Eur. J. Risk Regul. 2019, 10, 766–780. [Google Scholar] [CrossRef]
- Cimadori, I.; Di Concetto, A.; Grieger, K. The Protection of Selectively Bred and Gene Edited Farm Animals under EU Law. Eur. J. Risk Regul. 2025, 1–17. [Google Scholar] [CrossRef]
- Mahmood, U.; Li, X.; Fan, Y.; Chang, W.; Niu, Y.; Li, J.; Qu, C.; Lu, K. Multi-omics revolution to promote plant breeding efficiency. Front. Plant Sci. 2022, 13, 1062952. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Yang, W.; Cui, M.; Dai, B.; Dong, Y.; Yang, J.; Zhang, X.; Liu, D.; Liang, H. Comparison of gene editing efficiencies of CRISPR/Cas9 and TALEN for generation of MSTN knock-out cashmere goats. Theriogenology 2019, 132, 1–11. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Xue, C.; Greene, E.C. DNA repair pathway choices in CRISPR-Cas9-mediated genome editing. Trends Genet. 2021, 37, 639–656. [Google Scholar] [CrossRef]
- Zhang, J.; Khazalwa, E.M.; Abkallo, H.M.; Zhou, Y.; Nie, X.; Ruan, J.; Zhao, C.; Wang, J.; Xu, J.; Li, X. The advancements, challenges, and future implications of the CRISPR/Cas9 system in swine research. J. Genet. Genom. 2021, 48, 347–360. [Google Scholar] [CrossRef]
- Islam, M.A.; Rony, S.A.; Rahman, M.B.; Cinar, M.U.; Villena, J.; Uddin, M.J.; Kitazawa, H. Improvement of disease resistance in livestock: Application of immunogenomics and CRISPR/Cas9 technology. Animals 2020, 10, 2236. [Google Scholar] [CrossRef]
- Li, W.R.; Liu, C.X.; Zhang, X.M.; Chen, L.; Peng, X.R.; He, S.G.; Lin, J.P.; Han, B.; Wang, L.Q.; Huang, J.C. CRISPR/Cas9-mediated loss of FGF5 function increases wool staple length in sheep. FEBS J. 2017, 284, 2764–2773. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Scheben, A.; Edwards, D. Advances in integrating genomics and bioinformatics in the plant breeding pipeline. Agriculture 2018, 8, 75. [Google Scholar] [CrossRef]
- Chavhan, R.L.; Hinge, V.R.; Wankhade, D.J.; Deshmukh, A.S.; Mahajan, N.; Kadam, U.S. Bioinformatics for Molecular Breeding and Enhanced Crop Performance: Applications and Perspectives. Bioinform. Plant Res. Crop Breed. 2024, 22, 21–74. [Google Scholar]
- Benjelloun, B.; Alberto, F.J.; Streeter, I.; Boyer, F.; Coissac, E.; Stucki, S.; BenBati, M.; Ibnelbachyr, M.; Chentouf, M.; Bechchari, A. Characterizing neutral genomic diversity and selection signatures in indigenous populations of Moroccan goats (Capra hircus) using WGS data. Front. Genet. 2015, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Cai, Y.; Qi, M.; Liang, C.; Pan, L.; Liu, Y.; Feng, Y.; Cao, X.; Yang, Q.; Ren, G. LCORL and STC2 variants increase body size and growth rate in cattle and other animals. Genom. Proteom. Bioinform. 2025, 17, qzaf025. [Google Scholar] [CrossRef]
- Smith, S.; Turbill, C.; Suchentrunk, F. Introducing mother’s curse: Low male fertility associated with an imported mtDNA haplotype in a captive colony of brown hares. Mol. Ecol. 2010, 19, 36–43. [Google Scholar] [CrossRef]
- Chu, M.; He, Y.; Cheng, D.; Ye, S.; Fang, L.; Wang, J. Association between expression of reproductive seasonality and alleles of melatonin receptor 1A in goats. Anim. Reprod. Sci. 2007, 101, 276–284. [Google Scholar] [CrossRef]
- Makkar, H.P. Smart livestock feeding strategies for harvesting triple gain–the desired outcomes in planet, people and profit dimensions: A developing country perspective. Anim. Prod. Sci. 2016, 56, 519–534. [Google Scholar] [CrossRef]
- Wolfová, M.; Wolf, J. Strategies for defining traits when calculating economic values for livestock breeding: A review. Animal 2013, 7, 1401–1413. [Google Scholar] [CrossRef]
- Wolfová, M.; Wolf, J.; Přibyl, J.; Zahrádková, R.; Kica, J. Breeding objectives for beef cattle used in different production systems: 1. Model development. Livest. Prod. Sci. 2005, 95, 201–215. [Google Scholar] [CrossRef]
- Koots, K.; Gibson, J. Economic values for beef production traits from a herd level bioeconomic model. Can. J. Anim. Sci. Can. J. Anim. Sci. 1998, 78, 29–45. [Google Scholar] [CrossRef]
- Wolfová, M.; Wolf, J.; Kvapilík, J.; Kica, J. Selection for Profit in Cattle: I. Economic Weights for Purebred Dairy Cattle in the Czech Republic. J. Dairy. Sci. 2007, 90, 2442–2455. [Google Scholar] [CrossRef] [PubMed]
- Albera, A.; Carnier, P.; Groen, A.F. Definition of a breeding goal for the Piemontese breed: Economic and biological values and their sensitivity to production circumstances. Livest. Prod. Sci. 2004, 89, 66–77. [Google Scholar] [CrossRef]
- Misztal, I.; Aguilar, I.; Lourenco, D.; Ma, L.; Steibel, J.P.; Toro, M. Emerging issues in genomic selection. J. Anim. Sci. 2021, 99, skab092. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Van der Linden, A.; Oosting, S.; Wang, Y.; Ducro, B. Derivation of Economic Values for Breeding Objective Traits of Chinese Holstein Dairy Cows Using a Bio-Economic Model. J. Anim. Breed. Genet. 2025. [Google Scholar] [CrossRef]
- Zhang, B.; Ma, J.; Shen, L.; Li, Y.; Xie, S.; Li, H.; Li, J.; Li, X.; Wang, Z. Genomic Insights into Pigeon Breeding: GWAS for Economic Traits and the Development of a High-Throughput Liquid Phase Array Chip. Poult. Sci. 2025, 104, 104872. [Google Scholar] [CrossRef]
- Grosclaude, F.; Marie-Françoise, M.A.H.É.; Brignon, G.; Di Stasio, L.; Jeunet, R. A Mendelian polymorphism underlying quantitative variations of goat α s1-casein. Génét. Sél. Évol. 1987, 19, 399–412. [Google Scholar]
- Sztankoova, Z.; Senese, C.; Czernekova, V.; Dudkova, G.; Kott, T.; Matlova, V.; Soldat, J. Genomic analysis of the CSN2 and CSN3 loci in two Czech goat breeds. Animals 2005, 23, 67–70. [Google Scholar]
- Wang, J.; Zhou, H.; Luo, Y.; Zhao, M.; Gong, H.; Hao, Z.; Hu, J.; Hickford, J.G.H. Variation in the Caprine KAP24-1 Gene Affects Cashmere Fibre Diameter. Animals 2019, 9, 15. [Google Scholar] [CrossRef]
- Salo, A.M.; Cox, H.; Farndon, P.; Moss, C.; Grindulis, H.; Risteli, M.; Robins, S.P.; Myllylä, R. A connective tissue disorder caused by mutations of the lysyl hydroxylase 3 gene. Am. J. Hum. Genet. 2008, 83, 495–503. [Google Scholar] [CrossRef]
- Li, M.-H.; Li, K.; Kantanen, J.; Feng, Z.; Fan, B.; Zhao, S.-H. Allelic variations in exon 2 of caprine MHC class II DRB3 gene in Chinese indigenous goats. Small Rumin. Res. 2006, 66, 236–243. [Google Scholar] [CrossRef]
- Baghizadeh, A.; Bahaaddini, M.; Mohamadabadi, M.; Askari, N. Allelic variations in exon 2 of Caprine MHC class II DRB3 gene in Raeini Cashmere goat. Am. J. Agric. Environ. Sci. 2009, 6, 454–459. [Google Scholar]
- Paracha, H.; Hussain, T.; Tahir, M.; Yasmeen, A.; Pervez, M.; Sheikh, A.; Haider, A.; Ali, R.; Khan, W. Multifunctional DRB3, a MHC class II gene, as a useful biomarker in small ruminants: A review. J. Inf. Mol. Biol. 2015, 3, 19–23. [Google Scholar] [CrossRef]
- Freking, B.A.; Murphy, T.W.; Chitko-McKown, C.G.; Workman, A.M.; Heaton, M.P. Impact of Four Ovine TMEM154 Haplotypes on Ewes during Multiyear Lentivirus Exposure. Int. J. Mol. Sci. 2022, 23, 14966. [Google Scholar] [CrossRef]
- Weldenegodguad, M.; Popov, R.; Pokharel, K.; Ammosov, I.; Ming, Y.; Ivanova, Z.; Kantanen, J. Whole-Genome Sequencing of Three Native Cattle Breeds Originating from the Northernmost Cattle Farming Regions. Front. Genet. 2018, 9, 728. [Google Scholar] [CrossRef]
- Hiendleder, S.; Thomsen, H.; Reinsch, N.; Bennewitz, J.; Leyhe-Horn, B.; Looft, C.; Xu, N.; Medjugorac, I.; Russ, I.; Kühn, C. Mapping of QTL for body conformation and behavior in cattle. J. Hered. 2003, 94, 496–506. [Google Scholar] [CrossRef]
- Messer, L.A.; Wang, L.; Tuggle, C.K.; Yerle, M.; Chardon, P.; Pomp, D.; Womack, J.E.; Barendse, W.; Crawford, A.M.; Notter, D.R. Mapping of the melatonin receptor la (MTNR1A) gene in pigs, sheep, and cattle. Mamm. Genome 1997, 8, 368–370. [Google Scholar] [CrossRef]
- Guan, D.; Luo, N.; Tan, X.; Zhao, Z.; Huang, Y.; Na, R.; Zhang, J.; Zhao, Y. Scanning of selection signature provides a glimpse into important economic traits in goats (Capra hircus). Sci. Rep. 2016, 6, 36372. [Google Scholar] [CrossRef]
- Solberg Woods, L.C. QTL mapping in outbred populations: Successes and challenges. Physiol. Genom. 2014, 46, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, M.F.; Plastow, G.S. Applications of genomics to improve livestock in the developing world. Livest. Sci. 2014, 166, 76–83. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Keogh, K.; Carthy, T.R.; McClure, M.C.; Waters, S.M.; Kenny, D.A. Genome-wide association study of economically important traits in Charolais and Limousin beef cows. Animal 2021, 15, 100011. [Google Scholar] [CrossRef]
- Supakorn, C. The important candidate genes in goats—A review. Walailak J. Sci. Technol. 2009, 6, 17–36. [Google Scholar]
- Tambasco, D.; Paz, C.; Tambasco-Studart, M.; Pereira, A.; Alencar, M.; Freitas, A.; Coutinho, L.; Packer, I.; Regitano, L.D.A. Candidate genes for growth traits in beef cattle crosses Bos taurus× Bos indicus. J. Anim. Breed. Genet. 2003, 120, 51–56. [Google Scholar] [CrossRef]
- Womack, J.E. Advances in livestock genomics: Opening the barn door. Genome Res. 2005, 15, 1699–1705. [Google Scholar] [CrossRef]
- Rothschild, M.F. Identification of quantitative trait loci and interesting candidate genes in the pig: Progress and prospects. In Proceedings of the 6th World Congress on Genetics Applied to Livestock Production, Armstrong, Canada, 1–6 July 1998; pp. 403–409. [Google Scholar]
- Cheruiyot, E.K.; Negussie, E.; Tarekegn, G.M.; Haile-Mariam, M. Candidate Genes and Genomic Regions Relevant for Adaptation to Extreme Climates in Livestock. CABI Rev. 2025, 20, 0052. [Google Scholar] [CrossRef]
- Hill, W.G. Understanding and Using Quantitative Genetic Variation. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Kashi, Y.; Hallerman, E.; Soller, M. Marker-assisted selection of candidate bulls for progeny testing programmes. Anim. Sci. 1990, 51, 63–74. [Google Scholar] [CrossRef]
- Teneva, A. Molecular markers in animal genome analysis. Biotechnol. Anim. Husb. 2009, 25, 1267–1284. [Google Scholar]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef]
- Zhou, S.; Goldstein, S.; Place, M.; Bechner, M.; Patino, D.; Potamousis, K.; Ravindran, P.; Pape, L.; Rincon, G.; Hernandez-Ortiz, J.; et al. A clone-free, single molecule map of the domestic cow (Bos taurus) genome. BMC Genom. 2015, 16, 644. [Google Scholar] [CrossRef]
- Zimin, A.V.; Delcher, A.L.; Florea, L.; Kelley, D.R.; Schatz, M.C.; Puiu, D.; Hanrahan, F.; Pertea, G.; Van Tassell, C.P.; Sonstegard, T.S. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009, 10, R42. [Google Scholar] [CrossRef]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef]
- Warren, W.C.; Hillier, L.W.; Tomlinson, C.; Minx, P.; Kremitzki, M.; Graves, T.; Markovic, C.; Bouk, N.; Pruitt, K.D.; Thibaud-Nissen, F. A new chicken genome assembly provides insight into avian genome structure. G3 Genes. Genomes Genet. 2017, 7, 109–117. [Google Scholar] [CrossRef]
- Mason, A.S. Fourth Report on Chicken Genes and Chromosomes 2022. Cytogenet. Genome Res. 2022, 162, 405–528. [Google Scholar] [CrossRef]
- Fan, B.; Gorbach, D.M.; Rothschild, M.F. The Pig Genome Project Has Plenty to Squeal About. Cytogenet. Genome Res. 2011, 134, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef]
- Cutter, A.D.; Payseur, B.A. Genomic signatures of selection at linked sites: Unifying the disparity among species. Nat. Rev. Genet. 2013, 14, 262–274. [Google Scholar] [CrossRef]
- Kooverjee, B.B.; Soma, P.; Van Der Nest, M.A.; Scholtz, M.M.; Neser, F.W.C. Selection Signatures in South African Nguni and Bonsmara Cattle Populations Reveal Genes Relating to Environmental Adaptation. Front. Genet. 2022, 13, 909012. [Google Scholar] [CrossRef]
- Wang, Y.; Niu, Z.; Zeng, Z.; Jiang, Y.; Jiang, Y.; Ding, Y.; Tang, S.; Shi, H.; Ding, X. Using High-Density SNP Array to Reveal Selection Signatures Related to Prolificacy in Chinese and Kazakhstan Sheep Breeds. Animals 2020, 10, 1633. [Google Scholar] [CrossRef]
- Qanbari, S.; Simianer, H. Mapping signatures of positive selection in the genome of livestock. Livest. Sci. 2014, 166, 133–143. [Google Scholar] [CrossRef]
- Zhao, F.-P.; Wei, C.-H.; Zhang, L.; Liu, J.-S.; Wang, G.-K.; Tao, Z.; Du, L.-X. A genome scan of recent positive selection signatures in three sheep populations. J. Integr. Agric. 2016, 15, 162–174. [Google Scholar] [CrossRef]
- Brito, L.F.; Kijas, J.W.; Ventura, R.V.; Sargolzaei, M.; Porto-Neto, L.R.; Cánovas, A.; Feng, Z.; Jafarikia, M.; Schenkel, F.S. Genetic diversity and signatures of selection in various goat breeds revealed by genome-wide SNP markers. BMC Genom. 2017, 18, 229. [Google Scholar] [CrossRef]
- Li, X.; Su, R.; Wan, W.; Zhang, W.; Jiang, H.; Qiao, X.; Fan, Y.; Zhang, Y.; Wang, R.; Liu, Z. Identification of selection signals by large-scale whole-genome resequencing of cashmere goats. Sci. Rep. 2017, 7, 15142. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-S.; Elbeltagy, A.; Aboul-Naga, A.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.; Rothschild, M. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity 2016, 116, 255. [Google Scholar] [CrossRef] [PubMed]
- Talenti, A.; Bertolini, F.; Pagnacco, G.; Pilla, F.; Ajmone-Marsan, P.; Rothschild, M.F.; Crepaldi, P.; Consortium, I.G. The Valdostana goat: A genome-wide investigation of the distinctiveness of its selective sweep regions. Mamm. Genome 2017, 28, 114–128. [Google Scholar] [CrossRef]
- Yang, H.; Li, T.; Zhang, N.; Chen, J.; Zhang, Y.; Peng, S.; Zhou, L.; Ma, R.; Zhang, Z.; Liu, Q.; et al. Identification of Candidate Genes and Functional Pathways Associated with Body Size Traits in Hulunbuir Sheep Through GWAS Analysis. Genes 2025, 16, 410. [Google Scholar] [CrossRef]
- Johnston, S.E.; McEWAN, J.C.; Pickering, N.K.; Kijas, J.W.; Beraldi, D.; Pilkington, J.G.; Pemberton, J.M.; Slate, J. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol. Ecol. 2011, 20, 2555–2566. [Google Scholar] [CrossRef]
- Bahbahani, H.; Clifford, H.; Wragg, D.; Mbole-Kariuki, M.N.; Van Tassell, C.; Sonstegard, T.; Woolhouse, M.; Hanotte, O. Signatures of positive selection in East African Shorthorn Zebu: A genome-wide single nucleotide polymorphism analysis. Sci. Rep. 2015, 5, 11729. [Google Scholar] [CrossRef]
- Sigdel, A.; Abdollahi-Arpanahi, R.; Aguilar, I.; Peñagaricano, F. Whole genome mapping reveals novel genes and pathways involved in milk production under heat stress in US Holstein cows. Front. Genet. 2019, 10, 928. [Google Scholar] [CrossRef] [PubMed]
- Resende, A.; Gonçalves, J.; Muigai, A.W.T.; Pereira, F. Mitochondrial DNA variation of domestic sheep (Ovis aries) in Kenya. Anim. Genet. 2016, 47, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Cheruiyot, E.K.; Bett, R.C.; Amimo, J.O.; Zhang, Y.; Mrode, R.; Mujibi, F.D. Signatures of selection in admixed dairy cattle in Tanzania. Front. Genet. 2018, 9, 607. [Google Scholar] [CrossRef]
- Weikard, R.; Widmann, P.; Buitkamp, J.; Emmerling, R.; Kuehn, C. Revisiting the quantitative trait loci for milk production traits on BTA6. Anim. Genet. 2012, 43, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Gurgul, A.; Jasielczuk, I.; Semik-Gurgul, E.; Pawlina-Tyszko, K.; Stefaniuk-Szmukier, M.; Szmatoła, T.; Polak, G.; Tomczyk-Wrona, I.; Bugno-Poniewierska, M. A genome-wide scan for diversifying selection signatures in selected horse breeds. PLoS ONE 2019, 14, e0210751. [Google Scholar] [CrossRef]
- Nielsen, R.; Williamson, S.; Kim, Y.; Hubisz, M.J.; Clark, A.G.; Bustamante, C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005, 15, 1566–1575. [Google Scholar] [CrossRef]
- McRae, K.M.; McEwan, J.C.; Dodds, K.G.; Gemmell, N.J. Signatures of selection in sheep bred for resistance or susceptibility to gastrointestinal nematodes. BMC Genom. 2014, 15, 637. [Google Scholar] [CrossRef]
- Ramey, H.R.; Decker, J.E.; McKay, S.D.; Rolf, M.M.; Schnabel, R.D.; Taylor, J.F. Detection of selective sweeps in cattle using genome-wide SNP data. BMC Genom. 2013, 14, 382. [Google Scholar] [CrossRef]
- Álvarez, I.; Fernández, I.; Traoré, A.; Pérez-Pardal, L.; Menéndez-Arias, N.A.; Goyache, F. Genomic scan of selective sweeps in Djallonké (West African Dwarf) sheep shed light on adaptation to harsh environments. Sci. Rep. 2020, 10, 2824. [Google Scholar] [CrossRef]
- Saleh, A.A.; Hammoud, M.H.; Dabour, N.A.; Hafez, E.E.; Sharaby, M.A. BMPR-1B, BMP-15 and GDF-9 genes structure and their relationship with litter size in six sheep breeds reared in Egypt. BMC Res. Notes 2020, 13, 215. [Google Scholar] [CrossRef]
- Ben-Jemaa, S.; Mastrangelo, S.; Lee, S.-H.; Lee, J.H.; Boussaha, M. Genome-wide scan for selection signatures reveals novel insights into the adaptive capacity in local North African cattle. Sci. Rep. 2020, 10, 19466. [Google Scholar] [CrossRef]
- Hayes, B.J.; Pryce, J.; Chamberlain, A.J.; Bowman, P.J.; Goddard, M.E. Genetic architecture of complex traits and accuracy of genomic prediction: Coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 2010, 6, e1001139. [Google Scholar] [CrossRef]
- Smith, C. Improvement of metric traits through specific genetic loci. Anim. Sci. 2010, 9, 349–358. [Google Scholar] [CrossRef]
- Lande, R.; Thompson, R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 1990, 124, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Wiggans, G.R.; Cole, J.B.; Hubbard, S.M.; Sonstegard, T.S. Genomic Selection in Dairy Cattle: The USDA Experience. Annu. Rev. Anim. Biosci. 2017, 5, 309–327. [Google Scholar] [CrossRef]
- Knol, E.; Nielsen, B.; Knap, P. Genomic selection in commercial pig breeding. Anim. Front. 2016, 6, 15. [Google Scholar] [CrossRef]
- Wolc, A.; Arango, J.; Jankowski, T.; Dunn, I.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Preisinger, R.; Fernando, R.L.; Garrick, D.J.; et al. Genome-wide association study for egg production and quality in layer chickens. J. Anim. Breed. Genet. 2014, 131, 173–182. [Google Scholar] [CrossRef]
- Hickey, J.M.; Chiurugwi, T.; Mackay, I.; Powell, W. Genomic prediction unifies animal and plant breeding programs to form platforms for biological discovery. Nat. Genet. 2017, 49, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Schütz, E.; Scharfenstein, M.; Brenig, B. Implication of complex vertebral malformation and bovine leukocyte adhesion deficiency DNA-based testing on disease frequency in the Holstein population. J. Dairy Sci. 2008, 91, 4854–4859. [Google Scholar] [CrossRef] [PubMed]
- Georges, M.; Charlier, C.; Hayes, B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar] [CrossRef]
- Dekkers, J.C. Commercial application of marker- and gene-assisted selection in livestock: Strategies and lessons. J. Anim. Sci. 2004, 82, E313–E328. [Google Scholar] [CrossRef]
- Lowe, J.W.E.; Bruce, A. Genetics without genes? The centrality of genetic markers in livestock genetics and genomics. Hist. Philos. Life Sci. 2019, 41, 50. [Google Scholar] [CrossRef]
- Ragoussis, J. Genotyping Technologies for Genetic Research. Annu. Rev. Genom. Hum. Genet. 2009, 10, 117–133. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy. Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Misztal, I.; Lourenco, D.; Legarra, A. Current status of genomic evaluation. J. Anim. Sci. 2020, 98, skaa101. [Google Scholar] [CrossRef]
- Meuwissen, T.; Goddard, M. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 2010, 185, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Georges, M.; Nielsen, D.; Mackinnon, M.; Mishra, A.; Okimoto, R.; Pasquino, A.T.; Sargeant, L.S.; Sorensen, A.; Steele, M.R.; Zhao, X. Mapping quantitative trait loci controlling milk production in dairy cattle by exploiting progeny testing. Genetics 1995, 139, 907–920. [Google Scholar] [CrossRef]
- Seidel, G. Brief introduction to whole-genome selection in cattle using single nucleotide polymorphisms. Reprod. Fertil. Dev. 2009, 22, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Jafarikia, M.; Grossi, D.A.; Kijas, J.W.; Porto-Neto, L.R.; Ventura, R.V.; Salgorzaei, M.; Schenkel, F.S. Characterization of linkage disequilibrium, consistency of gametic phase and admixture in Australian and Canadian goats. BMC Genet. 2015, 16, 67. [Google Scholar] [CrossRef]
- Phua, S.; Hyndman, D.; Baird, H.; Auvray, B.; McEwan, J.; Lee, M.; Dodds, K. Towards genomic selection for facial eczema disease tolerance in the N ew Z ealand sheep industry. Anim. Genet. 2014, 45, 559–564. [Google Scholar] [CrossRef]
- Pickering, N.K.; Auvray, B.; Dodds, K.G.; McEwan, J.C. Genomic prediction and genome-wide association study for dagginess and host internal parasite resistance in New Zealand sheep. BMC Genom. 2015, 16, 958. [Google Scholar] [CrossRef] [PubMed]
- Larroque, H.; Robert-Granié, C. Comparison of joint versus purebred genomic evaluation in the French multi-breed dairy goat population. Genet. Sel. Evol. 2014, 46, 67. [Google Scholar] [CrossRef]
- Daetwyler, H.; Hickey, J.; Henshall, J.; Dominik, S.; Gredler, B.; Van Der Werf, J.; Hayes, B. Accuracy of estimated genomic breeding values for wool and meat traits in a multi-breed sheep population. Anim. Prod. Sci. 2010, 50, 1004–1010. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Swan, A.A.; van der Werf, J.H.; Hayes, B.J. Accuracy of pedigree and genomic predictions of carcass and novel meat quality traits in multi-breed sheep data assessed by cross-validation. Genet. Sel. Evol. 2012, 44, 33. [Google Scholar] [CrossRef] [PubMed]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the Bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- Van Marle-Köster, E.; Visser, C.; Berry, D. A review of genomic selection-Implications for the South African beef and dairy cattle industries. S. Afr. J. Anim. Sci. 2013, 43, 1–17. [Google Scholar] [CrossRef][Green Version]
- Montesinos-López, O.A.; Kismiantini; Montesinos-López, A. Two simple methods to improve the accuracy of the genomic selection methodology. BMC Genom. 2023, 24, 220. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.R.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef]
- Yadav, V.; Singh, N.; Sharma, S.; Lakhani, N.; Bhimte, A.; Khare, A.; Yousuf, S. Genomic selection and it’s application in livestock improvement. J. Entomol. Zool. Stud. 2018, 6, 1838–1844. [Google Scholar]
- Schaeffer, L. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 2006, 123, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Calus, M.P.L.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. BMC Genet. 2015, 16, 88. [Google Scholar] [CrossRef]
- Hayes, B. QTL Mapping, MAS, and Genomic Selection. A Short-Course; Animal Breeding & Genetics Department of Animal Science, Iowa State University: Victoria, Australia, 2007; Volume 1, pp. 3–4. [Google Scholar]
- Sharma, P.; Doultani, S.; Hadiya, K.; George, L.; Highland, H. Overview of marker-assisted selection in animal breeding. HISTORY 2024, 9, 303–318. [Google Scholar] [CrossRef]
- Hollifield, M.K. Improving Genomic Prediction Methods and Estimation of Genetic Parameters for Large Populations. Ph.D. Thesis, University of Georgia, Athens, GA, USA, 2024. [Google Scholar]
- Boichard, D.; Ducrocq, V.; Croiseau, P.; Fritz, S. Genomic selection in domestic animals: Principles, applications and perspectives. C. R. Biol. 2016, 339, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.B. Genomic selection signatures and animal breeding. J. Anim. Breed. Genet. 2021, 138, 1–3. [Google Scholar] [CrossRef]
- Boichard, D.; Fritz, S.; Croiseau, P.; Ducrocq, V.; Tribout, T.; Cuyabano, B.C.D. Erosion of estimated genomic breeding values with generations is due to long distance associations between markers and QTL. Genet. Sel. Evol. 2025, 57, 14. [Google Scholar] [CrossRef]
- Jabar, J.; Seyyed, H.H.; Mohsen, G.; Alireza, E. Trend of bias in prediction of genomic estimated breeding values due to selective genotyping in genomic selection schemes in consecutive generations. J. Livest. Sci. Technol. 2019, 7, 57–63. [Google Scholar] [CrossRef]
- Leite, N.G.; Bermann, M.; Tsuruta, S.; Misztal, I.; Lourenco, D. Marker effect p-values for single-step GWAS with the algorithm for proven and young in large genotyped populations. Genet. Sel. Evol. 2024, 56, 59. [Google Scholar] [CrossRef]
- Onogi, A.; Watanabe, T.; Ogino, A.; Kurogi, K.; Togashi, K. Genomic prediction with non-additive effects in beef cattle: Stability of variance component and genetic effect estimates against population size. BMC Genom. 2021, 22, 512. [Google Scholar] [CrossRef] [PubMed]
- Karaman, E.; Su, G.; Croue, I.; Lund, M.S. Genomic prediction using a reference population of multiple pure breeds and admixed individuals. Genet. Sel. Evol. 2021, 53, 46. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.Q.; Chen, B.; Ran, M.L.; Yang, G.M.; Zeng, C. The application of genomic selection in pig cross breeding. Yi Chuan 2020, 42, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, J. Ethical considerations of gene editing and genetic selection. J. Gen. Fam. Med. 2020, 21, 37–47. [Google Scholar] [CrossRef]
- Munsie, M.; Gyngell, C. Ethical issues in genetic modification and why application matters. Curr. Opin. Genet. Dev. 2018, 52, 7–12. [Google Scholar] [CrossRef]
- Xu, S.; Akhatayeva, Z.; Liu, J.; Feng, X.; Yu, Y.; Badaoui, B.; Esmailizadeh, A.; Kantanen, J.; Amills, M.; Lenstra, J.A.; et al. Genetic advancements and future directions in ruminant livestock breeding: From reference genomes to multiomics innovations. Sci. China Life Sci. 2025, 68, 934–960. [Google Scholar] [CrossRef]
- Allemailem, K.S.; Almatroudi, A.; Rahmani, A.H.; Alrumaihi, F.; Alradhi, A.E.; Alsubaiyel, A.M.; Algahtani, M.; Almousa, R.M.; Mahzari, A.; Sindi, A.A. Recent Updates of the CRISPR/Cas9 Genome Editing System: Novel Approaches to Regulate Its Spatiotemporal Control by Genetic and Physicochemical Strategies. Int. J. Nanomed. 2024, 31, 5335–5363. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, S.; Sheng, H.; Feng, X.; Wang, S.; Hu, Y.; Zhang, L.; Cai, B.; Ma, Y. Revolutionizing cattle breeding: Gene editing advancements for enhancing economic traits. Gene 2024, 927, 148595. [Google Scholar] [CrossRef]
- Karavolias, N.G.; Horner, W.; Abugu, M.N.; Evanega, S.N. Application of Gene Editing for Climate Change in Agriculture. Front. Sustain. Food Syst. 2021, 5, 685801. [Google Scholar] [CrossRef]
- Bhowmik, P.; Konkin, D.; Polowick, P.; Hodgins, C.L.; Subedi, M.; Xiang, D.; Yu, B.; Patterson, N.; Rajagopalan, N.; Babic, V.; et al. CRISPR/Cas9 gene editing in legume crops: Opportunities and challenges. Legume Sci. 2021, 3, e96. [Google Scholar] [CrossRef]
- Boettcher, P.; Hoffmann, I.; Baumung, R.; Drucker, A.; McManus, C.; Berg, P.; Stella, A.; Nilsen, L.; Moran, D.; Naves, M.; et al. Genetic resources and genomics for adaptation of livestock to climate change. Front. Genet. 2015, 5, 461. [Google Scholar] [CrossRef]
- Biscarini, F.; Nicolazzi, E.L.; Stella, A.; Boettcher, P.J.; Gandini, G. Challenges and opportunities in genetic improvement of local livestock breeds. Front. Genet. 2015, 6, 33. [Google Scholar] [CrossRef]
- Kalaignazhal, G.; Sejian, V.; Velayudhan, S.M.; Mishra, C.; Rebez, E.B.; Chauhan, S.S.; DiGiacomo, K.; Lacetera, N.; Dunshea, F.R. Applications of Next-Generation Sequencing Technologies and Statistical Tools in Identifying Pathways and Biomarkers for Heat Tolerance in Livestock. Vet. Sci. 2024, 11, 616. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jemaa, S.; Boussaha, M.; Mandonnet, N.; Bardou, P.; Naves, M. Uncovering structural variants in Creole cattle from Guadeloupe and their impact on environmental adaptation through whole genome sequencing. PLoS ONE 2024, 19, e0309411. [Google Scholar] [CrossRef]
- Armstrong, E.; Ciappesoni, G.; Iriarte, W.; Da Silva, C.; Macedo, F.; Navajas, E.; Brito, G.; San Julián, R.; Gimeno, D.; Postiglioni, A. Novel genetic polymorphisms associated with carcass traits in grazing Texel sheep. Meat Sci. 2018, 145, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Authority, E.F.S. Food Safety in the EU—Report; Publications Office of the European Union: Luxembourg, 2022. [Google Scholar]
- Zou, Y.-l.; Li, Z.-y.; Zou, Y.-j.; Hao, H.-y.; Hu, J.-x.; Li, N.; Li, Q.-y. Generation of pigs with a Belgian Blue mutation in MSTN using CRISPR/Cpf1-assisted ssODN-mediated homologous recombination. J. Integr. Agric. 2019, 18, 1329–1336. [Google Scholar] [CrossRef]
- Zou, Y.; Li, Z.; Zou, Y.; Hao, H.; Li, N.; Li, Q. An FBXO40 knockout generated by CRISPR/Cas9 causes muscle hypertrophy in pigs without detectable pathological effects. Biochem. Biophys. Res. Commun. 2018, 498, 940–945. [Google Scholar] [CrossRef]
- Process, D.A. Animal Drugs, Animal Biotechnology Products, and Animal Food Additives. Available online: https://www.fda.gov/animal-veterinary/development-approval-process (accessed on 12 December 2023).
- Wasmer, M. Roads Forward for European GMO Policy-Uncertainties in Wake of ECJ Judgment Have to be Mitigated by Regulatory Reform. Front. Bioeng. Biotechnol. 2019, 7, 132. [Google Scholar] [CrossRef] [PubMed]
- Gelinsky, E.; Hilbeck, A. European Court of Justice ruling regarding new genetic engineering methods scientifically justified: A commentary on the biased reporting about the recent ruling. Environ. Sci. Eur. 2018, 30, 52. [Google Scholar] [CrossRef]
- APET, A.-N. Genome Editing Policy Framework. Available online: https://amrh.nepad.org/sites/default/files/resourcefiles/Policy%20Framework%20for%20Applications%20of%20Genome%20Editing.pdf (accessed on 25 September 2022).
- Abkallo, H.M.; Arbuthnot, P.; Auer, T.O.; Berger, D.K.; Burger, J.; Chakauya, E.; Concordet, J.-P.; Diabate, A.; Di Donato, V.; Groenewald, J.-H.; et al. Making genome editing a success story in Africa. Nat. Biotechnol. 2024, 42, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Abkallo, H.M.; Hemmink, J.D.; Oduor, B.; Khazalwa, E.M.; Svitek, N.; Assad-Garcia, N.; Khayumbi, J.; Fuchs, W.; Vashee, S.; Steinaa, L. Co-Deletion of A238L and EP402R Genes from a Genotype IX African Swine Fever Virus Results in Partial Attenuation and Protection in Swine. Viruses 2022, 14, 2024. [Google Scholar] [CrossRef] [PubMed]


| Platform | Total Studies | Year Range | Dominant Topics |
|---|---|---|---|
| Elsevier | 285 | 1988–2025 | Genetic diversity, mitochondrial DNA analysis, gene polymorphisms (e.g., BMP15, GDF9 and PRLR), ovarian follicular development, milk protein synthesis (caseins, β-lactoglobulin), heat shock proteins (HSPs), IGF signalling, coat colour genetics (MC1R and KIT), QTL mapping, and CRISPR applications. |
| Springer | 142 | 1987–2025 | GS, domestication history, mitochondrial genomics, MHC-II diversity, heat stress adaptation, prion protein (PRNP) polymorphisms, GWAS for disease resistance, microbiome interactions, and transcriptome analysis. |
| MDPI | 89 | 2011–2025 | Molecular tools (SNPs, CRISPR-Cas9), epigenetics (DNA methylation), cashmere fibre traits (KAP genes), immune response genes (PTX3 and TLRs), gut microbiota studies, and GWASs for litter size and growth traits. |
| Frontiers | 47 | 2016–2025 | Coat colour genetics (FGF5 and TYRP1), immune system regulation, genomic prediction accuracy, CRISPR-mediated gene editing, climate resilience in livestock, and microbiome–host interactions. |
| Wiley | 35 | 1994–2025 | Follicle-stimulating hormone (FSH), GnRH receptors, candidate gene studies (POU1F1 and MSTN), lactation traits, keratin-associated proteins (KAPs), and myostatin (MSTN) polymorphisms. |
| BioMed Central | 28 | 2001–2025 | Mitochondrial genome diversity, SNP discovery, gene expression profiling (RNA-Seq), microbiome dynamics, and functional annotation of genomic regions. |
| PLOS | 19 | 2009–2025 | Genome-wide selection signatures, CRISPR applications in disease resistance, comparative genomics, parasite resistance (Haemonchus contortus), and transcriptomics under heat stress. |
| Oxford University Press | 15 | 1998–2025 | MHC class I/II evolution, phylogenetic studies, immune gene polymorphisms, and livestock adaptation to tropical environments. |
| Taylor & Francis | 12 | 2005–2025 | Candidate gene association studies, prolactin (PRL) gene variants, reproductive traits (litter size), and milk yield optimization. |
| Nature Research | 9 | 2002–2025 | Whole-genome sequencing (WGS), domestication genomics, functional studies of growth hormones (GH and IGF1), and evolutionary biology of ruminants. |
| Additional Sources | 755 | 1951–2025 | Foundational studies (domestication history, SNP surveys), breed-specific trait analysis (e.g., Booroola fecundity gene), disease resistance (SPP1, osteopontin), mitochondrial haplogroups, conference proceedings, and institutional reports, FAO/UN publications. |
| Species | Total QTLs (eQTLs/SNPs) | Publications | Genome Builds | Base Traits | Trait Variants | Key Traits Influenced |
|---|---|---|---|---|---|---|
| Cattle | 193,453 | 1206 | 5 | 558 | 417 | Growth, milk yield, disease resistance, and reproduction |
| Pig | 57,041 | 854 | 3 | 406 | 1088 | Meat quality, litter size, fat deposition, and immunity |
| Chicken | 29,116 | 416 | 4 | 246 | 246 | Egg production, growth rate, and feed efficiency |
| Sheep | 5417 | 289 | 4 | 178 | 264 | Wool quality, parasite resistance, and body size |
| Horse | 2482 | 129 | 2 | 71 | 14 | Athletic performance, coat colour, and skeletal traits |
| Goat | 2713 | 47 | 2 | 90 | 120 | Fibre quality, milk traits, and disease resistance |
| Rainbow Trout | 2201 | 23 | 2 | 35 | 6 | Growth rate, disease resistance, and stress tolerance |
| Species | Assembly Name | Breed/Strain | Accession Numbers | Key Features | Ref. |
|---|---|---|---|---|---|
| Cattle | ARS_UCD1.2 | Hereford | GCA_002263795.2 | 7× coverage, the combination of sequencing technologies | [123] |
| ARS_UCD2.0 | Hereford | GCA_002263795.4 | 31 chromosomes, 37,073 genes | ||
| Btau4.6 | Hereford | GCA_000000095.4 | 7-fold mixed assembly | [63,124] | |
| Btau5.0 | Hereford | GCA_000003205.6 | 95% genome coverage | ||
| UMD3.1 | Hereford | GCA_000001245.5 | Celera Assembler | [125] | |
| Chicken | GG4.0 | Red Junglefowl | GCA_000002315.2 | Initial draft assembly | [126] |
| GG5.0 | Red Junglefowl | GCA_000002315.3 | 70× PacBio coverage | [127] | |
| GRCg6a | Red Junglefowl | GCA_000002315.5 | 80× SMRT sequencing | [127] | |
| GRCg7b | White Leghorn | GCA_016699485.1 | Latest assembly | [128] | |
| Goat | CHIR1.0 | Yunnan Black | GCA_000317765.1 | Initial assembly | - |
| CHIR_ARS1 | San Clemente | GCA_001704415.1 | 50× PacBio coverage | [41] | |
| Horse | EC2.0 | - | GCA_000000165.1 | Initial draft | [64] |
| EC3.0 | - | GCA_002863925.1 | 88× coverage | ||
| Pig | SS10.2 | Duroc | GCA_000003025.4 | Initial assembly | [62,129] |
| SS11.1 | Duroc | GCA_000003025.6 | 65× PacBio reads | ||
| SS_MARC1 | Cross-bred | GCA_002844635.1 | Landrace–Duroc–Yorkshire | - | |
| Rainbow Trout | OM1.0 | Swanson | GCA_002163495.1 | Male genome | [130] |
| OM1.1 | - | GCA_013265735.3 | Doubled haploid | ||
| Sheep | OAR3.1 | Texel | GCA_000298745.1 | 75× Illumina | [44] |
| OAR4.0 | Texel | GCA_000298745.2 | Improved annotation | [61] | |
| OAR_rambo1 | Rambouillet | GCA_002742125.1 | 126× coverage | ||
| OAR_rambo2 | Rambouillet | GCA_016772045.1 | Latest assembly (includes PacBio reads) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassanine, N.N.A.M.; Saleh, A.A.; Essa, M.O.A.; Adam, S.Y.; Mohai Ud Din, R.; Rehman, S.U.; Ali, R.; Husien, H.M.; Wang, M. Candidate Genes, Markers, Signatures of Selection, and Quantitative Trait Loci (QTLs) and Their Association with Economic Traits in Livestock: Genomic Insights and Selection. Int. J. Mol. Sci. 2025, 26, 7688. https://doi.org/10.3390/ijms26167688
Hassanine NNAM, Saleh AA, Essa MOA, Adam SY, Mohai Ud Din R, Rehman SU, Ali R, Husien HM, Wang M. Candidate Genes, Markers, Signatures of Selection, and Quantitative Trait Loci (QTLs) and Their Association with Economic Traits in Livestock: Genomic Insights and Selection. International Journal of Molecular Sciences. 2025; 26(16):7688. https://doi.org/10.3390/ijms26167688
Chicago/Turabian StyleHassanine, Nada N. A. M., Ahmed A. Saleh, Mohamed Osman Abdalrahem Essa, Saber Y. Adam, Raza Mohai Ud Din, Shahab Ur Rehman, Rahmat Ali, Hosameldeen Mohamed Husien, and Mengzhi Wang. 2025. "Candidate Genes, Markers, Signatures of Selection, and Quantitative Trait Loci (QTLs) and Their Association with Economic Traits in Livestock: Genomic Insights and Selection" International Journal of Molecular Sciences 26, no. 16: 7688. https://doi.org/10.3390/ijms26167688
APA StyleHassanine, N. N. A. M., Saleh, A. A., Essa, M. O. A., Adam, S. Y., Mohai Ud Din, R., Rehman, S. U., Ali, R., Husien, H. M., & Wang, M. (2025). Candidate Genes, Markers, Signatures of Selection, and Quantitative Trait Loci (QTLs) and Their Association with Economic Traits in Livestock: Genomic Insights and Selection. International Journal of Molecular Sciences, 26(16), 7688. https://doi.org/10.3390/ijms26167688










