Epigenomics Nutritional Insights of Crocus sativus L.: Computational Analysis of Bioactive Molecules Targeting DNA Methyltransferases and Histone Deacetylases
Abstract
1. Introduction
2. Results
2.1. Evaluation of Saffron Compounds as Epigenetic Enzyme Ligands Through Docking
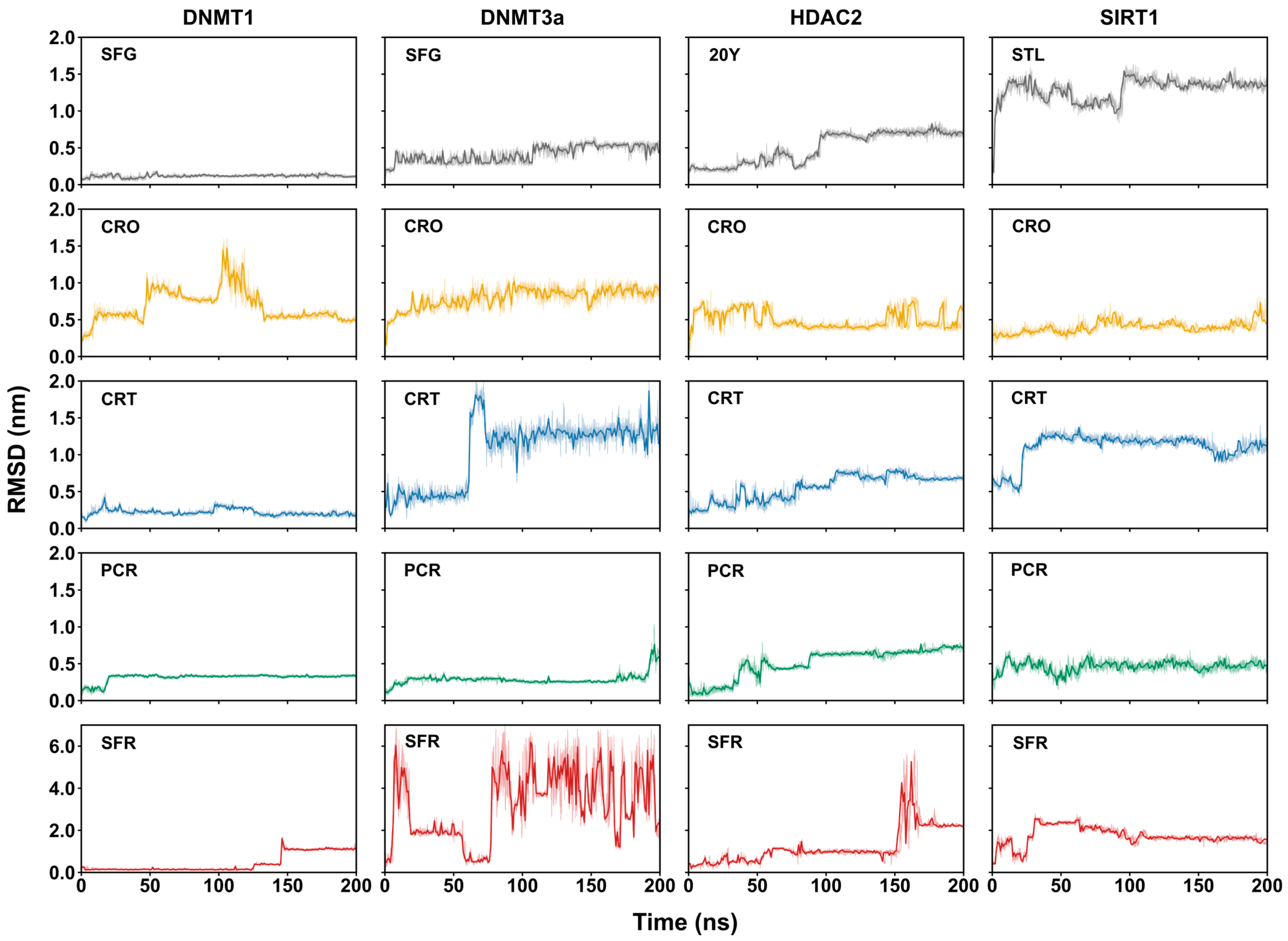
2.2. Structural Stability of Ligand–Epigenetic Target Complexes via Molecular Dynamics Simulations
2.2.1. Quality Assessment of the MD Simulations
2.2.2. Stability of Ligand-Binding Poses
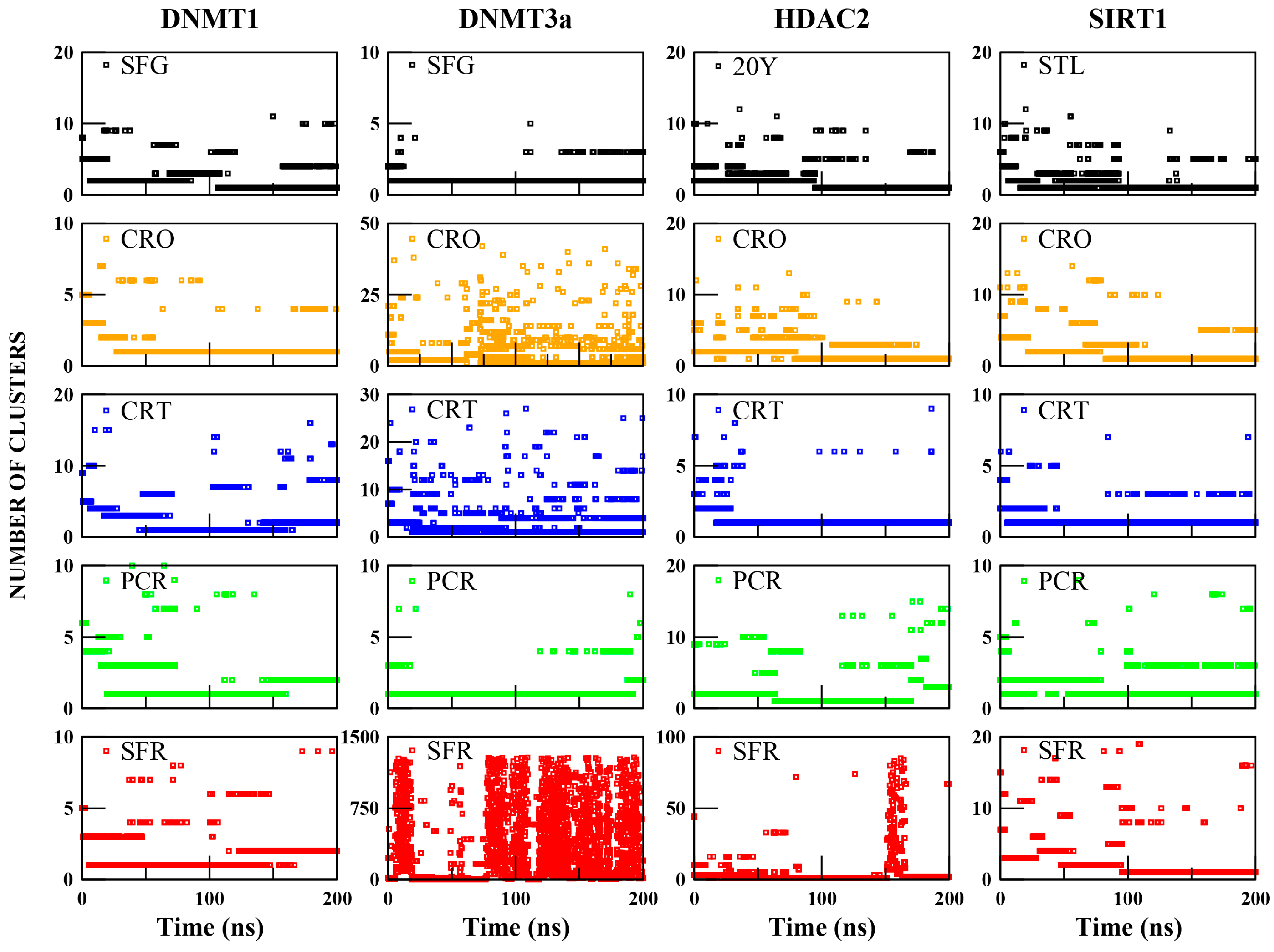
2.2.3. Binding Mode Dynamics and Conformational Variability
2.2.4. Crocin as a Promising DNMT1 Inhibitor Candidate

| DNMT1’s Residues | SFG | CRO | CRT | PCR | SFR |
|---|---|---|---|---|---|
| PRO 615 | - | 99.12 | - | - | - |
| LYS 617 | - | 88.53 | - | - | - |
| ALA 618 | - | 85.44 | - | - | - |
| PHE 1145 | 100.00 | 99.92 | - | 89.68 | - |
| SER 1146 | 99.93 | 100.00 | - | - | - |
| GLY 1147 | 98.21 | 87.24 | - | - | - |
| CYS 1148 | 95.95 | - | - | - | - |
| GLY 1149 | 97.45 | - | - | - | - |
| GLY 1150 | 99.97 | 99.41 | - | - | - |
| LEU 1151 | 95.81 | 97.69 | - | - | - |
| GLU 1168 | 100.00 | 94.53 | - | - | - |
| MET 1169 | 99.93 | 85.07 | 91.37 | 82.70 | - |
| TRP 1170 | 89.29 | - | - | - | - |
| GLU 1189 | 76.98 | - | - | - | - |
| ASP 1190 | 100.00 | 91.84 | - | - | - |
| CYS 1191 | 100.00 | - | - | - | - |
| ASN 1192 | - | 97.86 | - | - | - |
| GLY 1223 | - | 86.94 | - | 98.61 | - |
| PRO 1224 | - | - | - | 82.80 | - |
| PRO 1225 | - | - | - | 98.96 | - |
| CYS 1226 | - | - | - | 96.62 | - |
| GLN 1227 | - | - | - | 90.51 | 91.26 |
| ASN 1245 | - | 84.84 | - | - | - |
| LEU 1247 | 97.45 | - | - | - | - |
| ASN 1267 | - | - | - | 98.45 | - |
| VAL 1268 | - | - | - | 90.33 | - |
| ARG 1574 | - | - | 82.09 | - | - |
| ASN 1578 | 99.85 | 96.32 | - | 99.92 | - |
| ALA 1579 | 96.90 | 99.59 | - | - | - |
| VAL 1580 | 99.50 | 99.93 | - | - | - |
| Contacts (a) | 17 (17) | 18 (12) | 3 (1) | 11 (4) | 1 (0) |
| Residence time (b) | 200 ns | 199.93 ns | 0.00 ns | 0.00 ns | 0.00 ns |
2.2.5. Lack of Effective Binding of Saffron Compounds to DNMT3a
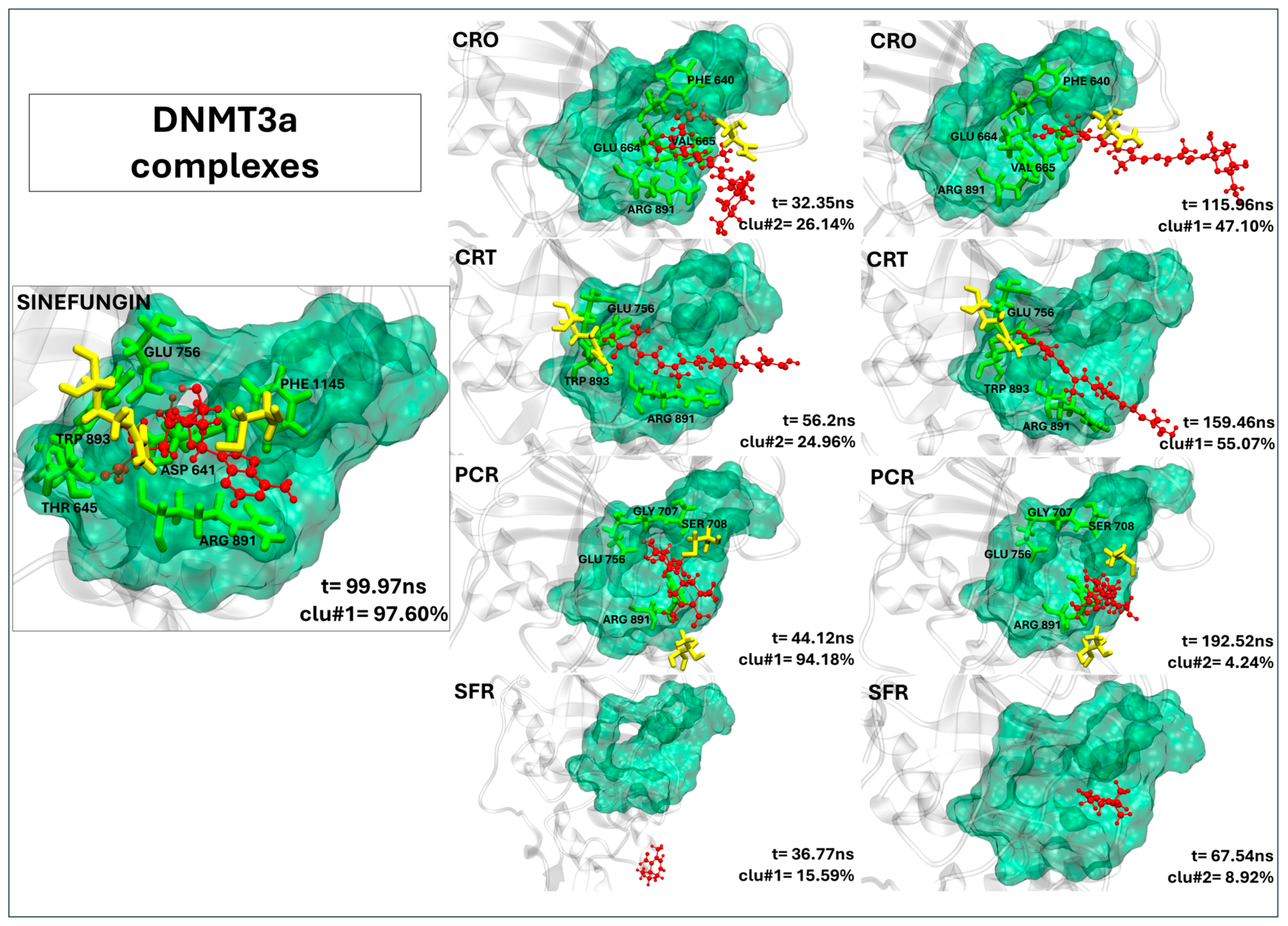
| DNMT3a’s Residues | SFG | CRO | CRT | PCR | SFR |
|---|---|---|---|---|---|
| PHE 640 | 99.65 | 95.12 | - | - | - |
| ASP 641 | 100.00 | - | - | - | - |
| ILE 643 | 98.59 | - | - | - | - |
| ALA 644 | 90.81 | - | - | - | - |
| THR 645 | 100.00 | - | - | - | - |
| GLU 664 | - | 99.83 | - | - | - |
| VAL 665 | - | 91.72 | - | - | - |
| GLY 707 | 99.14 | - | - | 91.01 | - |
| SER 708 | 99.98 | - | - | 94.95 | - |
| PRO 709 | 83.04 | - | - | - | - |
| CYS 710 | 91.56 | - | - | 83.14 | - |
| ASN 711 | - | 83.85 | - | - | - |
| GLU 756 | 100.00 | - | 98.18 | 84.22 | - |
| ASN 757 | 90.89 | - | - | - | |
| ARG 792 | 91.22 | - | 97.95 | - | - |
| THR 834 | - | - | - | 86.99 | - |
| ARG 891 | 99.53 | 97.13 | 98.34 | 91.44 | - |
| SER 892 | 99.25 | - | - | - | - |
| TRP 893 | 100.00 | - | 83.23 | - | - |
| Contacts (a) | 15 (12) | 5 (4) | 4 (3) | 6 (4) | 0 (0) |
| Residence time (b) | 194.31 ns | 0.00 ns | 40.42 ns | 0.00 ns | 0.00 ns |
2.2.6. Crocin and Crocetin Exhibit Stable and Persistent Binding Within the HDAC2 Active Site
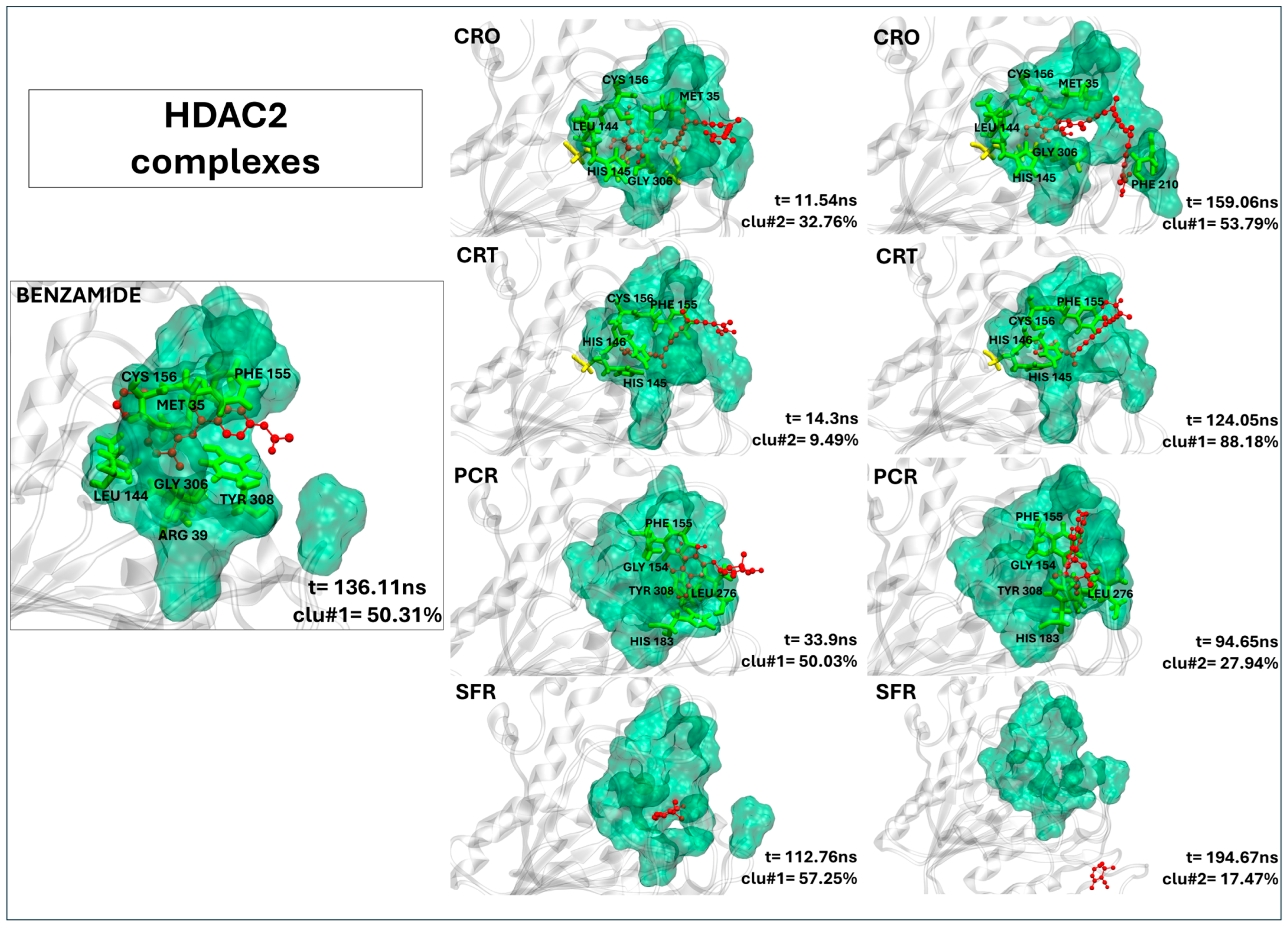
| HDAC2’s Residues | 20Y | CRO | CRT | PCR | SFR |
|---|---|---|---|---|---|
| MET 35 | 98.74 | 97.98 | 95.66 | - | - |
| ARG 39 | 89.98 | - | - | - | - |
| ASP 104 | - | - | 91.59 | - | - |
| GLY 143 | - | 90.34 | 92.04 | - | - |
| LEU 144 | 90.78 | 97.46 | - | - | - |
| HIS 145 | - | 85.45 | 98.14 | - | - |
| HIS 146 | - | - | 97.35 | - | - |
| GLY 154 | - | 97.53 | 99.69 | 93.04 | - |
| PHE 155 | 98.64 | 99.46 | 99.95 | 96.65 | - |
| CYS 156 | 93.29 | 95.75 | 92.62 | - | - |
| HIS 183 | - | - | - | 98.28 | - |
| PHE 210 | - | 88.18 | 99.95 | - | - |
| PRO 211 | - | - | 89.85 | - | - |
| LEU 276 | - | - | - | 94.36 | - |
| GLY 305 | - | 96.63 | - | - | - |
| GLY 306 | 95.80 | 99.67 | 96.98 | - | - |
| GLY 307 | - | 88.33 | - | - | - |
| TYR 308 | 95.17 | - | - | 88.64 | - |
| Contacts (a) | 7 (6) | 11 (8) | 11 (9) | 5 (5) | 0 (0) |
| Residence time (b) | 191.61 ns | 200.00 ns | 200.00 ns | 79.62 ns | 2.80 ns |
2.2.7. Crocin, Crocetin, and Picrocrocin Are Promising Candidates as SIRT1 Activators
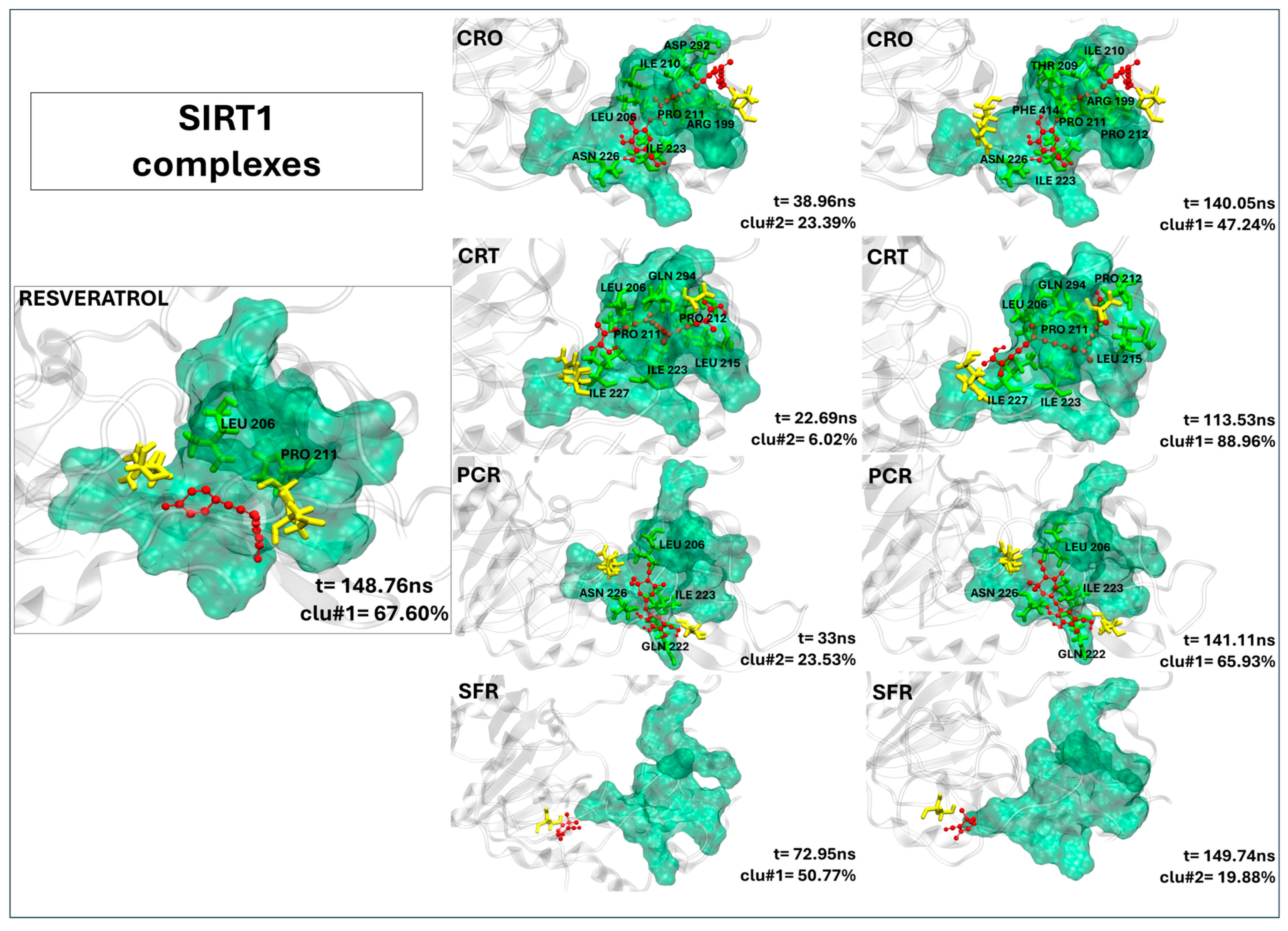
| SIRT1’s Residues | STL | CRO | CRT | PCR | SFR |
|---|---|---|---|---|---|
| ARG 199 | - | 97.32 | - | - | - |
| LEU 206 | 71.23 | 70.98 | 87.78 | 88.86 | - |
| THR 209 | - | 85.59 | 71.47 | - | - |
| ILE 210 | - | 99.92 | - | - | - |
| PRO 211 | 73.39 | 97.42 | 84.83 | - | - |
| PRO 212 | - | 94.17 | 92.52 | - | - |
| PRO 213 | - | 96.27 | - | - | |
| LEU 215 | - | - | 79.18 | - | - |
| THR 219 | - | - | - | 91.28 | - |
| GLN 222 (S2) | - | - | - | 96.48 | - |
| ILE 223 | - | 80.40 | 91.75 | 99.74 | - |
| ASN 226 (S2) | - | 77.78 | 78.98 | 98.82 | - |
| ILE 227 | - | 71.11 | 92.59 | - | - |
| ASP 292 (S3) | - | 86.70 | - | - | - |
| GLN 294 | - | 99.97 | 74.39 | - | - |
| ALA 295 | - | - | 73.05 | - | - |
| ASP 298 (S3) | - | 71.83 | - | - | - |
| PHE 414 | - | 90.03 | - | - | - |
| GLY 415 | 86.74 | - | - | - | - |
| GLU 416 | 71.04 | - | - | - | - |
| ARG 446 | 92.73 | 72.13 | 95.06 | 89.61 | - |
| SER 454 | - | - | - | - | 71.03 |
| Contacts (a) | 5 (2) | 15 (11) | 11 (9) | 6(4) | 1 (0) |
| Residence time (b) | 192.64 ns | 199.06 ns | 200.00 ns | 125.02 ns | 3.10 ns |
2.3. Binding Affinity Quantified by MM/PBSA Analysis
2.4. In Silico Toxicity Evaluation of the Candidate Saffron Ligands and in Silico ADME Assessment
3. Discussion
4. Materials and Methods
4.1. Protein Structures and Ligand Structures
4.2. Molecular Docking
4.3. Molecular Dynamics Simulations
4.4. Trajectory Analysis
4.5. MM/PBSA Calculations
4.6. Toxicity Prediction of the Ligands
4.7. ADME Profile of the Ligands
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 20Y | 4-(acetylamino)-N-[2-amino-5-(thiophen-2-yl) phenyl] benzamide |
| ACE | acetyl group |
| ACPYPE | AnteChamber PYthon Parser interfacE |
| AhR | aryl hydrocarbon receptor |
| AOP | adverse outcome pathways |
| AR | androgen receptor |
| AR-LBD | androgen receptor ligand-binding domain |
| ATAD5 | ATPase family AAA domain-containing protein 5 |
| BBB | Blood–brain barrier |
| CD | catalytic domain |
| CR | caloric restriction |
| CRO | beta-D-glucosyl trans-crocetin, crocin |
| CRT | crocetin |
| CYP450 | Cytochrome P450 |
| DNMT | DNA methyl transferase |
| EGCG | green tea epigallocatechin-3-gallate |
| ER | estrogen receptor alpha |
| ER-LBD | estrogen receptor ligand-binding domain |
| ESA | essential sirtuin-1 activity |
| FOXO | Forkhead box O |
| GHS | Globally Harmonized System |
| GI | gastrointestinal |
| HA | histone acetylation |
| HAT | histone acetyl transferase |
| HDAC | histone deacetylase |
| HSE | heat shock factor response element |
| LD50 | median lethal dose |
| MD | molecular dynamics |
| MMP | mitochondrial membrane potential |
| MM/PBSA | Molecular Mechanics/Poisson–Boltzmann Surface Area |
| NF-Kb | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NLRP3 | NOD-like receptors family pyrin domain-containing 3 |
| NME | N-methyl group |
| nrf2/ARE | nuclear factor (erythroid-derived 2)-like 2/antioxidant response element |
| NTD | N-terminal domain |
| p53 | tumor suppressor protein |
| P-gp | P-glycoprotein |
| PCR | picrocrocin |
| PME | particle mesh Ewald |
| PPAR-γ | peroxisome proliferator-activated receptor gamma |
| PTM | post-translational modification |
| RMSD | root mean square deviation |
| RMSF | root mean square fluctuation |
| SD | steepest descent |
| SFR | safranal |
| STL | resveratrol |
| TBI | traumatic brain injury |
References
- Tiffon, C. The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3425. [Google Scholar] [CrossRef]
- Ahluwalia, M.K. Nutrigenetics and Nutrigenomics-A Personalized Approach to Nutrition. Adv. Genet. 2021, 108, 277–340. [Google Scholar] [CrossRef]
- Kumari, A.; Bhawal, S.; Kapila, S.; Yadav, H.; Kapila, R. Health-Promoting Role of Dietary Bioactive Compounds through Epigenetic Modulations: A Novel Prophylactic and Therapeutic Approach. Crit. Rev. Food Sci. Nutr. 2022, 62, 619–639. [Google Scholar] [CrossRef]
- Leri, M.; Scuto, M.; Ontario, M.L.; Calabrese, V.; Calabrese, E.J.; Bucciantini, M.; Stefani, M. Healthy Effects of Plant Polyphenols: Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 1250. [Google Scholar] [CrossRef]
- Shah, D.; Gandhi, M.; Kumar, A.; Cruz-Martins, N.; Sharma, R.; Nair, S. Current Insights into Epigenetics, Noncoding RNA Interactome and Clinical Pharmacokinetics of Dietary Polyphenols in Cancer Chemoprevention. Crit. Rev. Food Sci. Nutr. 2021, 63, 1755–1791. [Google Scholar] [CrossRef]
- Monfoulet, L.-E.; Ruskovska, T.; Ajdžanović, V.; Havlik, J.; Vauzour, D.; Bayram, B.; Krga, I.; Corral-Jara, K.-F.; Kistanova, E.; Abadjieva, D.; et al. Molecular Determinants of the Cardiometabolic Improvements of Dietary Flavanols Identified by an Integrative Analysis of Nutrigenomic Data from a Systematic Review of Animal Studies. Mol. Nutr. Food Res. 2021, 65, 2100227. [Google Scholar] [CrossRef]
- Acharya, B.S.; Ghalehgolabbehbahani, A.; Hamido, S.; Zinati, G.; Bozzolo, A.; Archer, L.; Wendelberger, K.; Das, S.; Thapa, R.; Panday, D. Saffron (Crocus sativus L.): The Golden Spice—Management, Challenges, and Opportunities for Sustainable Production in the United States. J. Agric. Food Res. 2025, 21, 101970. [Google Scholar] [CrossRef]
- Khazdair, M.R.; Boskabady, M.H.; Hosseini, M.; Rezaee, R.; Tsatsakis, A.M. The Effects of Crocus Sativus (Saffron) and Its Constituents on Nervous System: A Review. Avicenna J. Phytomed 2015, 5, 376–391. [Google Scholar]
- Ghaffari, S.; Roshanravan, N. Saffron; An Updated Review on Biological Properties with Special Focus on Cardiovascular Effects. Biomed. Pharmacother. 2019, 109, 21–27. [Google Scholar] [CrossRef]
- Lambrianidou, A.; Koutsougianni, F.; Papapostolou, I.; Dimas, K. Recent Advances on the Anticancer Properties of Saffron (Crocus sativus L.) and Its Major Constituents. Molecules 2021, 26, 86. [Google Scholar] [CrossRef]
- Anaeigoudari, F.; Anaeigoudari, A.; Kheirkhah-Vakilabad, A. A Review of Therapeutic Impacts of Saffron (Crocus sativus L.) and Its Constituents. Physiol. Rep. 2023, 11, e15785. [Google Scholar] [CrossRef]
- Mokhtarian, R.; Rajabi, S.; Zahedian, S.; Jafarinejad-Farsangi, S.; Hadizadeh, M.; Sadeghinejad, M. The Effect of Saffron and Its Extracts on the Treatment of Breast Cancer: A Narrative Review-ScienceDirect. Ann. Pharm. Fr. 2024, 82, 629–640. [Google Scholar] [CrossRef]
- José Bagur, M.; Alonso Salinas, G.L.; Jiménez-Monreal, A.M.; Chaouqi, S.; Llorens, S.; Martínez-Tomé, M.; Alonso, G.L. Saffron: An Old Medicinal Plant and a Potential Novel Functional Food. Molecules 2018, 23, 30. [Google Scholar] [CrossRef]
- Elfardi, Y.R.; El Boukhari, R.; Fatimi, A.; Bouissane, L. The Multifaceted Therapeutic Potential of Saffron: An Overview Based on Research and Patents. Drugs Drug Candidates 2024, 3, 437–454. [Google Scholar] [CrossRef]
- Rashid, M.; Brim, H.; Ashktorab, H. Saffron, Its Active Components, and Their Association with DNA and Histone Modification: A Narrative Review of Current Knowledge. Nutrients 2022, 14, 3317. [Google Scholar] [CrossRef]
- Hoshyar, R.; Bathaie, S.Z.; Ashrafi, M. Interaction of Safranal and Picrocrocin with ctDNA and Their Preferential Mechanisms of Binding to GC- and AT-Rich Oligonucleotides. DNA Cell Biol. 2008, 27, 665–673. [Google Scholar] [CrossRef]
- Mykhailenko, O.; Kovalyov, V.; Goryacha, O.; Ivanauskas, L.; Georgiyants, V. Biologically Active Compounds and Pharmacological Activities of Species of the Genus Crocus: A Review. Phytochemistry 2019, 162, 56–89. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Q.; Shang, J.; Lu, L.; Chen, G. Crocin Inhibits the Migration, Invasion, and Epithelial-Mesenchymal Transition of Gastric Cancer Cells via miR-320/KLF5/HIF-1α Signaling. J. Cell. Physiol. 2019, 234, 17876–17885. [Google Scholar] [CrossRef]
- Al Aboud, N.M.; Tupper, C.; Jialal, I. Genetics, Epigenetic Mechanism. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Jirtle, R.L.; Skinner, M.K. Environmental Epigenomics and Disease Susceptibility. Nat. Rev. Genet. 2007, 8, 253–262. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Mizzen, C.A.; Allis, C.D. Linking Histone Acetylation to Transcriptional Regulation. Cell. Mol. Life Sci. CMLS 1998, 54, 6–20. [Google Scholar] [CrossRef]
- Marmorstein, R.; Zhou, M.-M. Writers and Readers of Histone Acetylation: Structure, Mechanism, and Inhibition. Cold Spring Harb. Perspect. Biol. 2014, 6, a018762. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Turner, B.M. Histone Acetylation and an Epigenetic Code. Bioessays 2000, 22, 836–845. [Google Scholar] [CrossRef]
- Hadley, M.; Noonepalle, S.; Banik, D.; Villagra, A. Functional Analysis of HDACs in Tumorigenesis. Methods Mol. Biol. 2019, 1983, 279–307. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, W.; Pan, H.; Feldser, H.G.; Lainez, E.; Miller, C.; Leung, S.; Zhong, Z.; Zhao, H.; Sweitzer, S.; et al. SIRT1 Activators Suppress Inflammatory Responses through Promotion of P65 Deacetylation and Inhibition of NF-κB Activity. PLoS ONE 2012, 7, e46364. [Google Scholar] [CrossRef]
- Lee, J.T.; Gu, W. SIRT1: Regulator of P53 Deacetylation. Genes Cancer 2013, 4, 112–117. [Google Scholar] [CrossRef]
- Wenzel, U. Nutrition, Sirtuins and Aging. Genes Nutr. 2006, 1, 85–93. [Google Scholar] [CrossRef]
- Bayod, S.; Del Valle, J.; Lalanza, J.F.; Sanchez-Roige, S.; de Luxán-Delgado, B.; Coto-Montes, A.; Canudas, A.M.; Camins, A.; Escorihuela, R.M.; Pallàs, M. Long-Term Physical Exercise Induces Changes in Sirtuin 1 Pathway and Oxidative Parameters in Adult Rat Tissues. Exp. Gerontol. 2012, 47, 925–935. [Google Scholar] [CrossRef]
- Bordone, L.; Guarente, L. Calorie Restriction, SIRT1 and Metabolism: Understanding Longevity. Nat. Rev. Mol. Cell Biol. 2005, 6, 298–305. [Google Scholar] [CrossRef]
- Boily, G.; Seifert, E.L.; Bevilacqua, L.; He, X.H.; Sabourin, G.; Estey, C.; Moffat, C.; Crawford, S.; Saliba, S.; Jardine, K.; et al. SirT1 Regulates Energy Metabolism and Response to Caloric Restriction in Mice. PLoS ONE 2008, 3, e1759. [Google Scholar] [CrossRef]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.-L.; et al. Small Molecule Activators of Sirtuins Extend Saccharomyces Cerevisiae Lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef]
- Borra, M.T.; Smith, B.C.; Denu, J.M. Mechanism of Human SIRT1 Activation by Resveratrol. J. Biol. Chem. 2005, 280, 17187–17195. [Google Scholar] [CrossRef]
- Evans, L.W.; Ferguson, B.S. Food Bioactive HDAC Inhibitors in the Epigenetic Regulation of Heart Failure. Nutrients 2018, 10, 1120. [Google Scholar] [CrossRef]
- Berger, A.; Venturelli, S.; Kallnischkies, M.; Böcker, A.; Busch, C.; Weiland, T.; Noor, S.; Leischner, C.; Weiss, T.S.; Lauer, U.M.; et al. Kaempferol, a New Nutrition-Derived Pan-Inhibitor of Human Histone Deacetylases. J. Nutr. Biochem. 2013, 24, 977–985. [Google Scholar] [CrossRef]
- Jang, Y.-G.; Hwang, K.-A.; Choi, K.-C. Rosmarinic Acid, a Component of Rosemary Tea, Induced the Cell Cycle Arrest and Apoptosis through Modulation of HDAC2 Expression in Prostate Cancer Cell Lines. Nutrients 2018, 10, 1784. [Google Scholar] [CrossRef]
- Bayat, S.; Mansoori Derakhshan, S.; Mansoori Derakhshan, N.; Shekari Khaniani, M.; Alivand, M.R. Downregulation of HDAC2 and HDAC3 via Oleuropein as a Potent Prevention and Therapeutic Agent in MCF-7 Breast Cancer Cells. J. Cell. Biochem. 2019, 120, 9172–9180. [Google Scholar] [CrossRef]
- Chouliaras, L.; van den Hove, D.L.A.; Kenis, G.; van Draanen, M.; Hof, P.R.; van Os, J.; Steinbusch, H.W.M.; Schmitz, C.; Rutten, B.P.F. Histone Deacetylase 2 in the Mouse Hippocampus: Attenuation of Age-Related Increase by Caloric Restriction. Curr. Alzheimer Res. 2013, 10, 868–876. [Google Scholar] [CrossRef][Green Version]
- Lyko, F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Lee, W.J.; Shim, J.-Y.; Zhu, B.T. Mechanisms for the Inhibition of DNA Methyltransferases by Tea Catechins and Bioflavonoids. Mol. Pharmacol. 2005, 68, 1018–1030. [Google Scholar] [CrossRef]
- Mukherjee, N.; Kumar, A.P.; Ghosh, R. DNA Methylation and Flavonoids in Genitourinary Cancers. Curr. Pharmacol. Rep. 2015, 1, 112–120. [Google Scholar] [CrossRef]
- Kadayifci, F.Z.; Zheng, S.; Pan, Y.-X. Molecular Mechanisms Underlying the Link between Diet and DNA Methylation. Int. J. Mol. Sci. 2018, 19, 4055. [Google Scholar] [CrossRef]
- Lautenschläger, M.; Sendker, J.; Hüwel, S.; Galla, H.J.; Brandt, S.; Düfer, M.; Riehemann, K.; Hensel, A. Intestinal Formation of Trans-Crocetin from Saffron Extract (Crocus sativus L.) and in Vitro Permeation through Intestinal and Blood Brain Barrier. Phytomedicine 2015, 22, 36–44. [Google Scholar] [CrossRef]
- Demurtas, O.C.; de Brito Francisco, R.; Diretto, G.; Ferrante, P.; Frusciante, S.; Pietrella, M.; Aprea, G.; Borghi, L.; Feeney, M.; Frigerio, L.; et al. ABCC Transporters Mediate the Vacuolar Accumulation of Crocins in Saffron Stigmas. Plant Cell 2019, 31, 2789–2804. [Google Scholar] [CrossRef]
- Innamorati, G.; Pierdomenico, M.; Benassi, B.; Arcangeli, C. The Interaction of DNMT1 and DNMT3A Epigenetic Enzymes with Phthalates and Perfluoroalkyl Substances: An in Silico Approach. J. Biomol. Struct. Dyn. 2023, 41, 1586–1602. [Google Scholar] [CrossRef]
- Krishnaswamy, V.K.D.; Alugoju, P.; Periyasamy, L. Multifaceted Targeting of Neurodegeneration with Bioactive Molecules of Saffron (Crocus Sativus): An In silico Evidence-Based Hypothesis. Med. Hypotheses 2020, 143, 109872. [Google Scholar] [CrossRef]
- Medoro, A.; Jafar, T.H.; Ali, S.; Trung, T.T.; Sorrenti, V.; Intrieri, M.; Scapagnini, G.; Davinelli, S. In Silico Evaluation of Geroprotective Phytochemicals as Potential Sirtuin 1 Interactors. Biomed. Pharmacother. 2023, 161, 114425. [Google Scholar] [CrossRef]
- Cao, D.; Wang, M.; Qiu, X.; Liu, D.; Jiang, H.; Yang, N.; Xu, R.-M. Structural Basis for Allosteric, Substrate-Dependent Stimulation of SIRT1 Activity by Resveratrol. Genes. Dev. 2015, 29, 1316–1325. [Google Scholar] [CrossRef]
- Ciccone, L.; Piragine, E.; Brogi, S.; Camodeca, C.; Fucci, R.; Calderone, V.; Nencetti, S.; Martelli, A.; Orlandini, E. Resveratrol-like Compounds as SIRT1 Activators. Int. J. Mol. Sci. 2022, 23, 15105. [Google Scholar] [CrossRef]
- Alonso, G.L.; Salinas, M.R.; Esteban-Infantes, F.J.; Sánchez-Fernández, M.A. Determination of Safranal from Saffron (Crocus sativus L.) by Thermal Desorption−Gas Chromatography. J. Agric. Food Chem. 1996, 44, 185–188. [Google Scholar] [CrossRef]
- Fernández-Albarral, J.A.; de Hoz, R.; Ramírez, A.I.; López-Cuenca, I.; Salobrar-García, E.; Pinazo-Durán, M.D.; Ramírez, J.M.; Salazar, J.J. Beneficial Effects of Saffron (Crocus sativus L.) in Ocular Pathologies, Particularly Neurodegenerative Retinal Diseases. Neural Regen. Res. 2020, 15, 1408–1416. [Google Scholar] [CrossRef]
- El Midaoui, A.; Ghzaiel, I.; Vervandier-Fasseur, D.; Ksila, M.; Zarrouk, A.; Nury, T.; Khallouki, F.; El Hessni, A.; Ibrahimi, S.O.; Latruffe, N.; et al. Saffron (Crocus sativus L.): A Source of Nutrients for Health and for the Treatment of Neuropsychiatric and Age-Related Diseases. Nutrients 2022, 14, 597. [Google Scholar] [CrossRef]
- Wang, C.-Z.; Ma, Q.; Kim, S.; Wang, D.H.; Shoyama, Y.; Yuan, C.-S. Effects of Saffron and Its Active Constituent Crocin on Cancer Management: A Narrative Review. Longhua Chin. Med. 2022, 5, 35. [Google Scholar] [CrossRef]
- Dobrek, L.; Głowacka, K. Depression and Its Phytopharmacotherapy—A Narrative Review. Int. J. Mol. Sci. 2023, 24, 4772. [Google Scholar] [CrossRef]
- Ashrafi, M.; Bathaie, S.Z.; Taghikhani, M.; Moosavi-Movahedi, A.A. The Effect of Carotenoids Obtained from Saffron on Histone H1 Structure and H1-DNA Interaction. Int. J. Biol. Macromol. 2005, 36, 246–252. [Google Scholar] [CrossRef]
- Bathaie, S.Z.; Bolhasani, A.; Hoshyar, R.; Ranjbar, B.; Sabouni, F.; Moosavi-Movahedi, A.-A. Interaction of Saffron Carotenoids as Anticancer Compounds with ctDNA, Oligo (dG.dC)15, and Oligo (dA.dT)15. DNA Cell Biol. 2007, 26, 533–540. [Google Scholar] [CrossRef]
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA Methyltransferases: A Novel Target for Prevention and Therapy. Front. Oncol. 2014, 4, 80. [Google Scholar] [CrossRef]
- Kritsi, E.; Christodoulou, P.; Tsiaka, T.; Georgiadis, P.; Zervou, M. A Computational Approach for the Discovery of Novel DNA Methyltransferase Inhibitors. Curr. Issues Mol. Biol. 2024, 46, 3394–3407. [Google Scholar] [CrossRef]
- Khan, M.; Hearn, K.; Parry, C.; Rashid, M.; Brim, H.; Ashktorab, H.; Kwabi-Addo, B. Mechanism of Antitumor Effects of Saffron in Human Prostate Cancer Cells. Nutrients 2024, 16, 114. [Google Scholar] [CrossRef]
- Fraga, M.F.; Agrelo, R.; Esteller, M. Cross-Talk between Aging and Cancer: The Epigenetic Language. Ann. N. Y Acad. Sci. 2007, 1100, 60–74. [Google Scholar] [CrossRef]
- Johnson, A.A.; Akman, K.; Calimport, S.R.G.; Wuttke, D.; Stolzing, A.; de Magalhães, J.P. The Role of DNA Methylation in Aging, Rejuvenation, and Age-Related Disease. Rejuvenation Res. 2012, 15, 483–494. [Google Scholar] [CrossRef]
- Pereira, B.; Correia, F.P.; Alves, I.A.; Costa, M.; Gameiro, M.; Martins, A.P.; Saraiva, J.A. Epigenetic Reprogramming as a Key to Reverse Ageing and Increase Longevity. Ageing Res. Rev. 2024, 95, 102204. [Google Scholar] [CrossRef]
- Fang, M.Z.; Chen, D.; Sun, Y.; Jin, Z.; Christman, J.K.; Yang, C.S. Reversal of Hypermethylation and Reactivation of p16INK4a, RARbeta, and MGMT Genes by Genistein and Other Isoflavones from Soy. Clin. Cancer Res. 2005, 11 (Pt 1), 7033–7041. [Google Scholar] [CrossRef]
- Nandakumar, V.; Vaid, M.; Katiyar, S.K. (−)-Epigallocatechin-3-Gallate Reactivates Silenced Tumor Suppressor Genes, Cip1/P21 and p 16 INK4a, by Reducing DNA Methylation and Increasing Histones Acetylation in Human Skin Cancer Cells. Carcinogenesis 2011, 32, 537–544. [Google Scholar] [CrossRef]
- Xiao, L.; Sun, R.; Han, Y.; Xia, L.; Lin, K.; Fu, W.; Zhong, K.; Ye, Y. NAMPT-NAD+ Is Involved in the Senescence-delaying Effects of Saffron in Aging Mice. Exp. Ther. Med. 2024, 27, 1–12. [Google Scholar] [CrossRef]
- Naeimifar, A.; Ahmad Nasrollahi, S.; Samadi, A.; Talari, R.; Sajad Ale-nabi, S.; Massoud Hossini, A.; Firooz, A. Preparation and Evaluation of Anti-Wrinkle Cream Containing Saffron Extract and Avocado Oil. J. Cosmet. Dermatol. 2020, 19, 2366–2373. [Google Scholar] [CrossRef]
- Falsini, B.; Piccardi, M.; Minnella, A.; Savastano, C.; Capoluongo, E.; Fadda, A.; Balestrazzi, E.; Maccarone, R.; Bisti, S. Influence of Saffron Supplementation on Retinal Flicker Sensitivity in Early Age-Related Macular Degeneration. Invest. Ophthalmol. Vis. Sci. 2010, 51, 6118–6124. [Google Scholar] [CrossRef]
- Bassett, S.A.; Barnett, M.P.G. The Role of Dietary Histone Deacetylases (HDACs) Inhibitors in Health and Disease. Nutrients 2014, 6, 4273–4301. [Google Scholar] [CrossRef]
- Wang, L.; Ahn, Y.J.; Asmis, R. Inhibition of Myeloid HDAC2 Upregulates Glutaredoxin 1 Expression, Improves Protein Thiol Redox State and Protects against High-Calorie Diet-Induced Monocyte Dysfunction and Atherosclerosis. Atherosclerosis 2021, 328, 23–32. [Google Scholar] [CrossRef]
- Mir, M.A.; Ganai, S.A.; Mansoor, S.; Jan, S.; Mani, P.; Masoodi, K.Z.; Amin, H.; Rehman, M.U.; Ahmad, P. Isolation, Purification and Characterization of Naturally Derived Crocetin Beta-d-Glucosyl Ester from Crocus sativus L. against Breast Cancer and Its Binding Chemistry with ER-Alpha/HDAC2. Saudi J. Biol. Sci. 2020, 27, 975–984. [Google Scholar] [CrossRef]
- Al-Hrout, A.; Chaiboonchoe, A.; Khraiwesh, B.; Murali, C.; Baig, B.; El-Awady, R.; Tarazi, H.; Alzahmi, A.; Nelson, D.R.; Greish, Y.E.; et al. Safranal Induces DNA Double-Strand Breakage and ER-Stress-Mediated Cell Death in Hepatocellular Carcinoma Cells. Sci. Rep. 2018, 8, 16951. [Google Scholar] [CrossRef]
- Colapietro, A.; Mancini, A.; Vitale, F.; Martellucci, S.; Angelucci, A.; Llorens, S.; Mattei, V.; Gravina, G.L.; Alonso, G.L.; Festuccia, C. Crocetin Extracted from Saffron Shows Antitumor Effects in Models of Human Glioblastoma. Int. J. Mol. Sci. 2020, 21, 423. [Google Scholar] [CrossRef]
- Iside, C.; Scafuro, M.; Nebbioso, A.; Altucci, L. SIRT1 Activation by Natural Phytochemicals: An Overview. Front. Pharmacol. 2020, 11, 1225. [Google Scholar] [CrossRef]
- Prasetyawan, S.; Safitri, A.; Atho’illah, M.F.; Rahayu, S. Computational Evaluation of Bioactive Compounds in Curcuma Zanthorrhiza Targeting SIRT1 and NFκB. BioTechnologia 2023, 104, 171–182. [Google Scholar] [CrossRef]
- Nezamdoost, Z.; Saghebjoo, M.; Hoshyar, R.; Hedayati, M.; Keska, A. High-Intensity Training and Saffron: Effects on Breast Cancer-Related Gene Expression. Med. Sci. Sports Exerc. 2020, 52, 1470–1476. [Google Scholar] [CrossRef]
- Abedimanesh, N.; Motlagh, B.; Abedimanesh, S.; Bathaie, S.Z.; Separham, A.; Ostadrahimi, A. Effects of Crocin and Saffron Aqueous Extract on Gene Expression of SIRT1, AMPK, LOX1, NF-κB, and MCP-1 in Patients with Coronary Artery Disease: A Randomized Placebo-Controlled Clinical Trial. Phytother. Res. 2020, 34, 1114–1122. [Google Scholar] [CrossRef]
- Shaheen, M.J.; Bekdash, A.M.; Itani, H.A.; Borjac, J.M. Saffron Extract Attenuates Neuroinflammation in rmTBI Mouse Model by Suppressing NLRP3 Inflammasome Activation via SIRT1. PLoS ONE 2021, 16, e0257211. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. Modeller: Generation and Refinement of Homology-Based Protein Structure Models. Meth. Enzymol. 2003, 374, 461–491. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminformat. 2011, 3, 33. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Kagami, L.; Wilter, A.; Diaz, A.; Vranken, W. The ACPYPE Web Server for Small-Molecule MD Topology Generation. Bioinformatics 2023, 39, btad350. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Banerjee, P.; Kemmler, E.; Dunkel, M.; Preissner, R. ProTox 3.0: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2024, 52, W513–W520. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
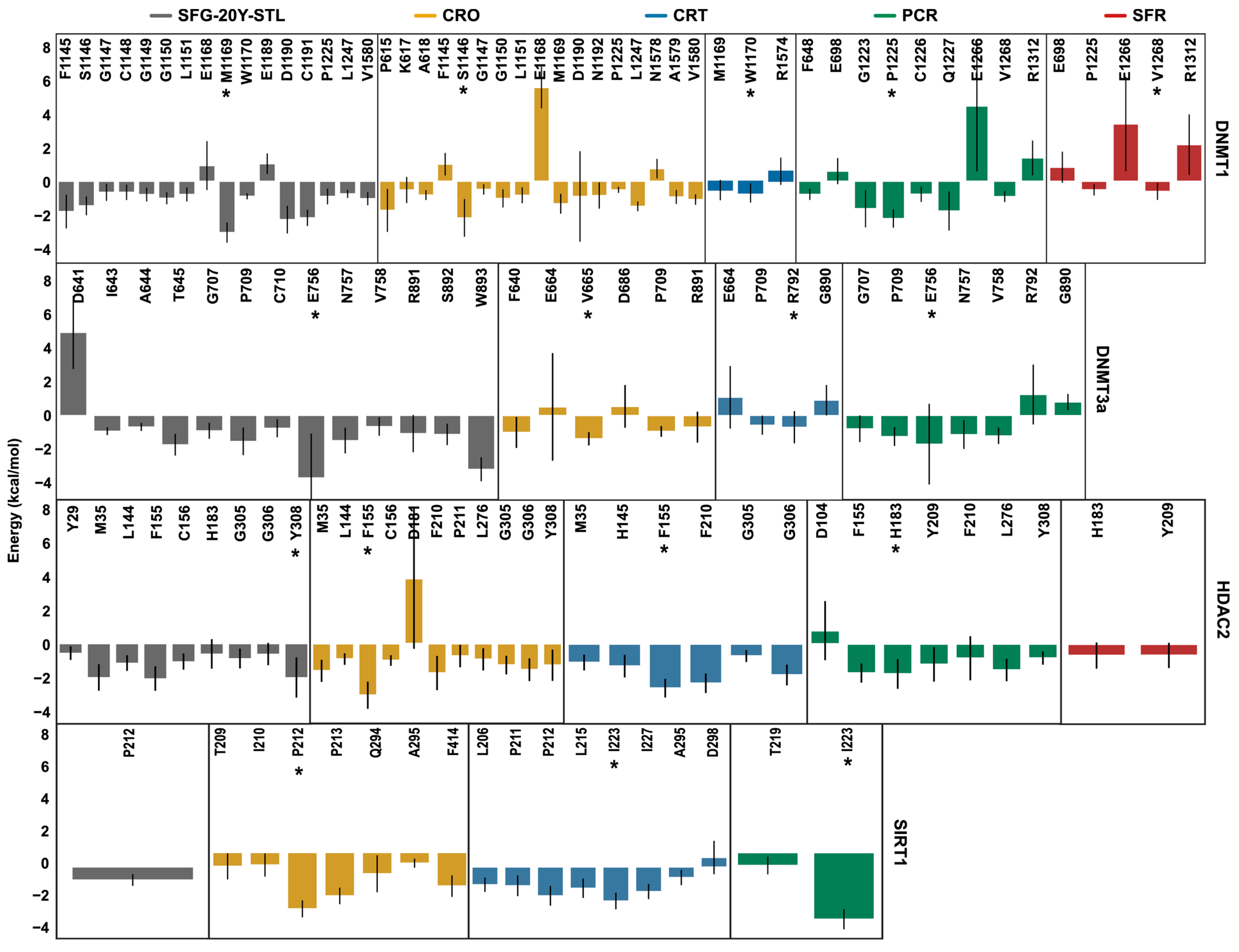
| Ligand | DNMT1 | DNMT3a | HDAC2 | SIRT1 |
|---|---|---|---|---|
| SFG/20Y/STL (a) | −9.3 | −8.1 | −8.4 | −8.0 |
| CRO (b) | −8.9 | −9.2 | −7.3 | −8.5 |
| CRT | −7.8 | −8.3 | −7.5 | −8.1 |
| PCR | −8.2 | −7.2 | −6.1 | −7.3 |
| SFR | −6.0 | −5.2 | −5.6 | −5.2 |
| Ligand | DNMT1 | DNMT3a | HDAC2 | SIRT1 |
|---|---|---|---|---|
| SFG/20Y/STL (a) | 463.60 ± 25.82 | 367.27 ± 31.44 | 149.04 ± 20.29 | 74.56 ± 22.52 |
| CRO (b) | 371.31 ± 30.59 | 174.17 ± 50.88 | 241.10 ± 24.48 | 202.24 ± 30.84 |
| CRT | 62.62 ± 39.87 | 157.67 ± 55.52 | 192.74 ± 19.97 | 144.34 ± 27.16 |
| PCR | 167.31 ± 18.01 | 119.02 ± 34.71 | 137.67 ± 32.20 | 121.69 ± 18.66 |
| SFR | 43.44 ± 17.29 | 9.11 ± 25.27 | 19.00 ± 19.20 | 6.08 ± 15.65 |
| Ligand | DNMT1 | DNMT3a | HDAC2 | SIRT1 |
|---|---|---|---|---|
| SFG/20Y/STL(a) | −47.17 ± 6.64 | −42.82 ± 9.22 | −42.48 ± 5.96 | −25.80 ± 2.66 |
| CRO (b) | −42.39 ± 8.24 | −21.96 ± 7.43 | −45.00 ± 8.14 | −50.17 ± 4.50 |
| CRT | −21.31 ± 6.38 | −21.47 ± 4.53 | −47.23 ± 4.12 | −34.49 ± 5.62 |
| PCR | −25.69 ± 7.66 | −20.03 ± 7.56 | −25.67 ± 5.70 | −23.31 ± 2.15 |
| SFR | −9.8 ± 4.50 | −3.61 ± 4.17 | −14.42 ± 6.01 | −16.51 ± 3.16 |
| SFG | 20Y | STL | CRO | CRT | PCR | SFR | |
|---|---|---|---|---|---|---|---|
| Predicted Toxicity Class | |||||||
| LD50 (mg/kg) | 1000 | 1500 | 1560 | 5600 | 4300 | 55 | 5000 |
| Class | IV | IV | IV | VI | V | III | V |
| Organ Toxicity | |||||||
| Hepatotoxicity | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
| Neurotoxicity | Active | Active | Inactive | Inactive | Inactive | Inactive | Active |
| Nephrotoxicity | Active | Inactive | Active | Inactive | Active | Active | Inactive |
| Respiratory toxicity | Active | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| Cardiotoxicity | Inactive | Inactive | Active | Inactive | Inactive | Active | Inactive |
| Endpoint Toxicity | |||||||
| Carcinogenicity | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| Immunotoxicity | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| Mutagenicity | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
| Cytotoxicity | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| Nuclear Receptor Pathway | |||||||
| AhR | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
| AR | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Inactive |
| AR-LBD | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| Aromatase | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| ER | Inactive | Active | Active | Inactive | Inactive | Inactive | Inactive |
| ER-LBD | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Inactive |
| PPAR-Gamma | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| Stress Response Pathway | |||||||
| nrf2/ARE | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| HSE | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| MMP | Inactive | Active | Active | Inactive | Inactive | Inactive | Inactive |
| p53 | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| ATAD5 | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Inactive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piergentili, A.; Saraceni, P.R.; Demurtas, O.C.; Benassi, B.; Arcangeli, C. Epigenomics Nutritional Insights of Crocus sativus L.: Computational Analysis of Bioactive Molecules Targeting DNA Methyltransferases and Histone Deacetylases. Int. J. Mol. Sci. 2025, 26, 7575. https://doi.org/10.3390/ijms26157575
Piergentili A, Saraceni PR, Demurtas OC, Benassi B, Arcangeli C. Epigenomics Nutritional Insights of Crocus sativus L.: Computational Analysis of Bioactive Molecules Targeting DNA Methyltransferases and Histone Deacetylases. International Journal of Molecular Sciences. 2025; 26(15):7575. https://doi.org/10.3390/ijms26157575
Chicago/Turabian StylePiergentili, Alessia, Paolo Roberto Saraceni, Olivia Costantina Demurtas, Barbara Benassi, and Caterina Arcangeli. 2025. "Epigenomics Nutritional Insights of Crocus sativus L.: Computational Analysis of Bioactive Molecules Targeting DNA Methyltransferases and Histone Deacetylases" International Journal of Molecular Sciences 26, no. 15: 7575. https://doi.org/10.3390/ijms26157575
APA StylePiergentili, A., Saraceni, P. R., Demurtas, O. C., Benassi, B., & Arcangeli, C. (2025). Epigenomics Nutritional Insights of Crocus sativus L.: Computational Analysis of Bioactive Molecules Targeting DNA Methyltransferases and Histone Deacetylases. International Journal of Molecular Sciences, 26(15), 7575. https://doi.org/10.3390/ijms26157575








