Ribosomal RNA-Specific Antisense DNA and Double-Stranded DNA Trigger rRNA Biogenesis and Insecticidal Effects on the Insect Pest Coccus hesperidum
Abstract
1. Introduction
2. Results
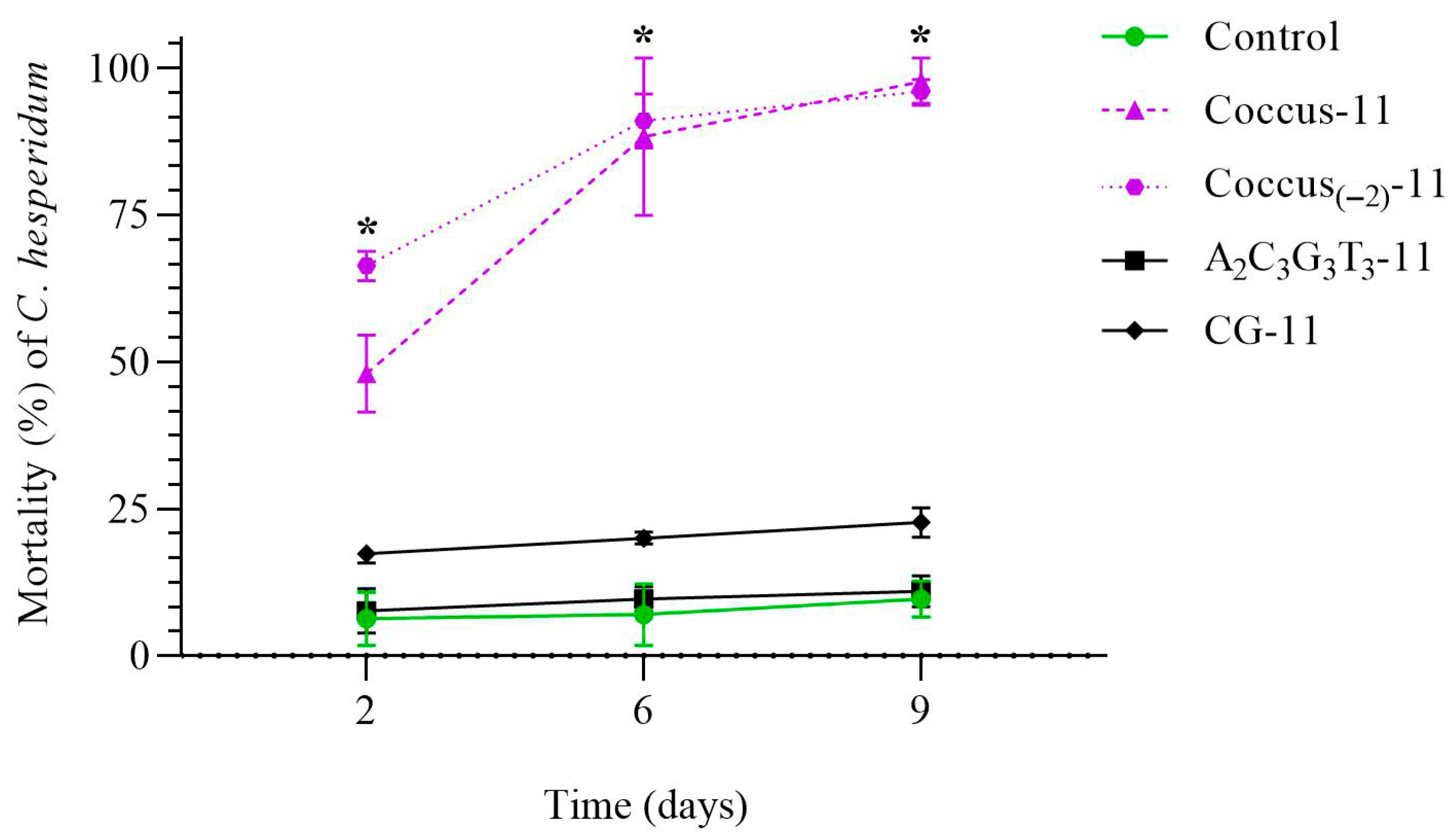
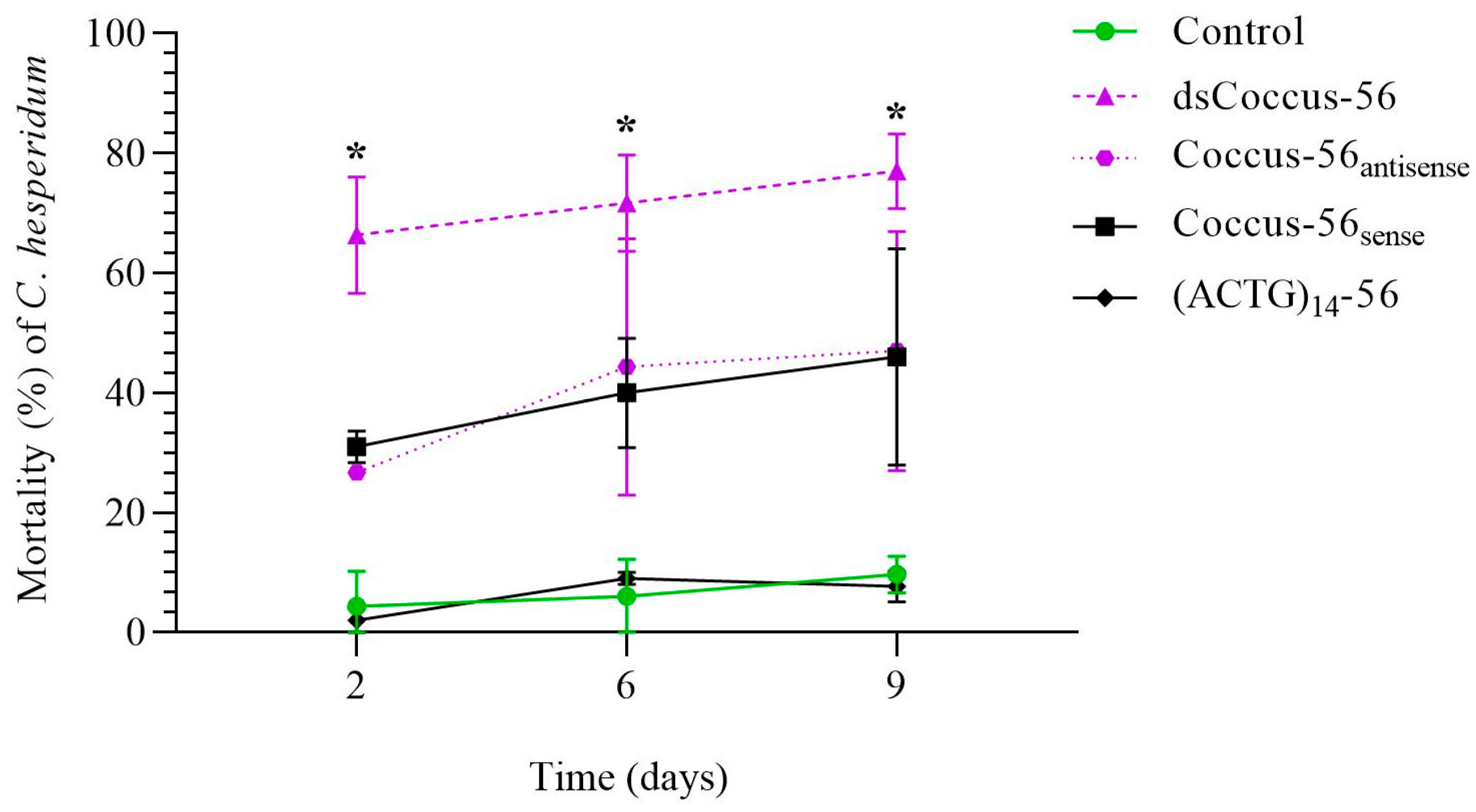
2.1. Mortality of C. hesperidum After Treatment with Single-Stranded and Double-Stranded DNA Sequences in the Natural Habitat
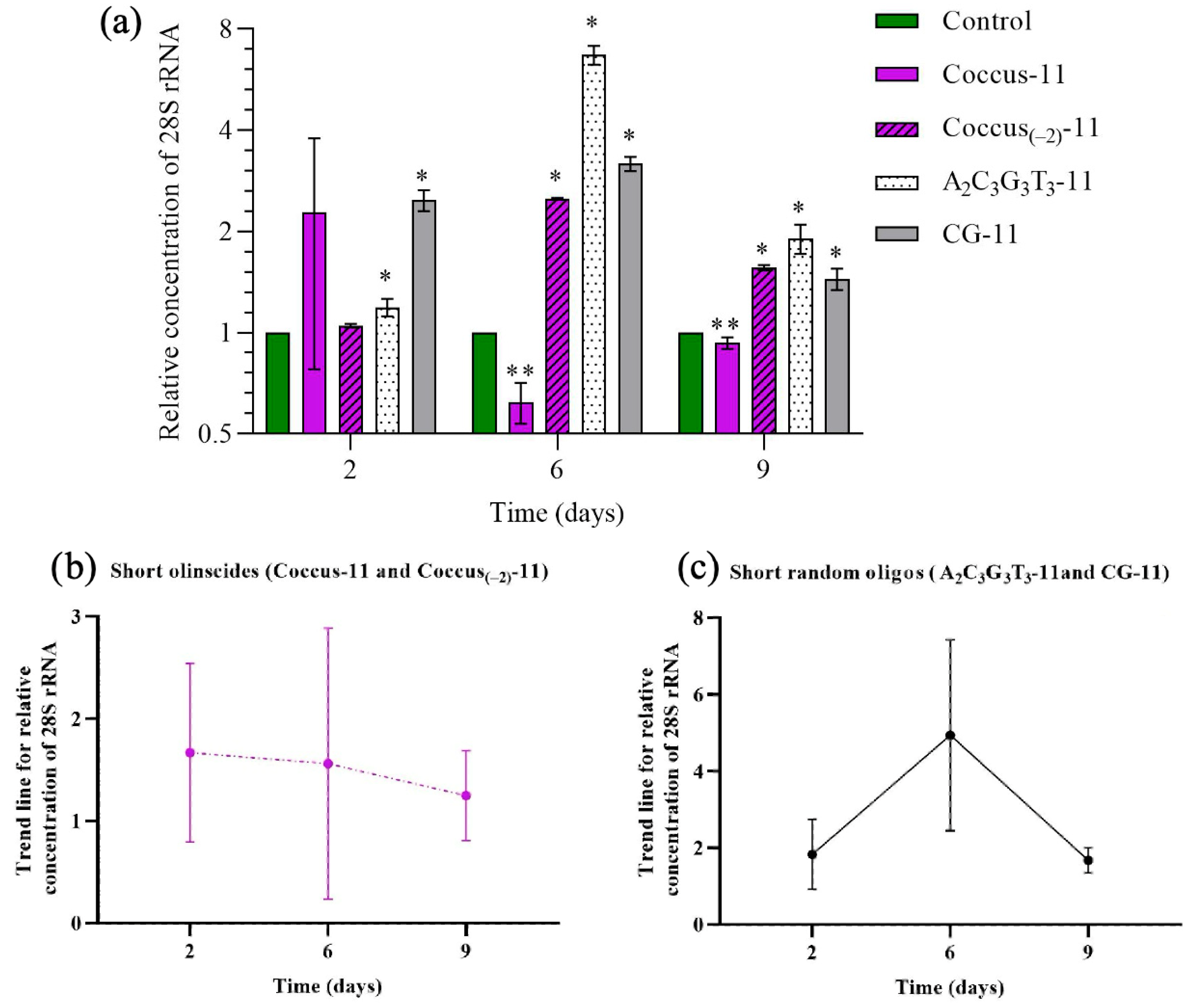
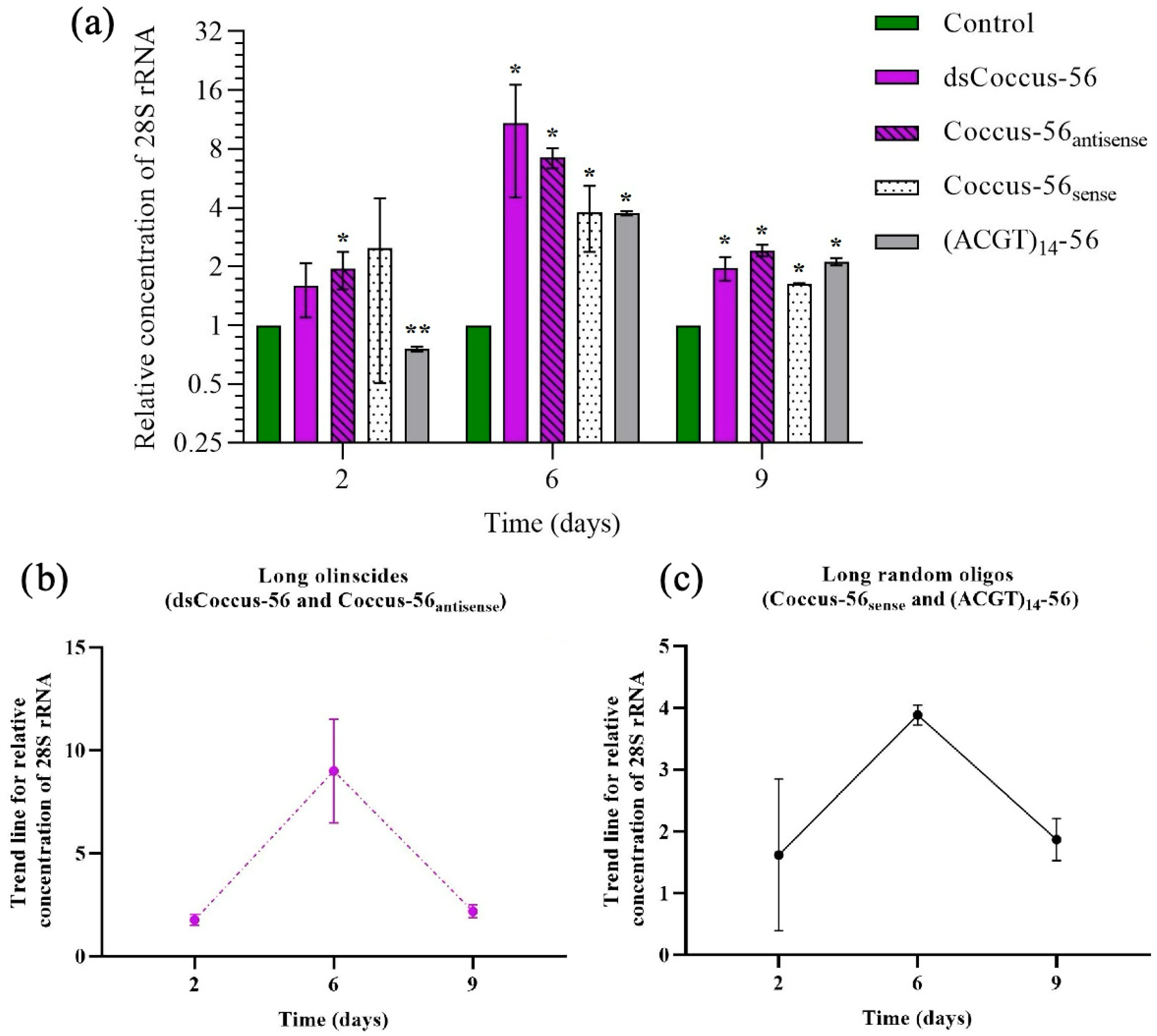
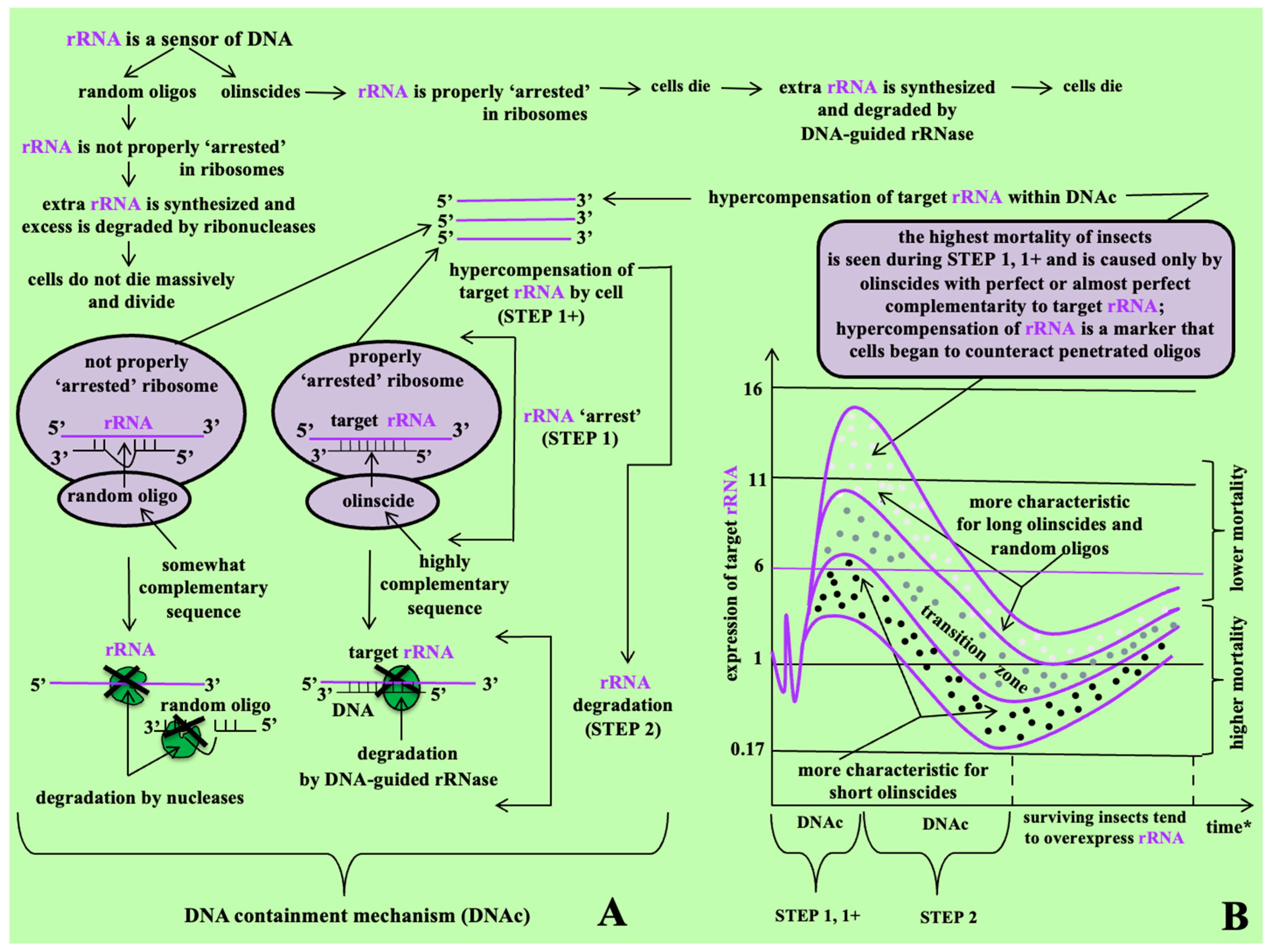
2.2. Target rRNA Expression of C. hesperidum During DNAc
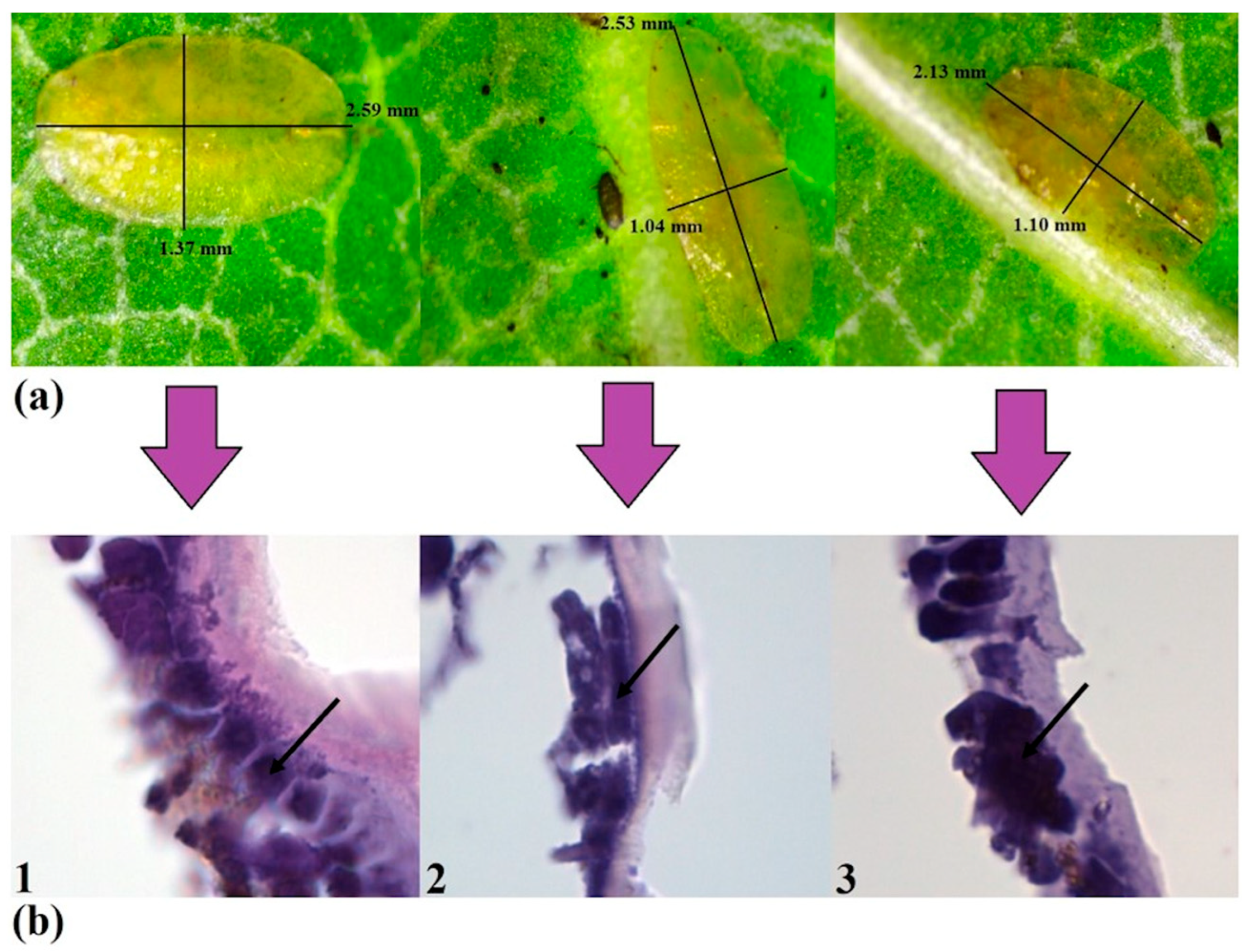
2.3. Histological Studies
2.4. Oligonucleotide Insecticides Are Predominantly Contact Insecticides
2.5. Differential Gene Expression Analysis (DGE) After Contact Application of Coccus-11
3. Discussion
3.1. Sequences of DNA Oligos Highly Complementary to rRNA (Olinscides)
3.2. Sequences of DNA Oligos Somewhat Complementary to rRNA (Random Oligos)
3.3. Horizons of Fundamental Understanding and Practical Application of DNAc Mechanism
4. Materials and Methods
4.1. Origin of C. hesperidum L.
4.2. Sequences and Applied Short (Coccus-11; Coccus(−2)-11) and Long (dsCoccus-56; Coccus-56antisense) Olinscides
4.3. Sequences and Applied Short (A2C3G3T3-11; CG-11) and Long (Coccus-56sense; (ACGT)14-56) Random Oligos
4.4. Synthesis of Oligonucleotides
4.5. Evaluation of 28S rRNA Expression of C. hesperidum
4.6. Histochemical Assay
4.7. Differential Gene Expression (DGE) Analysis
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, H.; Li, J.; Zhang, C.; Chen, J. Hemipteran-transmitted plant viruses: Research progress and control strategies. Front. Agric. Sci. Eng. 2022, 9, 98–109. [Google Scholar] [CrossRef]
- Wu, W.; Shan, H.W.; Li, J.M.; Zhang, C.X.; Chen, J.P.; Mao, Q. Roles of Bacterial Symbionts in Transmission of Plant Virus by Hemipteran Vectors. Front. Microbiol. 2022, 13, 805352. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; An, J.J.; Dang, Z.H.; Pan, W.L.; Gao, Z.L. Systemic control efficacy of neonicotinoids seeds dressing on English grain aphid (Hemiptera: Aphididae). J. Asia-Pac. Entomol. 2018, 21, 430–435. [Google Scholar] [CrossRef]
- Bacci, L.; Convertini, S.; Rossaro, B. A review of sulfoxaflor, a derivative of biological acting substances as a class of insecticides with a broad range of action against many insect pests. J. Entomol. Acarol. Res. 2018, 50, 7836. [Google Scholar] [CrossRef]
- Zhou, C.S.; Lv, H.H.; Guo, X.H.; Cao, Q.; Zhang, R.X.; Ma, Y. Transcriptional analysis of Bemisia tabaci MEAM1 cryptic species under the selection pressure of neonicotinoids imidacloprid, acetamiprid and thiamethoxam. BMC Genom. 2022, 23, 15. [Google Scholar] [CrossRef]
- Elmquist, J.; Biddinger, D.; Phan, N.T.; Moural, T.W.; Zhu, F.; Hoover, K. Potential risk to pollinators from neonicotinoid applications to host trees for management of spotted lanternfly, Lycorma delicatula (Hemiptera: Fulgoridae). J. Econ. Entomol. 2023, 116, 368–378. [Google Scholar] [CrossRef]
- Calvo-Agudo, M.; González-Cabrera, J.; Picó, Y.; Calatayud-Vernich, P.; Urbaneja, A.; Dicke, M.; Tena, A. Neonicotinoids in excretion product of phloem-feeding insects kill beneficial insects. Proc. Natl. Acad. Sci. USA 2019, 116, 16817–16822. [Google Scholar] [CrossRef]
- Siddiqui, J.A.; Fan, R.; Naz, H.; Bamisile, B.S.; Hafeez, M.; Ghani, M.I.; Wei, Y.; Xu, Y.; Chen, X. Insights into insecticide-resistance mechanisms in invasive species: Challenges and control strategies. Front. Physiol. 2023, 13, 1112278. [Google Scholar] [CrossRef]
- Bavithra, C.M.L.; Murugan, M.; Pavithran, S.; Naveena, K. Enthralling genetic regulatory mechanisms meddling insecticide resistance development in insects: Role of transcriptional and post-transcriptional events. Front. Mol. Biosci. 2023, 10, 1257859. [Google Scholar] [CrossRef]
- Oberemok, V.V. Method of Elimination of Phyllophagous Insects from Order Lepidoptera. Ukraine Patent UA 36 445, 27 October 2008. [Google Scholar]
- Oberemok, V.V.; Laikova, E.V.; Andreeva, O.A.; Gal’chisnky, N.V. Oligonucleotide insecticides and RNA-based insecticides: 16 years of experience in contact using of the next generation pest control agents. J. Plant. Dis. Prot. 2024. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Gal’chinsky, N.V. Oligonucleotide insecticides (contact unmodified antisense DNA biotechnology) and RNA biocontrols (double-stranded RNA technology): Newly born fraternal twins in plant protection. bioRxiv 2024. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Novikov, I.A.; Yatskova, E.V.; Bilyk, A.I.; Sharmagiy, A.K.; Gal’chinsky, N.V. Potent and selective ‘genetic zipper’ method for DNA-programmable plant protection: Innovative oligonucleotide insecticides against Trioza alacris Flor. Chem. Biol. Technol. Agric. 2024, 11, 144. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Laikova, K.V.; Gal’chinsky, N.V. Contact Unmodified Antisense DNA (CUAD) Biotechnology: List of Pest Species Successfully Targeted by Oligonucleotide Insecticides. Front. Agron. 2024, 6, 1415314. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Puzanova, Y.V.; Gal’chinsky, N.V. The ‘genetic zipper’ method offers a cost-effective solution for aphid control. Front. Insect Sci. 2024, 4, 1467221. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Useinov, R.Z.; Skorokhod, O.A.; Gal’chinsky, N.V.; Novikov, I.A.; Makalish, T.P.; Yatskova, E.V.; Sharmagiy, A.K.; Golovkin, I.O.; Gninenko, Y.V.; et al. Oligonucleotide Insecticides for Green Agriculture: Regulatory Role of Contact DNA in Plant-Insect Interactions. Int. J. Mol. Sci. 2022, 23, 15681. [Google Scholar] [CrossRef]
- Kumar, H.; Gal’chinsky, N.; Sweta, V.; Negi, N.; Filatov, R.; Chandel, A.; Ali, J.; Oberemok, V.; Laikova, K. Perspectives of RNAi, CUADb and CRISPR/Cas as Innovative Antisense Technologies for Insect Pest Control: From Discovery to Practice. Insects 2025, 16, 746. [Google Scholar] [CrossRef]
- Gal’chinsky, N.V.; Yatskova, E.V.; Novikov, I.A.; Sharmagiy, A.K.; Plugatar, Y.V.; Oberemok, V.V. Mixed insect pest populations of Diaspididae species under control of oligonucleotide insecticides: 3′-end nucleotide matters. Pestic. Biochem. Physiol. 2024, 200, 105838. [Google Scholar] [CrossRef]
- Barrangou, R.; Birmingham, A.; Wiemann, S.; Beijersbergen, R.L.; Hornung, V.; Smith, A.V. Advances in CRISPR-Cas9 genome engineering: Lessons learned from RNA interference. Nucleic Acids Res. 2015, 43, 3407–3419. [Google Scholar] [CrossRef]
- Liu, H.; Chen, W.; Li, Y.; Sun, L.; Chai, Y.; Chen, H.; Nie, H.; Huang, C. CRISPR/Cas9 Technology and Its Utility for Crop Improvement. Int. J. Mol. Sci. 2022, 23, 10442. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell. Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Laikova, K.V.; Zaitsev, A.S.; Shumskykh, M.N.; Kasich, I.N.; Gal’chinsky, N.V.; Bekirova, V.V.; Makarov, V.V.; Agranovsky, A.A.; Gushchin, V.A.; et al. Molecular Alliance of Lymantria dispar Multiple Nucleopolyhedrovirus and a Short Unmodified Antisense Oligonucleotide of Its Anti-Apoptotic IAP-3 Gene: A Novel Approach for Gypsy Moth Control. Int. J. Mol. Sci. 2017, 18, 2446. [Google Scholar] [CrossRef]
- Rawlinson, S.M.; Zhao, T.; Rozario, A.M.; Rootes, C.L.; McMillan, P.J.; Purcell, A.W.; Woon, A.; Marsh, G.A.; Lieu, K.G.; Wang, L.F.; et al. Viral regulation of host cell biology by hijacking of the nucleolar DNA-damage response. Nat. Commun. 2018, 9, 3057. [Google Scholar] [CrossRef]
- Kondo, T.; Watson, G.W. Encyclopedia of Scale Insect Pests; CABI: Wallingford, UK, 2022; pp. 249–251. [Google Scholar] [CrossRef]
- Boukouvala, M.C.; Kavallieratos, N.G.; Skourti, A.; Pons, X.; Alonso, C.L.; Eizaguirre, M.; Fernandez, E.B.; Solera, E.D.; Fita, S.; Bohinc, T.; et al. Lymantria dispar (L.) (Lepidoptera: Erebidae): Current Status of Biology, Ecology, and Management in Europe with Notes from North America. Insects 2022, 13, 854. [Google Scholar] [CrossRef]
- Manju, M.; Nirosha, V.; Tullika, T.; Mankhanniang, G. DNA insecticides: An emerging tool in pest management. Agriallis 2022, 4, 29–35. [Google Scholar]
- Du, Q.; Thonberg, H.; Wang, J.; Wahlestedt, C.; Liang, Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005, 33, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Luige, O.; Karalė, K.; Bose, P.P.; Bollmarkm, M.; Tedebark, U.; Murtola, M.; Strömberg, R. Influence of sequence variation on the RNA cleavage activity of Zn2+-dimethyl-dppz-PNA-based artificial enzymes. RSC Adv. 2022, 12, 5398–5406. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.G.; Sanghvi, Y.S. Liquid-Phase Oligonucleotide Synthesis: Past, Present, and Future Predictions. Curr. Protoc. Nucleic Acid. Chem. 2019, 77, e82. [Google Scholar] [CrossRef]
- Brzezinska, J.; Trzciński, S.; Strzelec, J.; Chmielewski, M.K. From CPG to hybrid support: Review on the approaches in nucleic acids synthesis in various media. Bioorg. Chem. 2023, 140, 106806. [Google Scholar] [CrossRef]
- Gal’chinsky, N.V.; Yatskova, E.V.; Novikov, I.A.; Useinov, R.Z.; Kouakou, N.J.; Kouame, K.F.; Kra, K.D.; Sharmagiy, A.K.; Plugatar, Y.V.; Laikova, K.V.; et al. Icerya purchasi Maskell (Hemiptera: Monophlebidae) Control Using Low Carbon Footprint Oligonucleotide Insecticides. Int. J. Mol. Sci. 2023, 24, 11650. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Sharma, M.; Chandel, A. DNA Insecticides: Future of Crop Protection. Agric. Food e-Newsl. 2022, 4, 551. [Google Scholar]
- Patil, V.; Jangra, S.; Ghosh, A. Advances in Antisense Oligo Technology for Sustainable Crop Protection. Crit. Rev. Plant Sci. 2024, 43, 405–427. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Gal’chinsky, N.V.; Useinov, R.Z.; Novikov, I.A.; Puzanova, Y.V.; Filatov, R.I.; Kouakou, N.J.; Kouame, K.F.; Kra, K.D.; Laikova, K.V. Four most pathogenic superfamilies of insect pests of suborder sternorrhyncha: Invisible superplunderers of plant vitality. Insects 2023, 14, 462. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Laikova, K.V.; Gal’chinsky, N.V.; Useinov, R.Z.; Novikov, I.A.; Temirova, Z.Z.; Shumskykh, M.N.; Krasnodubets, A.M.; Repetskaya, A.I.; Dyadichev, V.V.; et al. DNA insecticide developed from the Lymantria dispar 5.8S ribosomal RNA gene provides a novel biotechnology for plant protection. Sci. Rep. 2019, 9, 6197. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Laikova, K.; Shumskykh, M.; Kenyo, I.; Kasich, I.; Deri, K.; Seidosmanova, E.; Krasnodubets, K.; Bekirova, V.; Gal’chinsky, N. A primary attempt of Leptinotarsa decemlineata control using contact DNA insecticide based on short antisense oligonucleotide of its CYP6B gene. J. Plant. Prot. Res. 2018, 58, 106–108. [Google Scholar]
- Gavrilova, D.; Grizanova, E.; Novikov, I.; Laikova, E.; Zenkova, A.; Oberemok, V.; Dubovskiy, I. Antisense DNA acaricide targeting pre-rRNA of two-spotted spider mite Tetranychus urticae as efficacy-enhancing agent of fungus Metarhizium robertsii. J. Invertebr. Pathol. 2025, 211, 108297. [Google Scholar] [CrossRef]
- Oberemok, V.V. Development of DNA Insecticides: A Modern Approach. Available online: https://www.researchgate.net/publication/380727180_Development_of_DNA_insecticides_a_modern_approach_Razrabotka_DNK-insekticidov_sovremennyj_podhod (accessed on 25 May 2024).
- Lomate, P.; Bonning, B. Distinct properties of proteases and nucleases in the gut, salivary gland and saliva of southern green stink bug, Nezara viridula. Sci. Rep. 2016, 6, 27587. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Laikova, K.V.; Repetskaya, A.I.; Kenyo, I.M.; Gorlov, M.V.; Kasich, I.N.; Krasnodubets, A.M.; Gal’chinsky, N.V.; Fomochkina, I.I.; Zaitsev, A.S.; et al. A Half-Century History of Applications of Antisense Oligonucleotides in Medicine, Agriculture and Forestry: We Should Continue the Journey. Molecules 2018, 23, 1302. [Google Scholar] [CrossRef]
- Zaitsev, A.; Omel’chenko, O.; Nyadar, P.; Oberemok, V. Influence of DNA oligonucleotides used as insecticides on biochemical parameters of Quercus robur and Malus domestica. Bull. Transilvania Univ. Brasov. 2015, 8, 37–46. [Google Scholar]
- Oberemok, V.V.; Nyadar, P.; Zaytsev, O.; Levchenko, N.; Shiyntum, H.; Omelchenko, O. Pioneer evaluation of the possible side effects of the DNA insecticides on wheat (Triticum aestivum L.). Int. J. Biochem. Biophys. 2013, 1, 57–63. [Google Scholar] [CrossRef]
- Nyadar, P.M.; Oberemok, V.; Omelchenko, A.; Kerimova, S.; Seidosmanova, E.; Krasnodubiets, A.; Shumskykh, M.; Bekirova, V. DNA insecticides: The effect of concentration on non-target plant organisms such as wheat (Triticum aestivum L.). J. Plant Prot. Res. 2019, 59, 60–68. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Laikova, V.K.; Zaitsev, S.A.; Nyadar, M.P.; Shumskykh, N.M.; Gninenko, I.Y. DNA insecticides based on iap3 gene fragments of cabbage looper and gypsy moth nuclear polyhedrosis viruses show selectivity for non-target insects. Arch. Biol. Sci. 2015, 67, 785–792. [Google Scholar] [CrossRef]
- Aliakbarpour, H.; Salmah, M.R.C.; Salehi, L. Determination of suitable host plant for rearing of Coccus hesperidum (Homoptera: Coccidae). Entomol. Fenn. 2010, 21, 84–89. [Google Scholar] [CrossRef]
- Lin, J.; Zhou, D.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef]
- Yi, X.; Tesmer, V.M.; Savre-Train, I.; Shay, J.W.; Wright, W.E. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 1999, 19, 3989–3997. [Google Scholar] [CrossRef]
- Bachellerie, J.P.; Cavaille, J.; Huttenhofer, A. The expanding snoRNA world. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef]
- Will, C.L.; Luhrmann, R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 2001, 13, 290–301. [Google Scholar] [CrossRef]
- Ortiz-Hidalgo, C.; Pina-Oviedo, S. Hematoxylin: Mesoamerica’s Gift to Histopathology. Palo de Campeche (Logwood Tree), Pirates’ Most Desired Treasure, and Irreplaceable Tissue Stain. Int. J. Surg. Pathol. 2019, 27, 4–14. [Google Scholar] [CrossRef]
- Tudi, M.; Daniel Ruan, H.; Wang, L.; Lyu, J.; Sadler, R.; Connell, D.; Chu, C.; Phung, D.T. Agriculture Development, Pesticide Application and Its Impact on the Environment. Int. J. Environ. Res. Public Health. 2021, 18, 1112. [Google Scholar] [CrossRef]
- Szaflarski, W.; Leśniczak-Staszak, M.; Sowiński, M.; Ojha, S.; Aulas, A.; Dave, D.; Malla, S.; Anderson, P.; Ivanov, P.; Lyons, S.M. Early rRNA processing is a stress-dependent regulatory event whose inhibition maintains nucleolar integrity. Nucleic Acids Res. 2022, 50, 1033–1051. [Google Scholar] [CrossRef]
- Lockhart, A.; Pires, V.B.; Bento, F.; Kellner, V.; Luke-Glaser, S.; Yakoub, G.; Ulrich, H.D.; Luke, B. RNase H1 and H2 Are Differentially Regulated to Process RNA-DNA Hybrids. Cell Rep. 2019, 29, 2890–2900.e5. [Google Scholar] [CrossRef]
- Dörner, K.; Ruggeri, C.; Zemp, I.; Kutay, U. Ribosome biogenesis factors-from names to functions. EMBO J. 2023, 42, e112699. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Osellame, L.D.; Blacker, T.S.; Duchen, M.R. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 2, 711–723. [Google Scholar] [CrossRef]
- Jonckheere, A.I.; Smeitink, J.A.; Rodenburg, R.J. Mitochondrial ATP synthase: Architecture, function and pathology. J. Inherit. Metab. Dis. 2012, 35, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Lipid Metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040576. [Google Scholar] [CrossRef]
- Kim, J.W.; Dang, C.V. Multifaceted roles of glycolytic enzymes. Trends Biochem. Sci. 2005, 30, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.-Y.; Zhang, Y.-H.; Xiao, A.-F.; Zhang, A.-H.; Fang, B.-S. Key Enzymes in Fatty Acid Synthesis Pathway for Bioactive Lipids Biosynthesis. Front. Nutr. 2022, 9, 851402. [Google Scholar] [CrossRef] [PubMed]
- Hyjek, M.; Figiel, M.; Nowotny, M. RNases H: Structure and mechanism. DNA Repair 2019, 84, 102672. [Google Scholar] [CrossRef]
- Puzanova, Y.V.; Novikov, I.A.; Bilyk, A.I.; Sharmagiy, A.K.; Plugatar, Y.V.; Oberemok, V.V. Perfect Complementarity Mechanism for Aphid Control: Oligonucleotide Insecticide Macsan-11 Selectively Causes High Mortality Rate for Macrosiphoniella sanborni Gillette. Int. J. Mol. Sci. 2023, 24, 11690. [Google Scholar] [CrossRef]
- Wang, W.; Nag, S.; Zhang, X.; Wang, M.H.; Wang, H.; Zhou, J.; Zhang, R. Ribosomal proteins and human diseases: Pathogenesis, molecular mechanisms, and therapeutic implications. Med. Res. Rev. 2015, 35, 225–285. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Pagano, M.; Russo, G.; Russo, A. Ribosome Biogenesis and Cancer: Overview on Ribosomal Proteins. Int. J. Mol. Sci. 2021, 22, 5496. [Google Scholar] [CrossRef]
- Bononi, A.; Agnoletto, C.; De Marchi, E.; Marchi, S.; Patergnani, S.; Bonora, M.; Giorgi, C.; Missiroli, S.; Poletti, F.; Rimessi, A.; et al. Protein kinases and phosphatases in the control of cell fate. Enzyme Res. 2011, 2011, 329098. [Google Scholar] [CrossRef]
- Pang, K.; Wang, W.; Qin, J.X.; Shi, Z.D.; Hao, L.; Ma, Y.Y.; Xu, H.; Wu, Z.X.; Pan, D.; Chen, Z.S.; et al. Role of protein phosphorylation in cell signaling, disease, and the intervention therapy. MedComm 2022, 3, e175. [Google Scholar] [CrossRef]
- Wang, X.-W.; Blanc, S. Insect transmission of plant single-stranded DNA viruses. Annu. Rev. Entomol. 2021, 66, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Gai, X.; Xin, D.; Wu, D.; Wang, X.; Chen, L.; Wang, Y.; Ma, K.; Li, Q.; Li, P.; Yu, X. Pre-ribosomal RNA reorganizes DNA damage repair factors in nucleus during meiotic prophase and DNA damage response. Cell Res. 2022, 32, 254–268. [Google Scholar] [CrossRef]
- Oberemok, V.V. Ecological Basis for Controlling the Number of Leaf-Eating Insects Using DNA Insecticides. Available online: https://www.dissercat.com/content/ekologicheskie-osnovy-kontrolya-chislennosti-listogryzushchikh-nasekomykh-s-primeneniem-dnk (accessed on 25 May 2024).
- Cerio, R.J.; Vandergaast, R.; Friesen, P.D. Host insect inhibitor-of-apoptosis SfIAP functionally replaces baculovirus IAP but is differentially regulated by its N-terminal leader. J. Virol. 2010, 84, 11448–11460. [Google Scholar] [CrossRef]
- Guo, Y.; Ji, N.; Bai, L.; Ma, J.; Li, Z. Aphid viruses: A brief view of a Long history. Front. Insect. Sci. 2022, 2, 846716. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.M.; Selvam, S.; Fuchs, G. Fatal attraction: The roles of ribosomal proteins in the viral life cycle. Wiley Interdiscip. Rev. RNA 2021, 12, e1613. [Google Scholar] [CrossRef]
- Iuchi, H.; Kawasaki, J.; Kubo, K.; Fukunaga, T.; Hokao, K.; Yokoyama, G.; Ichinose, A.; Suga, K.; Hamada, M. Bioinformatics approaches for unveiling virus-host interactions. Comput. Struct. Biotechnol. J. 2023, 21, 1774–1784. [Google Scholar] [CrossRef]
- Evseev, P.V.; Landysheva, Y.G.; Landyshev, N.N.; Ignatov, A.N. Presence of rRNA-like regions in Genbank viral sequences. In Proceedings of the 2021 IEEE Ural-Siberian Conference on Computational Technologies in Cognitive Science, Genomics and Biomedicine (CSGB), Novosibirsk-Yekaterinburg, Russia, 26–28 May 2021; pp. 310–314. [Google Scholar] [CrossRef]
- Deng, Z.L.; Münch, P.C.; Mreches, R.; McHardy, A.C. Rapid and accurate identification of ribosomal RNA sequences via deep learning. Nucleic Acids Res. 2022, 50, e60. [Google Scholar] [CrossRef]
- Salvetti, A.; Greco, A. Viruses and the nucleolus: The fatal attraction. Biochim. Biophys. Acta. 2024, 1842, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Araújo, M.F.; Castanheira, E.M.S.; Sousa, S.F. The Buzz on Insecticides: A Review of Uses, Molecular Structures, Targets, Ad-verse Effects, and Alternatives. Molecules 2023, 28, 3641. [Google Scholar] [CrossRef] [PubMed]
- Lazarević-Pašti, T.; Milanković, V.; Tasić, T.; Petrović, S.; Leskovac, A. With or Without You?-A Critical Review on Pesticides in Food. Foods 2025, 14, 1128. [Google Scholar] [CrossRef] [PubMed]
- Nayak, M.K.; Daglish, G.J.; Phillips, T.W.; Ebert, P.R. Resistance to the fumigant phosphine and its management in insect pests of stored products: A global perspective. Annu. Rev. Entomol. 2020, 65, 333–350. [Google Scholar] [CrossRef]
- Sparks, T.C.; Storer, N.; Porter, A.; Slater, R.; Nauen, R. Insecticide resistance management and industry–The origins and evolution of the Insecticide Resistance Action Committee (IRAC) and the mode of action classification scheme. Pest Manag. Sci. 2021, 77, 2609–2619. [Google Scholar] [CrossRef]
- Barathi, S.; Sabapathi, N.; Kandasamy, S.; Lee, J. Present status of insecticide impacts and eco-friendly approaches for remediation-a review. Environ. Res. 2024, 240, 117432. [Google Scholar] [CrossRef]
- Gul, H.; Gadratagi, B.G.; Güncan, A.; Tyagi, S.; Ullah, F.; Desneux, N.; Liu, X. Fitness costs of resistance to insecticides in insects. Front. Physiol. 2023, 14, 1238111. [Google Scholar] [CrossRef]
- Lehmann, P.; Ammunét, T.; Barton, M.; Battisti, A.; Eigenbrode, S.D.; Jepsen, J.U.; Kalinkat, G.; Neuvonen, S.; Niemelä, P.; Terblanche, J.S.; et al. Complex responses of global insect pests to climate warming. Front. Ecol. Environ. 2020, 18, 141–150. [Google Scholar] [CrossRef]
- Skendžić, S.; Zovko, M.; Živković, I.P.; Lešić, V.; Lemić, D. The Impact of Climate Change on Agricultural Insect Pests. Insects 2021, 12, 440. [Google Scholar] [CrossRef]
- Hawkins, N.J.; Bass, C.; Dixon, A.; Neve, P. The evolutionary origins of pesticide resistance. Biol. Rev. Camb. Philos. Soc. 2019, 94, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Yainna, S.; Nègre, N.; Silvie, P.J.; Brévault, T.; Tay, W.T.; Gordon, K.; dAlençon, E.; Walsh, T.; Nam, K. Geographic Monitoring of Insecticide Resistance Mutations in Native and Invasive Populations of the Fall Armyworm. Insects 2021, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Lavine, L.; O’Neal, S.; Lavine, M.; Foss, C.; Walsh, D. Insecticide Resistance and Management Strategies in Urban Ecosystems. Insects 2016, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X.; Zhou, A.; Zhang, J.; Ge, X.; Moussian, B.; Yan, C.; Gao, S.; Wang, Y. Trends and emerging hotspots in RNAi-based arthropod pest control: A comprehensive bibliometric analysis. J. Insect Physiol. 2025, 161, 104754. [Google Scholar] [CrossRef]
- Qi, H.-Y.; Zhang, D.-D.; Liu, B.; Chen, J.-Y.; Han, D.; Wang, D. Leveraging RNA interference technology for selective and sustainable crop protection. Front. Plant Sci. 2024, 15, 1502015. [Google Scholar] [CrossRef]
- De Schutter, K.; Taning, C.N.T.; Van Daele, L.; Van Damme, E.J.M.; Dubruel, P.; Smagghe, G. RNAi-Based Biocontrol Products: Market Status, Regulatory Aspects, and Risk Assessment. Front. Insect Sci. 2022, 1, 818037. [Google Scholar] [CrossRef]
- Islam, M.T.; Sherif, S.M. RNAi-Based Biofungicides as a Promising Next-Generation Strategy for Controlling Devastating Gray Mold Diseases. Int. J. Mol. Sci. 2020, 21, 2072. [Google Scholar] [CrossRef]
- Parker, K.M.; Barragán Borrero, V.; van Leeuwen, D.M.; Lever, M.A.; Mateescu, B.; Sander, M. Environmental Fate of RNA Interference Pesticides: Adsorption and Degradation of Double-Stranded RNA Molecules in Agricultural Soils. Environ. Sci. Technol. 2019, 53, 3027–3036. [Google Scholar] [CrossRef]
- Menezes, P.S.; Yan, Y.; Yang, Y.; Mitter, N.; Mahony, T.J.; Mody, K.T. RNAi-Based Biocontrol of Pests to Improve the Productivity and Welfare of Livestock Production. Appl. Biosci. 2022, 1, 229–243. [Google Scholar] [CrossRef]
- Fletcher, S.J.; Reeves, P.T.; Hoang, B.T.; Mitter, N. A Perspective on RNAi-Based Biopesticides. Front. Plant Sci. 2020, 11, 51. [Google Scholar] [CrossRef]
- Basso, M.F.; Vásquez, D.D.N.; Campos-Pinto, E.R.; Pinheiro, D.H.; Cruz, B.; Maktura, G.C.; Guidelli, G.V.; Marques-Souza, H.; Grossi-de-Sa, M.F. Progress and Opportunities of In Planta and Topical RNAi for the Biotechnological Control of Agricultural Pests. Agronomy 2025, 15, 859. [Google Scholar] [CrossRef]
- Joaquim, M.E.S.; Belchior, G.G.; José, M.O.M.A.; Zapata, F.; Jiang, C.; Fischer, J.; Berger, G.U. Dissipation of DvSnf7 double-stranded RNA in Brazilian soils. Agric. Environ. Lett. 2019, 4, 190016. [Google Scholar] [CrossRef]
- Koo, J.; Palli, S.R. Recent advances in understanding of the mechanisms of RNA interference in insects. Insect Mol. Biol. 2024, 34, 491–504. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E.A. Programmable dual-RNA-guided DNA en-donuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Kumari, R.; Saha, T.; Kumar, P.; Singh, A.K. CRISPR/Cas9-mediated genome editing technique to control fall armyworm (Spodoptera frugiperda) in crop plants with special reference to maize. Physiol. Mol. Biol. Plants 2024, 30, 1161–1173. [Google Scholar] [CrossRef]
- Gouda, M.N.R.; Jeevan, H.; Shashank, H.G. CRISPR/Cas9: A cutting-edge solution for combatting the fall armyworm, Spodoptera frugiperda. Mol. Biol. Rep. 2023, 51, 13. [Google Scholar] [CrossRef]
- El-Awaad, E.; Merzendorfer, H. CRISPR/Cas: An emerging molecular technology for biological control of fall armyworm. New Plant Prot. 2025, 2, e26. [Google Scholar] [CrossRef]
- Komal, J.; Desai, H.R.; Samal, I.; Mastinu, A.; Patel, R.D.; Kumar, P.V.D.; Majhi, P.K.; Mahanta, D.K.; Bhoi, T.K. Unveiling the Genetic Symphony: Harnessing CRISPR-Cas Genome Editing for Effective Insect Pest Management. Plants 2023, 12, 3961. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, L.; Wei, L.; Wang, Y.; Han, Z. Advancements and Future Prospects of CRISPR-Cas-Based Population Replacement Strategies in Insect Pest Management. Insects 2024, 15, 653. [Google Scholar] [CrossRef] [PubMed]
- Moon, T.T.; Maliha, I.J.; Khan, A.A.M.; Chakraborty, M.; Uddin, M.S.; Amin, M.R.; Islam, T. CRISPR-Cas Genome Editing for Insect Pest Stress Management in Crop Plants. Stresses 2022, 2, 493–514. [Google Scholar] [CrossRef]
- Singh, S.; Rahangdale, S.; Pandita, S.; Saxena, G.; Upadhyay, S.K.; Mishra, G.; Verma, P.C. CRISPR/Cas9 for Insect Pests Management: A Comprehensive Review of Advances and Applications. Agriculture 2022, 12, 1896. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, M.; Zeng, J.; Sun, H.; Wei, X.; Jiang, H.; Shentu, X.; Sun, D. CRISPR/Cas Technology in Insect Insecticide Resistance. Insects 2025, 16, 345. [Google Scholar] [CrossRef]
- Kumar, R.; Shanker, R.; Singh, P.; Yadav, M.K.; Chaudhary, V.; Kumar, M. Genome editing towards pests and disease management in agricultural crops: Recent developments, challenges and future prospects. Physiol. Mol. Plant Pathol. 2024, 134, 102402. [Google Scholar] [CrossRef]
- Smith, D.R. Agarose gel electrophoresis. Methods Mol. Biol. 1993, 18, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Khehra, N.; Padda, I.S.; Swift, C.J. Polymerase Chain Reaction (PCR). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK589663/ (accessed on 12 June 2025).

| Adaxial Side of the Leaf | Abaxial Side of the Leaf | ||||
|---|---|---|---|---|---|
| Control | A2C3G3T3-11 | Coccus-11 | Control | A2C3G3T3-11 | Coccus-11 |
| 7.2 ± 1.9% | 7.9 ± 5.5% | 31.8 ± 7.1% * | 7.5 ± 2.4% | 16.6 ± 6.8% | 85.3± 8.9% * |
| Protein | Expression (Upregulated or Downregulated) | p-Value * |
|---|---|---|
| Ribosomal proteins | ||
| 60S subunit of ribosome RP-L19e; RP-L8e; RP-LP1; RP-L6e; RP-L34e; RP-L10e; RP-L4e; RP-L27e; L13; RP-L32e; RP-L17e | upregulated | p < 0.05 |
| 40S subunit of ribosome RP-S18e; RP-S3Ae; RP-S8e; RP-S9e; RP-S11e; RP-S30e; RP-S3e; RP-S24e; RP-S15e; RP-S4e; RP-S21e | upregulated | p < 0.05 |
| Ribosome biogenesis proteins | ||
| NOP53; UTP30; NSA2; MAK21; BRX1; WDR12 | upregulated | p < 0.04 |
| Proteins involved in regulation of ribosome biogenesis | ||
| mTOR; RAPTOR | downregulated | p < 0.01 |
| RNase H1 | ||
| rnhA | upregulated | p < 0.001 |
| Glycolytic enzymes | ||
| Pyruvate kinase; pyruvate carboxylase; 6-phosphofructokinase 1 | downregulated | p < 0.02 |
| Enzymes involved in production of energy from lipids | ||
| PNLIP; LYPLA3; LIPA; SPLA2; PLA2G | upregulated | p < 0.001 |
| Mitochondrial ATP synthase complex (complex V) proteins | ||
| ATPeF1B; ATPeF1G; ATPeF0D; ATPeF0E; ATPeF0F; ATPeF0F; ATPeFG; ATPeF0O | upregulated | p < 0.05 |
| Mitochondrial proteins involved in energy production | ||
| pckA; ACADS; ACADSB; GCDH, ACOX1, ADHFE1; subunits of cytochrome c oxidase (VIIc; COX4; Vb; COX5A; Via; VIc/VIIs; COX5B); subunits of NADH dehydrogenase (NDUFA2; NDUFB10; NDUFA13; NDUFS4; NDUFS5; NDUFA12; NDUFV3; NDUFAB1; NDUFV2; NDUFA5; NDUFS8; NDUFB4; NDUFS6; NDUFS7; NDUFA10; NDUFA7; NDUFB11; NDUFS3; NDUFA8; NDUFAF4; NDUFA11; NDUFB6) | upregulated | p < 0.05 |
| Kinases | ||
| PINK1; MAP2K4; PLK1; WEE1; TLK; ADRBK; MAP2K1; BUB1; FLT1; dgkA; AAK; PIP5K; TNK2; STK24; CDK12; EPS8; MUSK; SRPK1; STK11; MAPKAPK2; PDPK1; FRK | downregulated | p < 0.01 |
| RNAi pathway enzymes | ||
| DICER1, Argonaute 2, DROSHA | downregulated | p < 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oberemok, V.; Gal’chinsky, N.; Novikov, I.; Sharmagiy, A.; Yatskova, E.; Laikova, E.; Plugatar, Y. Ribosomal RNA-Specific Antisense DNA and Double-Stranded DNA Trigger rRNA Biogenesis and Insecticidal Effects on the Insect Pest Coccus hesperidum. Int. J. Mol. Sci. 2025, 26, 7530. https://doi.org/10.3390/ijms26157530
Oberemok V, Gal’chinsky N, Novikov I, Sharmagiy A, Yatskova E, Laikova E, Plugatar Y. Ribosomal RNA-Specific Antisense DNA and Double-Stranded DNA Trigger rRNA Biogenesis and Insecticidal Effects on the Insect Pest Coccus hesperidum. International Journal of Molecular Sciences. 2025; 26(15):7530. https://doi.org/10.3390/ijms26157530
Chicago/Turabian StyleOberemok, Vol, Nikita Gal’chinsky, Ilya Novikov, Alexander Sharmagiy, Ekaterina Yatskova, Ekaterina Laikova, and Yuri Plugatar. 2025. "Ribosomal RNA-Specific Antisense DNA and Double-Stranded DNA Trigger rRNA Biogenesis and Insecticidal Effects on the Insect Pest Coccus hesperidum" International Journal of Molecular Sciences 26, no. 15: 7530. https://doi.org/10.3390/ijms26157530
APA StyleOberemok, V., Gal’chinsky, N., Novikov, I., Sharmagiy, A., Yatskova, E., Laikova, E., & Plugatar, Y. (2025). Ribosomal RNA-Specific Antisense DNA and Double-Stranded DNA Trigger rRNA Biogenesis and Insecticidal Effects on the Insect Pest Coccus hesperidum. International Journal of Molecular Sciences, 26(15), 7530. https://doi.org/10.3390/ijms26157530







