Transcriptional Analysis of Spodoptera frugiperda Sf9 Cells Infected with Daphnis nerii Cypovirus-23
Abstract
1. Introduction
2. Results
2.1. Viral Infection of Cells
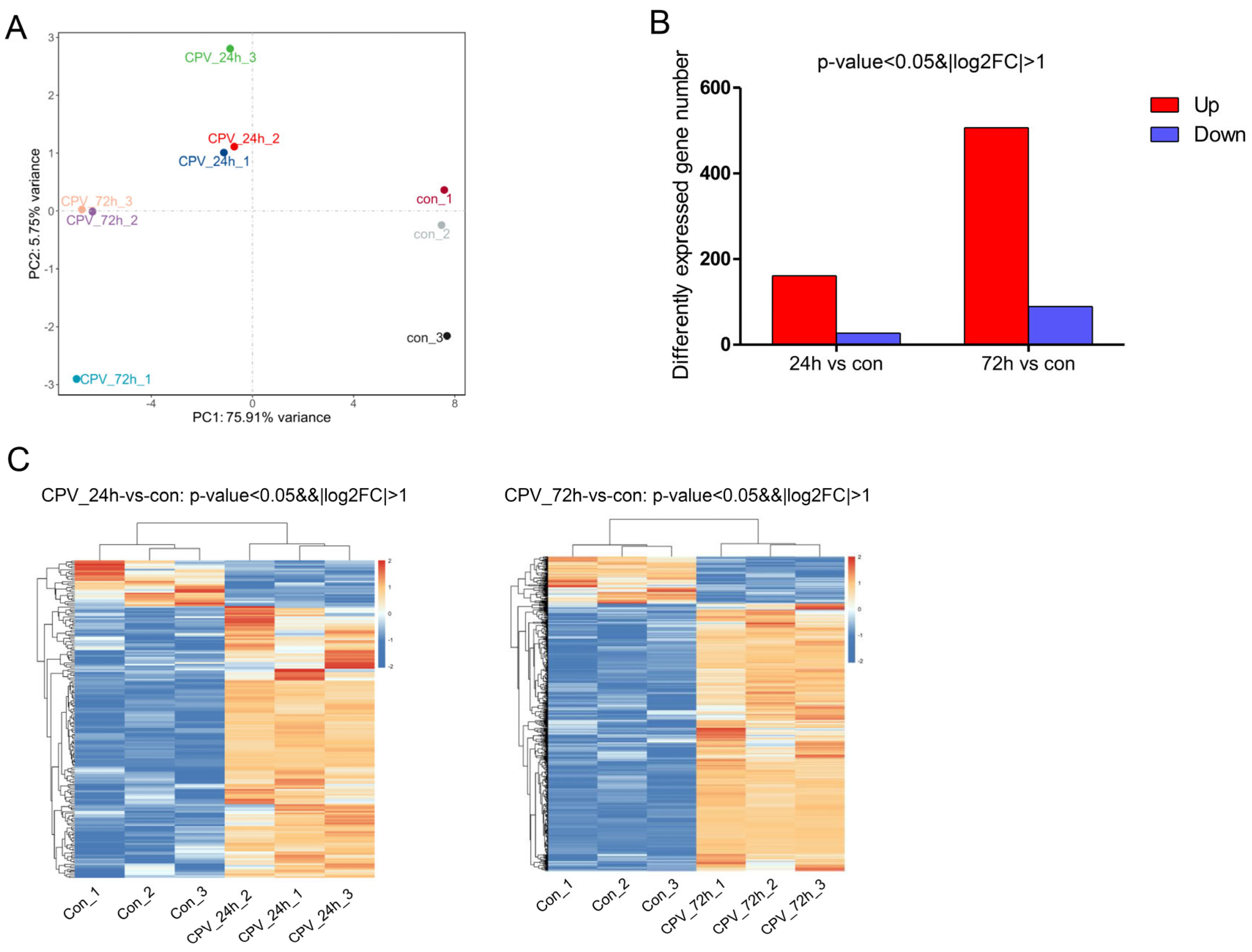
2.2. Overview of Transcriptome Sequencing
2.3. Effects of Viral Infection on the Transcriptome Expression of Sf9 Cells
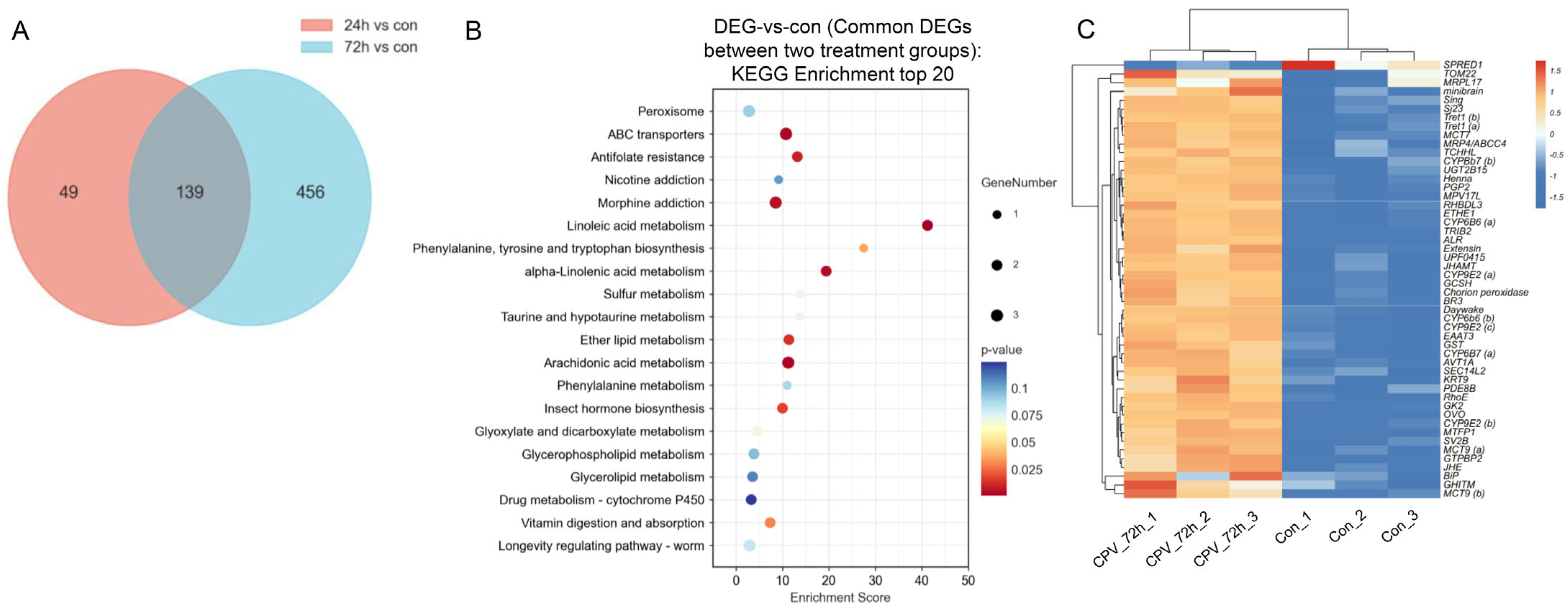
2.4. Analysis of DEGs
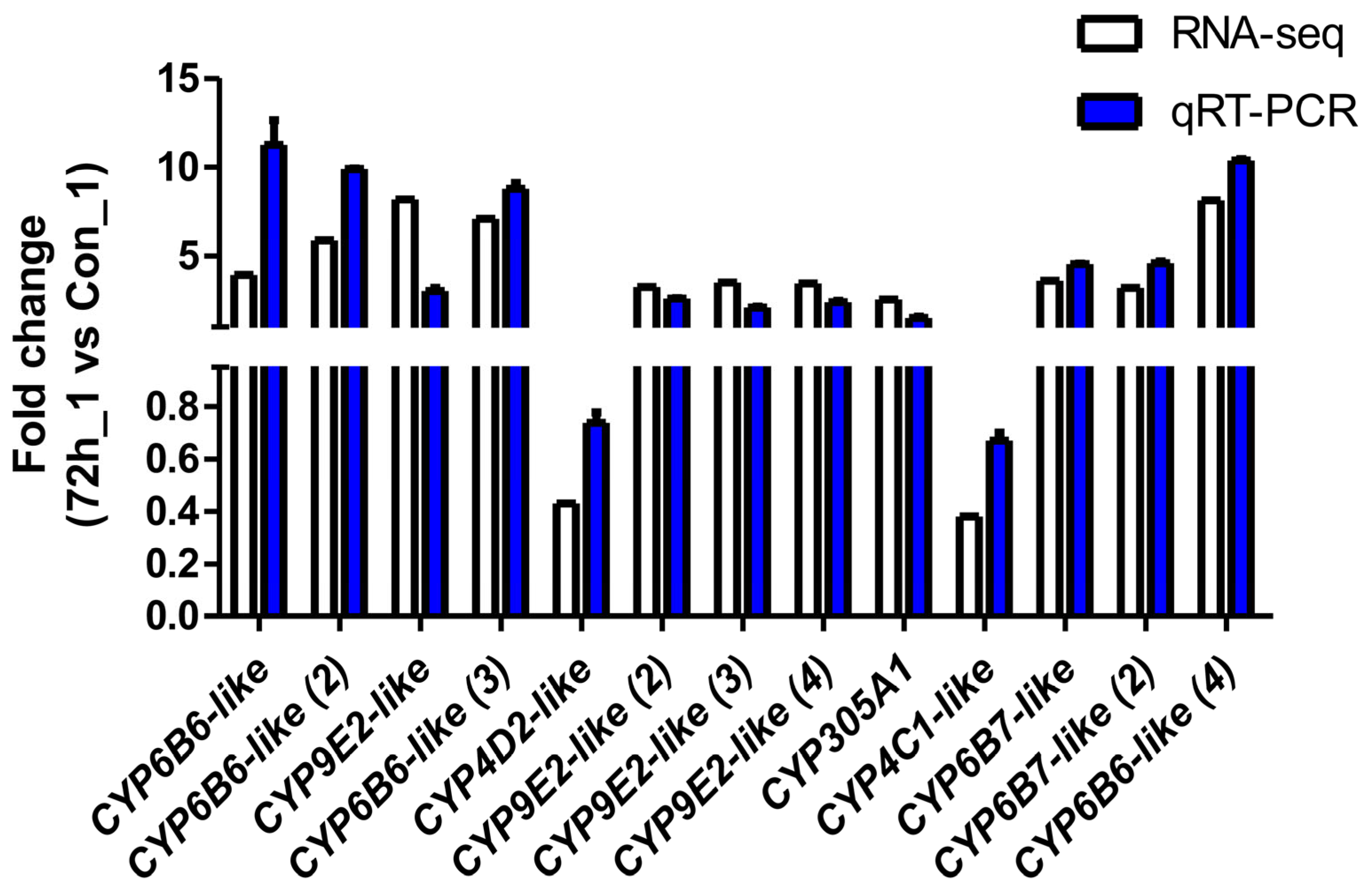
2.5. qRT–PCR Validation of DEGs
3. Discussion
4. Materials and Methods
4.1. Cell Line, Virus Stock, Inhibitors, and Antibodies
4.2. Virus Inoculation
4.3. RNA Extraction, Library Preparation, and RNA-Seq
4.4. RNA-Seq Data Analysis
4.5. Quantitative Real-Time PCR
4.6. Cell Treatments
4.7. Western Blotting
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kuang, W.D.; Yan, C.H.; Zhan, Z.G.; Guan, L.M.; Wang, J.C.; Chen, J.H.; Li, J.H.; Ma, G.Q.; Zhou, X.; Jin, L. Transcriptional responses of Daphnis nerii larval midgut to oral infection by Daphnis nerii cypovirus-23. Virol. J. 2021, 18, 250. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, C.; Gao, J.; Abbas, M.N.; Kausar, S.; Qian, C.; Wang, L.; Wei, G.; Zhu, B.J.; Liu, C.L. Comparative mitochondrial genome analysis of Daphnis nerii and other lepidopteran insects reveals conserved mitochondrial genome organization and phylogenetic relationships. PLoS ONE 2017, 12, e0178773. [Google Scholar] [CrossRef]
- Green, T.B.; White, S.; Rao, S.; Mertens, P.P.; Adler, P.H.; Becnel, J.J. Biological and molecular studies of a cypovirus from the black fly Simulium ubiquitum (Diptera: Simuliidae). J. Invertebr. Pathol. 2007, 95, 26–32. [Google Scholar] [CrossRef]
- Zhou, Y.; Qin, T.; Xiao, Y.; Qin, F.; Lei, C.; Sun, X. Genomic and biological characterization of a new cypovirus isolated from Dendrolimus punctatus. PLoS ONE 2014, 9, e0113201. [Google Scholar] [CrossRef] [PubMed]
- Kuang, W.D.; Zhan, Z.G.; Guan, L.M.; Wang, J.C.; Yan, C.H.; Chen, J.H.; Li, J.H.; Zhou, X.; Jin, L. Study on proliferation characteristics of Daphnis nerii cypovirus-23 in Sf9 Cells. J. Agric. Biotechnol. 2021, 29, 772–779. (In Chinese) [Google Scholar]
- Nai, Y.S.; Lo, C.M. Persistent PnV (Perina nuda virus) infection in a heterologous Lymantria xylina cell line, NTU-LY. Biocontrol Sci. Technol. 2020, 30, 929–940. [Google Scholar] [CrossRef]
- Erez, T.; Chejanovsky, N. Infection of a lepidopteran cell line with deformed wing virus. Viruses 2020, 12, 739. [Google Scholar] [CrossRef]
- Agosto, J.A.M.; McCabe, E.R.B. Conserved family of glycerol kinase loci in Drosophila melanogaster. Mol. Genet. Metab. 2006, 88, 334–345. [Google Scholar] [CrossRef]
- Vatanparast, M.; Sajjadian, S.M.; Park, Y. Glycerol biosynthesis plays an essential role in mediating cold tolerance the red imported fire ant, Solenopsis invicta. Arch. Insect Biochem. Physiol. 2022, 109, e21861. [Google Scholar] [CrossRef]
- Majumdar, S.; Chowdhury, D.R.; Chakraborty, B.C.; Chowdhury, A.; Datta, S.; Banerjee, S. MiR-451a attenuates hepatic steatosis and hepatitis C virus replication by targeting glycerol kinase. J. Transl. Med. 2025, 23, 322. [Google Scholar] [CrossRef]
- Kikawada, T.; Saito, A.; Kanamori, Y.; Nakahara, Y.; Iwata, K.I.; Tanaka, D.; Watanabe, M.; Okuda, T. Trehalose transporter 1, a facilitated and high-capacity trehalose transporter, allows exogenous trehalose uptake into cells. Proc. Natl. Acad. Sci. USA 2007, 104, 11585–11590. [Google Scholar] [CrossRef]
- Marten, A.D.; Haslitt, D.P.; Martin, C.A.; Karthikeyan, A.; Swanson, D.H.; Kalera, K.; Johnson, U.G.; Swarts, B.M.; Conway, M.J. Trehalose supports the growth of Aedes aegypti cells and modifies gene expression and dengue virus type 2 replication. PLoS Pathog. 2025, 21, e1012795. [Google Scholar] [CrossRef]
- Kaur, A.; Singh, S.; Sharma, S.C. Unlocking Trehalose’s versatility: A comprehensive Journey from biosynthesis to therapeutic applications. Exp. Cell Res. 2024, 442, 114250. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, H.D.; Wang, Y.X.; Guo, Z.X.; Liu, Y.X.; Huang, Z.H.; Zhu, L.B.; Liu, M.H.; Liu, S.H.; Xu, J.P. Trehalose hydrolysis and transport-related genes promote Bombyx mori nucleopolyhedrovirus proliferation through the phosphoinositide 3-kinase-Akt signalling pathway in BmN cell. Dev. Comp. Immunol. 2023, 140, 104625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Liu, X.J.; Shiotsuki, T.; Wang, Z.S.; Xu, X.; Huang, Y.P.; Li, M.W.; Li, K.; Tan, A.J. Depletion of juvenile hormone esterase extends larval growth in Bombyx mori. Insect Biochem. Mol. Biol. 2017, 81, 72–79. [Google Scholar] [CrossRef]
- Reis, M.A.; Noriega, D.D.; Alves, G.D.; Coelho, R.R.; Grossi-de-Sa, M.F.; Antonino, J.D. Why is oral-induced RNAi inefficient in Diatraea saccharalis? A possible role for DsREase and other nucleases. Pestic. Biochem. Physiol. 2022, 186, 105166. [Google Scholar] [CrossRef]
- Saito, Y.; Kamita, S.G.; Hammock, B.D.; Kunimi, Y.; Inoue, M.N.; Nakai, M. Juvenile hormone (JH) esterase activity but not JH epoxide hydrolase activity is downregulated in larval Adoxophyes honmai following nucleopolyhedroviruses infection. J. Insect Physiol. 2015, 80, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Q.; Dai, J.P.; Zhang, G.Z.; Singh, D.; Zhang, X.X.; Liang, Z.P. Mamestra brassicae multiple nucleopolyhedroviruses prevents pupation of Helicoverpa armigera by regulating juvenile hormone titer. Insects 2024, 15, 202. [Google Scholar] [CrossRef]
- Takatsuka, J.; Nakai, M.; Shinoda, T. A virus carries a gene encoding juvenile hormone acid methyltransferase, a key regulatory enzyme in insect metamorphosis. Sci. Rep. 2017, 7, 13522. [Google Scholar] [CrossRef]
- Gu, Y.F.; Yang, X.Y.; Liu, S.H.; Chen, X.W.; Liu, R.; Gao, J.L.; Zhong, Y.H.; Li, X.Y.; Han, W.S. RNAi-mediated knockdown of juvenile hormone acid methyltransferase depresses reproductive performance in female. Pestic. Biochem. Physiol. 2025, 211, 106420. [Google Scholar] [CrossRef]
- Zhu, S.M.; Liu, F.F.; Chen, X.Y.; Xia, S.S.; Wu, Y.T.; Tang, W.; Ren, C.H.; Wang, J.; Li, S. Inter-organelle communication dynamically orchestrates juvenile hormone biosynthesis and female reproduction. Natl. Sci. Rev. 2025, 12, nwaf022. [Google Scholar] [CrossRef]
- Wang, A.Y.; Yang, Y.X.; Zhang, Y.; Xue, C.; Cheng, Y.J.; Zhang, Y.F.; Zhang, W.J.; Zhao, M.; Zhang, J.H. Insecticide-induced sublethal effect in the fall armyworm is mediated by miR-9993/miR-2a-3p-FPPS/JHAMT-JH molecular module. Pestic. Biochem. Physiol. 2025, 210, 106400. [Google Scholar] [CrossRef]
- Shi, Z.K.; Wen, D.; Chang, M.M.; Sun, X.M.; Wang, Y.H.; Cheng, C.H.; Zhang, L.Q.; Zheng, A.H.; Zou, Z. Juvenile hormone-sensitive ribosomal activity enhances viral replication in Aedes aegypti. Msystems 2021, 6, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.; Liu, D.; Yuan, Y.Y.; Li, M.; Qiu, X.H. CYP6B6 is involved in esfenvalerate detoxification in the polyphagous lepidopteran pest, Helicoverpa armigera. Pestic. Biochem. Physiol. 2017, 138, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Xiong, T.F.; Yu, M.Y.; Li, M.; Reilly, C.A.; Qiu, X.H. CYP6B6 mediated adaptation to capsaicinoids in the generalist Helicoverpa armigera and specialist H. assulta: Transcriptional response and metabolic detoxification. Int. J. Biol. Macromol. 2025, 286, 138286. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, J.Y.; Wu, P.Z.; Zhang, Y.; Qiu, L.H. Functional study on candidate regulators mediating the expression of CYP6B7 induced by fenvalerate in a susceptible strain of Helicoverpa armigera. Pestic. Biochem. Physiol. 2024, 202, 105918. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zheng, J.Y.; Wu, P.Z.; Zhang, Y.; Qiu, L.H. A comparative study of transcriptional regulation mechanism of Cytochrome P450 CYP6B7 between resistant and susceptible strains of Helicoverpa armigera. J. Agric. Food Chem. 2023, 71, 9314–9323. [Google Scholar] [CrossRef]
- Manjon, C.; Troczka, B.J.; Zaworra, M.; Beadle, K.; Randall, E.; Hertlein, G.; Singh, K.S.; Zimmer, C.T.; Homem, R.A.; Lueke, B.; et al. Unravelling the molecular determinants of bee sensitivity to neonicotinoid insecticides. Curr. Biol. 2018, 28, 1137–1143.e5. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Y.; Liu, Z.Y.; Zhu, Y.; Xiao, Y.; Xiao, W.F.; Tang, L.; Dong, Z.Q.; Pan, M.H.; Lu, C.; Chen, P. MicroRNA bmo-miR-31-5p inhibits apoptosis and promotes BmNPV proliferation by targeting the CYP9e2 gene of Bombyx mori. Pest Manag. Sci. 2024, 80, 4564–4574. [Google Scholar] [CrossRef]
- Li, X.Y.; Liu, Z.D.; Lv, X.X.; Liu, X.C.; Li, Y.F.; Tian, Z.; Zhang, Y.L.; Liu, J.Y. Molecular mechanism of λ-cyhalothrin detoxification by a delta-class Glutathione S-Transferase (PxGSTD3) from. J. Agric. Food Chem. 2025, 73, 6559–6566. [Google Scholar] [CrossRef]
- Haas, G.; Seiler, M.; Nguyen, J.; Troxler, L.; Pennarun, S.; Lefebvre, E.; Benamrouche, Y.; Loizeau, L.; Reinbolt, C.; Liang, M.; et al. Regulation of detoxifying enzymes expression and restriction of picorna-like virus infection by natural polysaccharide extracts in. Virology 2025, 607, 110513. [Google Scholar] [CrossRef]
- Gui, Z.Z.; Hou, C.X.; Liu, T.; Qin, G.X.; Li, M.W.; Jin, B. Effects of insect viruses and pesticides on Glutathione S-Transferase activity and gene expression in Bombyx mori. J. Econ. Entomol. 2009, 102, 1591–1598. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Dalton, T.P. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 2006, 6, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.H.; Kong, W.W.; Ling, B.; Wang, Z.Y.; Zhang, Y.; Guo, Z.X.; Liu, S.H.; Xu, J.P. Bmo-miR-3351 modulates glutathione content and inhibits BmNPV proliferation by targeting BmGSTe6 in Bombyx mori. Insect Sci. 2024, 31, 1378–1396. [Google Scholar] [CrossRef]
- Fan, C.C.; Cui, Z.X.; Yang, T.Y.; Sun, L.L.; Cao, C.W. UDP-glucuronosyltransferase is involved in susceptibility of Chironomus kiiensis Tokunaga, 1936 (Diptera: Chironomidae) to insecticides. Ecotoxicol. Environ. Saf. 2023, 263, 115353. [Google Scholar] [CrossRef]
- Zhu, F.F.; Han, J.Y.; Hong, J.D.; Cai, F.C.; Tang, Q.; Yu, Q.; Ma, S.S.; Liu, X.Y.; Huo, S.H.; Chen, K.P. Characterization of the UDP-glycosyltransferase UGT33D1 in silkworm. Insect Mol. Biol. 2024, 33, 697–707. [Google Scholar] [CrossRef]
- Shang, F.; Ding, B.Y.; Niu, J.Z.; Lu, J.M.; Xie, X.C.; Li, C.Z.; Zhang, W.; Pan, D.; Jiang, R.X.; Wang, J.J. microRNA maintains nutrient homeostasis in the symbiont-host interaction. Proc. Natl. Acad. Sci. USA 2024, 121, e2406925121. [Google Scholar] [CrossRef]
- Huang, H.; Lu-Bo, Y.; Haddad, G.G. A Drosophila ABC transporter regulates lifespan. PLoS Genet. 2014, 10, e1004844. [Google Scholar] [CrossRef]
- Jorajuria, S.; Dereuddre-Bosquet, N.; Naissant-Storck, K.; Dormont, D.; Clayette, P. Differential expression levels of MRP1, MRP4, and MRP5 in response to human immunodeficiency virus infection in human macrophages. Antimicrob. Agents Chemother. 2004, 48, 1889–1891. [Google Scholar] [CrossRef]
- Zhu, M.; Pan, J.; Zhang, M.T.; Tong, X.Y.; Zhang, Y.S.; Zhang, Z.Y.; Liang, Z.; Zhang, X.; Hu, X.L.; Xue, R.Y.; et al. Bombyx mori cypovirus (BmCPV) induces PINK1-Parkin mediated mitophagy via interaction of VP4 with host Tom40. Dev. Comp. Immunol. 2022, 126, 104244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, M.; Pan, J.; Qiu, Q.; Tong, X.; Hu, X.; Gong, C. BmCPV replication is suppressed by the activation of the NF-kappaB/autophagy pathway through the interaction of vsp21 translated by vcircRNA_000048 with ubiquitin carboxyl-terminal hydrolase. Insect Biochem. Mol. Biol. 2023, 156, 103947. [Google Scholar] [CrossRef]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Tu, S.Y.; Ding, L.; Jin, M.L.; Chen, H.C.; Zhou, H.B. The role of autophagy in viral infections. J. Biomed. Sci. 2023, 30, 5. [Google Scholar] [CrossRef] [PubMed]
- Nardacci, R.; Ciccosanti, F.; Marsella, C.; Ippolito, G.; Piacentini, M.; Fimia, G.M. Role of autophagy in HIV infection and pathogenesis. J. Intern. Med. 2017, 281, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Kreutzberger, A.J.B.; Sanyal, A.; Saminathan, A.; Bloyet, L.M.; Stumpf, S.; Liu, Z.M.; Ojha, R.; Patjas, M.T.; Geneid, A.; Scanavachi, G.; et al. SARS-CoV-2 requires acidic pH to infect cells. Proc. Natl. Acad. Sci. USA 2022, 119, 422–432. [Google Scholar] [CrossRef]
- Doyle, C.A.; Busey, G.W.; Iobst, W.H.; Kiessling, V.; Renken, C.; Doppalapudi, H.; Stremska, M.E.; Manjegowda, M.C.; Arish, M.; Wang, W.M.; et al. Endosomal fusion of pH-dependent enveloped viruses requires ion channel TRPM7. Nat. Commun. 2024, 15, 8479. [Google Scholar] [CrossRef]
- Li, Y.H.; Li, C.Y.; Zhao, C.C.; Wu, J.Y.; Zhu, Y.; Wang, F.; Zhong, J.P.; Yan, Y.; Jin, Y.L.; Dong, W.R.; et al. Coronavirus M protein promotes mitophagy over virophagy by recruiting PDPK1 to phosphorylate SQSTM1 at T138. Nat. Commun. 2024, 15, 8927. [Google Scholar] [CrossRef]
- Pesti, I.; Barczánfalvi, G.; Dulka, K.; Kata, D.; Farkas, E.; Gulya, K. Bafilomycin 1A affects p62/SQSTM1 autophagy marker protein level and autophagosome puncta formation oppositely under various inflammatory conditions in cultured rat microglial cells. Int. J. Mol. Sci. 2024, 25, 8265. [Google Scholar] [CrossRef]
- Tan, X.Y.; Wang, X.; Liu, Q.S.; Xie, X.Q.; Li, Y.; Li, B.Q.; Li, Z.Q.; Xia, Q.Y.; Zhao, P. Inhibition of silkworm vacuolar-type ATPase activity by its inhibitor Bafilomycin A1 induces caspase-dependent apoptosis in an embryonic cell line of silkworm. Arch. Insect Biochem. Physiol. 2018, 99, e21507. [Google Scholar] [CrossRef]
- Mauvezin, C.; Neufeld, T.P. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 2015, 11, 1437–1438. [Google Scholar] [CrossRef]
- Song, B.Y.W.; Korolkova, O. Bafilomycin A1 inhibits HIV-1 infection by disrupting lysosomal cholesterol transport. Viruses 2024, 16, 1374. [Google Scholar] [CrossRef]
- Jin, M.H.; Shan, Y.X.; Peng, Y.; Wang, W.H.; Zhang, H.H.; Liu, K.Y.; Heckel, D.G.; Wu, K.M.; Tabashnik, B.E.; Xiao, Y.T. Downregulation of a transcription factor associated with resistance to Bt toxin Vip3Aa in the invasive fall armyworm. Proc. Natl. Acad. Sci. USA 2023, 120, e2306932120. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Gene_ID | Gene Name | Abbreviation of Gene Name | Average Expression of CPV_72h | Fold Change of CPV_72h vs. Con | Impact of Gene on Virus Replication |
|---|---|---|---|---|---|
| LOC118281152 | GTP-binding protein 2-like | GTPBP2 | 79.60941971 | 2.939862906 | Unknown |
| LOC118281850 | tribbles homolog 2-like | TRIB2 | 83.6385479 | 3.116434931 | Unknown |
| LOC118276660 | glycerol kinase-like | GK2 | 27.26451044 | 2.891990222 | Related to HCV replication |
| LOC118263046 | juvenile hormone esterase-like | JHE | 44.06177965 | 2.213259729 | Inhibits MbMNPV replication |
| LOC118278849 | UDP-glucuronosyltransferase 2B15-like | UGT2B15 | 52.08919252 | 3.208530207 | May promote virus replication |
| LOC118272911 | FAD-linked sulfhydryl oxidase ALR-like | ALR | 72.24821537 | 3.384447759 | Unknown |
| LOC118264059 | cytochrome P450 9e2-like | CYP9E2 (a) | 65.3765198 | 3.155621533 | Promotes BmNPV replication |
| LOC118273268 | growth hormone-inducible transmembrane protein-like | GHITM | 39.35624854 | 2.069179782 | Unknown |
| LOC118275035 | mitochondrial fission process protein 1-like | MTFP1 | 39.59756816 | 3.266534569 | Unknown |
| LOC118268094 | UPF0415 protein C7orf25 homolog | UPF0415 | 30.07996836 | 2.787196551 | Unknown |
| LOC118264054 | cytochrome P450 9e2-like | CYP9E2 (b) | 25.4264185 | 2.914285755 | Promotes BmNPV replication |
| LOC118276320 | phosphoglycolate phosphatase 2-like | PGP2 | 46.20605215 | 4.101708059 | Unknown |
| LOC118274261 | synaptic vesicle glycoprotein 2B-like | SV2B | 15.33200925 | 2.282949456 | Unknown |
| LOC118267668 | persulfide dioxygenase ETHE1, mitochondrial-like | ETHE1 | 27.24655442 | 3.715194818 | Unknown |
| LOC118276246 | chorion peroxidase-like | Chorion peroxidase | 8.672305521 | 3.575164367 | Unknown |
| LOC118271617 | rho-related GTP-binding protein RhoE-like | RhoE | 8.074911214 | 3.360901616 | Unknown |
| LOC118263839 | balbiani ring protein 3-like | BR3 | 12.93191276 | 2.824491018 | Unknown |
| LOC118276116 | circadian clock-controlled protein daywake-like | Daywake | 26.71343267 | 5.011601593 | Unknown |
| LOC118264074 | rhomboid-related protein 3-like | RHBDL3 | 6.692910582 | 3.302604069 | Unknown |
| LOC118268821 | protein henna-like | Henna | 8.512926579 | 2.48340151 | Unknown |
| LOC118275579 | monocarboxylate transporter 9-like | MCT9 (a) | 7.291586508 | 2.356887011 | Unknown |
| LOC118267228 | protein singles bar-like | Sing | 14.17416077 | 3.521595539 | Unknown |
| LOC118279835 | cytochrome P450 6B7-like | CYP6B7 (a) | 6.115680087 | 2.885802312 | Unknown |
| LOC118262727 | cytochrome P450 9e2-like | CYP9E2 (c) | 7.514131089 | 7.340344336 | Unknown |
| LOC118268856 | glycine cleavage system H protein-like | GCSH | 17.70414895 | 4.804584058 | Unknown |
| LOC118262982 | endoplasmic reticulum chaperone BiP | BiP | 4.831579472 | 2.149337031 | Unknown |
| LOC118264312 | amino acid transporter AVT1A-like | AVT1A | 6.275652722 | 4.412106553 | Unknown |
| LOC118263447 | transcriptional regulator ovo-like | OVO | 2.995905576 | 2.958866706 | Unknown |
| LOC118279736 | cytochrome P450 6B7-like | CYP6B7 (b) | 5.224798912 | 3.233611488 | Unknown |
| LOC118263048 | cytochrome P450 6B6-like | CYP6B6 (a) | 7.812977386 | 6.348864958 | Unknown |
| LOC118268820 | extensin-like | Extensin | 4.414752748 | 4.159429339 | Unknown |
| LOC118269313 | mitochondrial import receptor subunit TOM22 homolog | TOM22 | 7.870868019 | 2.250525698 | Unknown |
| LOC118271571 | facilitated trehalose transporter Tret1-like | Tret1 (a) | 4.258888116 | 2.285612846 | Promotes BmNPV replication |
| LOC118267488 | serine/threonine-protein kinase minibrain-like | Minibrain | 8.060249965 | 2.327461242 | Unknown |
| LOC118279019 | facilitated trehalose transporter Tret1-like | Tret1 (b) | 4.17484285 | 2.427275019 | Promotes BmNPV replication |
| LOC118274614 | monocarboxylate transporter 7-like | MCT7 | 5.706508283 | 2.048856356 | Unknown |
| LOC118261931 | glutathione S-transferase 1-like | GST | 14.72725987 | 5.251990067 | Promotes BmNPV replication |
| LOC118276669 | sprouty-related, EVH1 domain-containing protein 1-like | SPRED1 | 1.72739488 | 0.41233409 | Unknown |
| LOC118266697 | SEC14-like protein 2 | SEC14L2 | 3.912579123 | 3.121802027 | Unknown |
| LOC118267308 | mpv17-like protein | MPV17L | 8.924007294 | 2.829362881 | Unknown |
| LOC118276314 | monocarboxylate transporter 9-like | MCT9 (b) | 3.561204881 | 2.266039059 | Unknown |
| LOC118265759 | high affinity cAMP-specific and IBMX-insensitive 3′,5′-cyclic phosphodiesterase 8-like | PDE8B | 0.91473512 | 2.468483098 | Unknown |
| LOC118276616 | 23 kDa integral membrane protein-like | Sj23 | 2.825832687 | 2.633377326 | Unknown |
| LOC118262642 | cytochrome P450 6B6-like | CYP6B6 (b) | 4.842834846 | 5.279997561 | Unknown |
| LOC118267867 | 39S ribosomal protein L17, mitochondrial-like | MRPL17 | 9.329088079 | 2.393602125 | Unknown |
| LOC118265812 | excitatory amino acid transporter 3-like | EAAT3 | 2.680965274 | 4.855375004 | Unknown |
| LOC118265567 | juvenile hormone acid O-methyltransferase-like | JHAMT | 2.885832192 | 4.064307053 | May promote ZIKV infection |
| LOC118262225 | multidrug resistance-associated protein 4-like | MRP4/ABCC4 | 1.162929203 | 3.585355174 | Promotes HIV-1 replication |
| LOC118280766 | keratin, type I cytoskeletal 9-like | KRT9 | 2.440427412 | 2.243261847 | Unknown |
| LOC118270457 | trichohyalin-like | TCHHL | 2.221009547 | 3.830439946 | Unknown |
| LOC118273096 | sequestosome-1-like | p62/SQSTM1 | 97.47736978 | 2.15174043 | Plays a dual role in viral infection |
| LOC118273082 | sequestosome-1-like | p62/SQSTM1 | 82.74454193 | 2.035871822 | Plays a dual role in viral infection |
| No. | Primer Name | Primer Sequence (5′ to 3′) | Tm (°C) | Gene ID | Target Gene |
|---|---|---|---|---|---|
| 1 | S1-RTPCR-F | GTGCTGATGGTCTGCTAA | 49.6 | N/A | DnCPV-23 S1 |
| 2 | S1-RTPCR-R | TGATTGATGACGACATTGAG | 51.5 | ||
| 3 | 1893F | GTATTAATCCTACTGTACCACTACG | 51.7 | LOC118261893 | CYP6B6-like |
| 4 | 1893R | CTCTTCTTTGCTACGAGATTAGG | 52.4 | ||
| 5 | 2642F | TACTAGAGGTGAGGTGAGTGATAT | 53.4 | LOC118262642 | CYP6B6-like |
| 6 | 2642R | CAGCGTGTAGTTCTTTAGGATATG | 52.7 | ||
| 7 | 2727F | TTCGAGACAGTATCATCAGGAATG | 53.7 | LOC118262727 | CYP9E2-like |
| 8 | 2727R | GACACAACCATATCCATATAGACC | 52.2 | ||
| 9 | 3048F | CTCAAGCATTCTTCTTCTTCTTAGC | 53.3 | LOC118263048 | CYP6B6-like |
| 10 | 3048R | GTCTACTTCATCCTGTACCTTCTT | 53.2 | ||
| 11 | 3400F | GGTACTTCAGTAGTGGTGAATATC | 52 | LOC118263400 | CYP4D2-like |
| 12 | 3400R | GAGATAGTGATCTTGAGTTCCATC | 52.1 | ||
| 13 | 4054F | GTCTACCAGTGTTCACCTTTATTAG | 52.5 | LOC118264054 | CYP9E2-like |
| 14 | 4054R | CATTAGTGACCTTCGCTATGAGAT | 53.8 | ||
| 15 | 4059F | GATCAACATCCTCATGGAAGTTAAG | 53 | LOC118264059 | CYP9E2-like |
| 16 | 4059R | CGATATGTGACTCTTCAACAGTTG | 53.2 | ||
| 17 | 4410F | CAAGATTGTCAGGAACGATATGATC | 53 | LOC118264410 | CYP9E2-like |
| 18 | 4410R | CGATATGTGACTCTTCAACAGTTG | 53.2 | ||
| 19 | 6520F | TAGACGCAGTAATAGGAGATAGAC | 52.4 | LOC118266520 | CYP305A1-like |
| 20 | 6520R | CTATAGGGTAGTATCTATGGACTTC | 50.5 | ||
| 21 | 0774F | GAGAGTAATTATCGTCGGTTGAAG | 52.6 | LOC118270774 | CYP4C1-like |
| 22 | 0774R | CATAGTCCATAGTCGGGTAGTATT | 52.7 | ||
| 23 | 9736F | GAGGTACTCTCAGTCTCTAAGTA | 51.7 | LOC118279736 | CYP6B7-like |
| 24 | 9736R | CGTTATACGAGGCTATACATAGG | 51.6 | ||
| 25 | 9835F | AGTGAGACCTCAAGAGATAATGAC | 53.2 | LOC118279835 | CYP6B7-like |
| 26 | 9835R | CTTGTAGTCTCTCTTCCTCAGTAT | 52.5 | ||
| 27 | 2155F | CTGTAGCGTTATATGATCTCTTCC | 52.1 | LOC118282155 | CYP6B6-like |
| 28 | 2155R | GTCCAATAGGCTTGTAGTTTCTCT | 53.9 | ||
| 29 | gapdhF | GTGCCAAGAAGGTCATCAT | 52.2 | N/A | GAPDH |
| 30 | gapdhR | GAGAGGAGCGAGACAGTT | 53.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuang, W.; Yang, J.; Wang, J.; Yan, C.; Chen, J.; Liu, X.; Yang, C.; Zhan, Z.; Guan, L.; Li, J.; et al. Transcriptional Analysis of Spodoptera frugiperda Sf9 Cells Infected with Daphnis nerii Cypovirus-23. Int. J. Mol. Sci. 2025, 26, 7487. https://doi.org/10.3390/ijms26157487
Kuang W, Yang J, Wang J, Yan C, Chen J, Liu X, Yang C, Zhan Z, Guan L, Li J, et al. Transcriptional Analysis of Spodoptera frugiperda Sf9 Cells Infected with Daphnis nerii Cypovirus-23. International Journal of Molecular Sciences. 2025; 26(15):7487. https://doi.org/10.3390/ijms26157487
Chicago/Turabian StyleKuang, Wendong, Jian Yang, Jinchang Wang, Chenghua Yan, Junhui Chen, Xinsheng Liu, Chunhua Yang, Zhigao Zhan, Limei Guan, Jianghuai Li, and et al. 2025. "Transcriptional Analysis of Spodoptera frugiperda Sf9 Cells Infected with Daphnis nerii Cypovirus-23" International Journal of Molecular Sciences 26, no. 15: 7487. https://doi.org/10.3390/ijms26157487
APA StyleKuang, W., Yang, J., Wang, J., Yan, C., Chen, J., Liu, X., Yang, C., Zhan, Z., Guan, L., Li, J., Deng, T., Yang, F., Ma, G., & Jin, L. (2025). Transcriptional Analysis of Spodoptera frugiperda Sf9 Cells Infected with Daphnis nerii Cypovirus-23. International Journal of Molecular Sciences, 26(15), 7487. https://doi.org/10.3390/ijms26157487






