Genome-Editing Tools for Lactic Acid Bacteria: Past Achievements, Current Platforms, and Future Directions
Abstract
1. Introduction
- Classical homologous recombination and transposon mutagenesis—single- or double-crossover events with suicide vectors, ISS1/Tn917 insertions and elaborate counter-selection schemes provided the first knockouts but left antibiotic scars and required weeks of culturing.
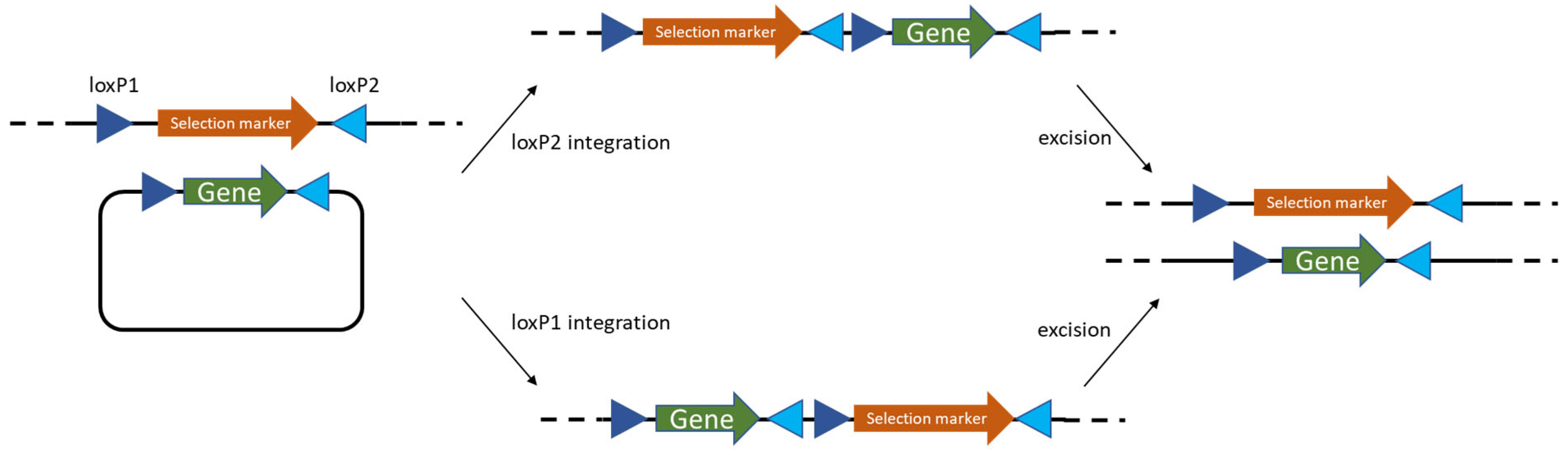
- Phage-derived recombineering and site-specific recombinases—prophage RecT/RecE systems, often paired with Cre/lox, cut editing times to days and enabled marker recycling; efficiencies > 50% for single-gene edits are now routine.
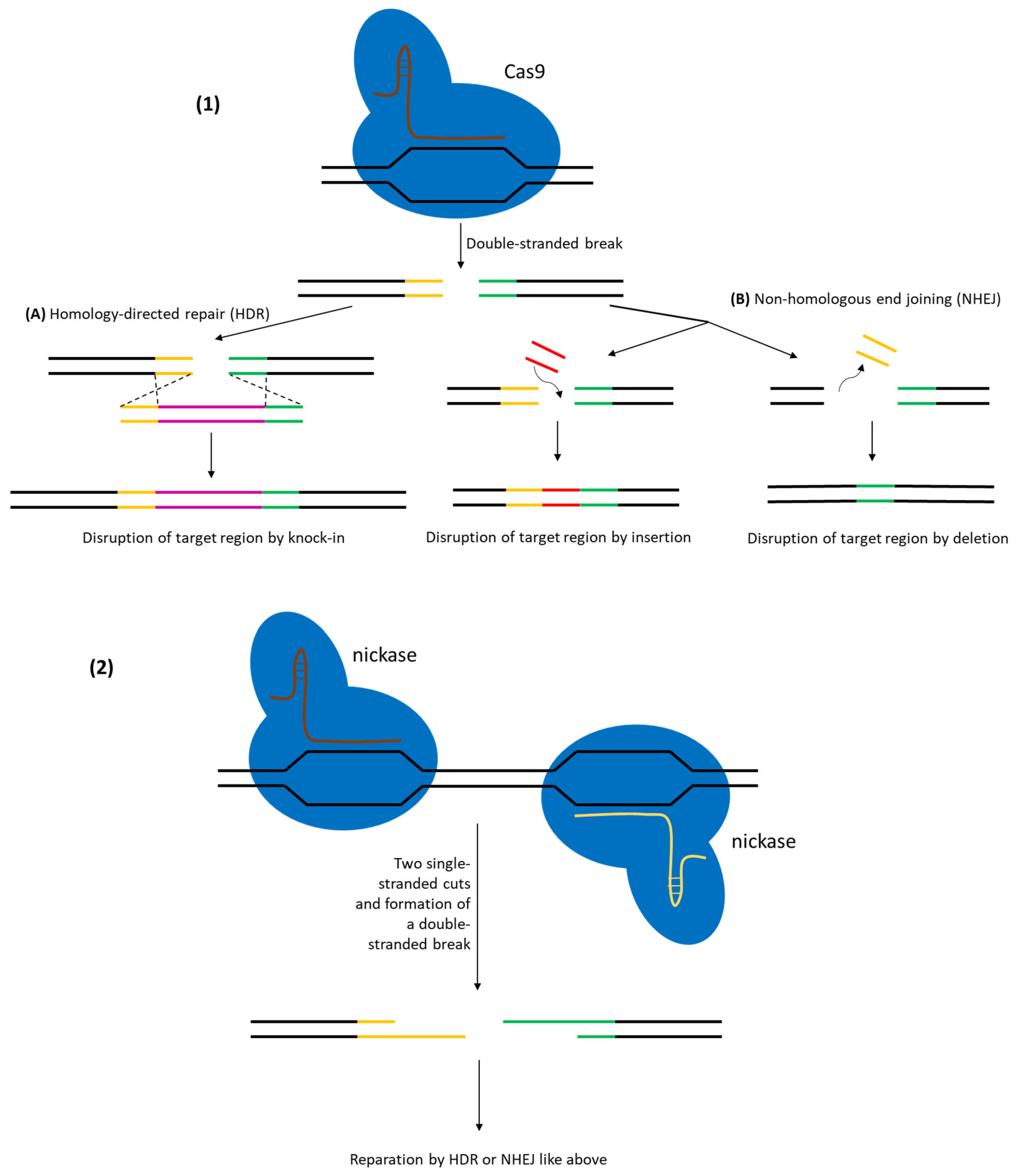
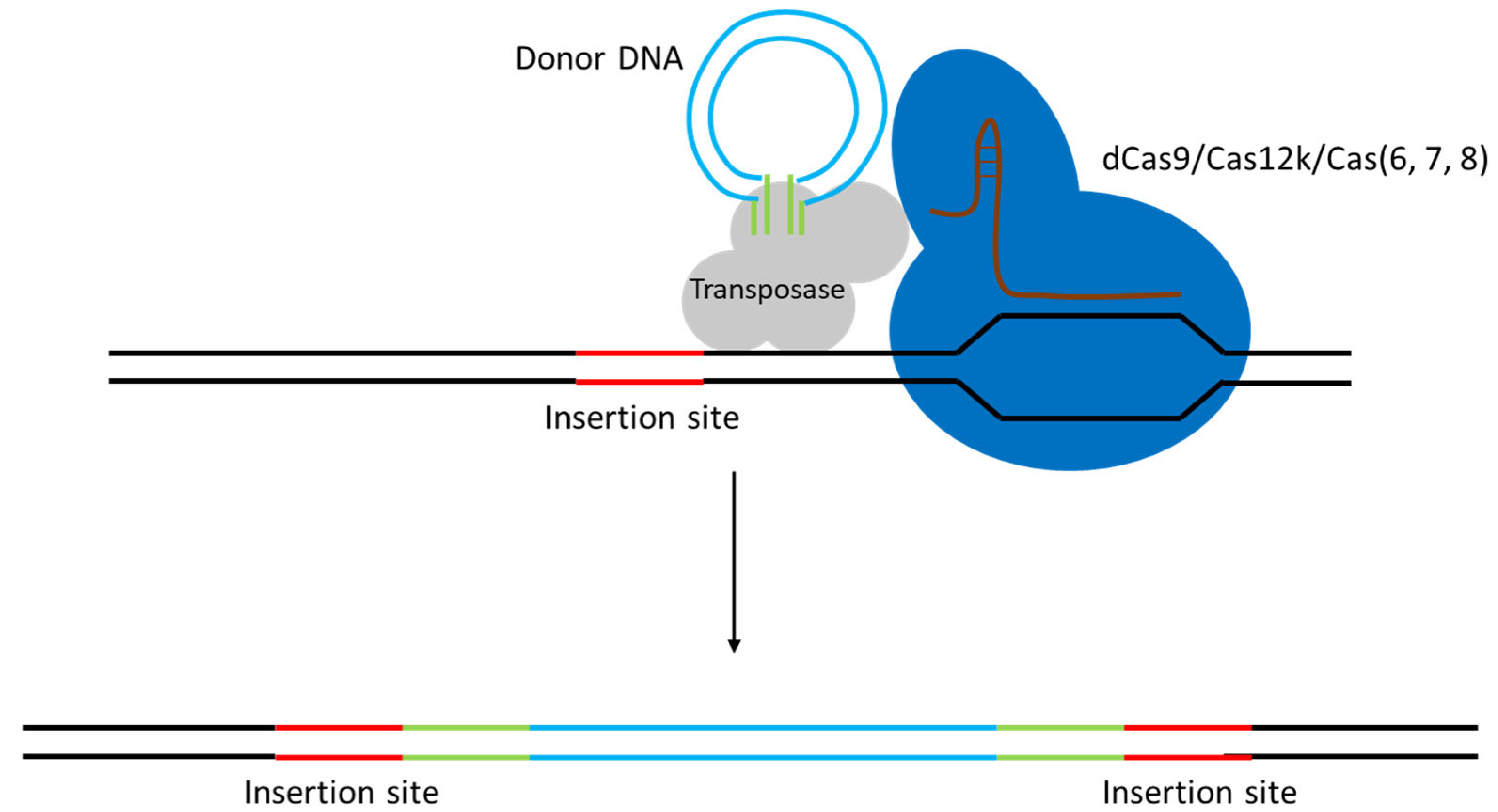
- CRISPR-based platforms—Cas9 (wild-type or nickase) used as a lethal counter-selector, dCas9 for CRISPR interference, endogenous Cas systems repurposed for scar-less editing, and most recently CRISPR-guided transposases that shuttle payloads up to 10 kb without homologous arms.
2. Instruments for Genetic Engineering of Lactic Acid Bacteria: Recombination and Recombineering Systems
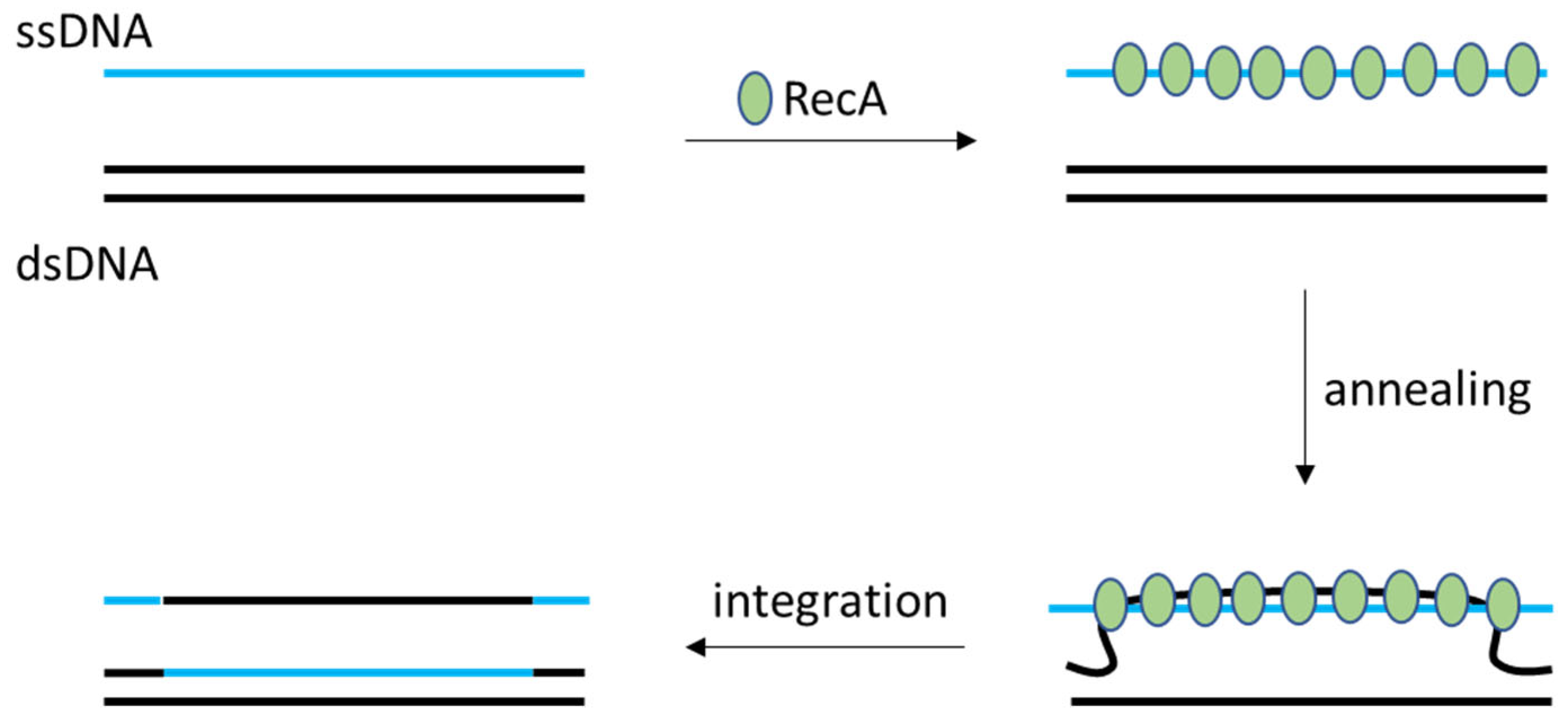
2.1. LAB’s Native Homologous Recombination Systems (RecA-Type)
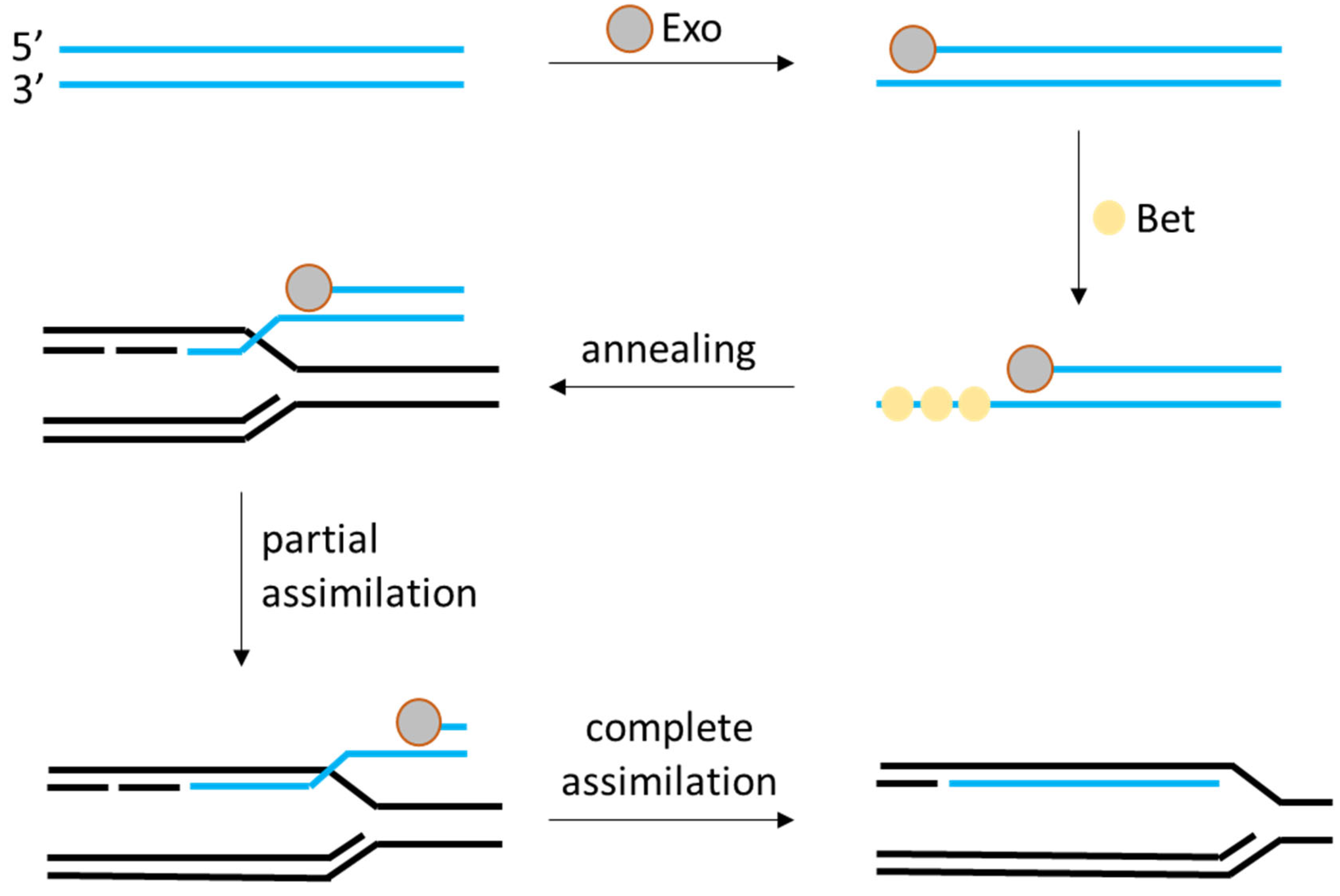
2.2. Random Insertion and Phage-Derived Recombineering Tools
3. CRISPR/Cas Systems for Genetic Engineering in Lactic Acid Bacteria
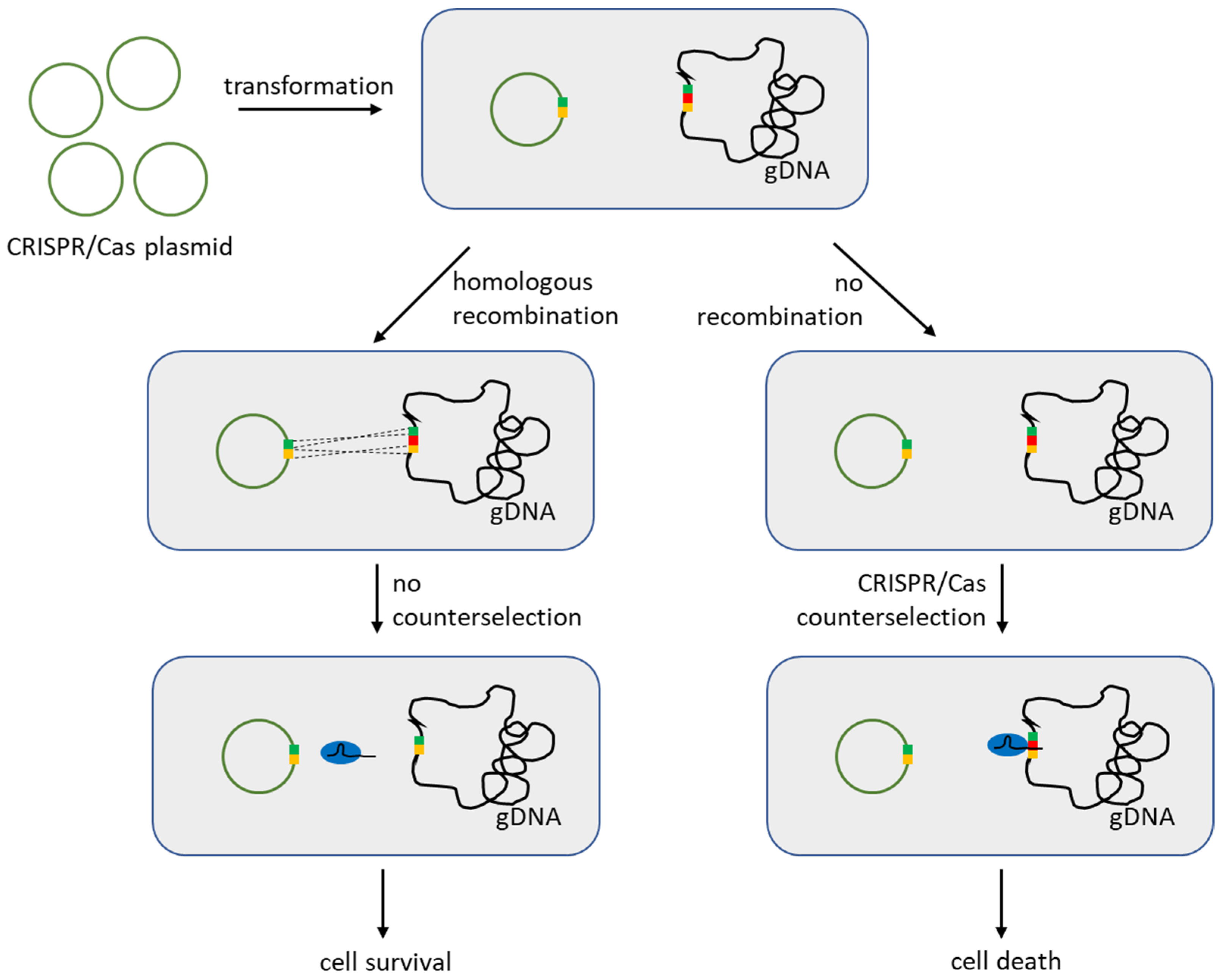
3.1. Reverse Selection Using CRISPR/Cas in LAB
3.2. Gene Knockouts Using CRISPR/Cas in LAB
3.3. DNA Sequence Integration Using CRISPR/Cas in LAB
3.4. RNA Interference Using CRISPR/Cas in LAB
3.5. Using LAB’s Own CRISPR/Cas Systems for Genetic Engineering
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kameni, A.-P.T.; Couture-Tosi, E.; Saint-Girons, I.; Picardeau, M. Inactivation of the Spirochete recA Gene Results in a Mutant with Low Viability and Irregular Nucleoid Morphology. J. Bacteriol. 2002, 184, 452–458. [Google Scholar] [CrossRef][Green Version]
- Leenhouts, K.J.; Kok, J.; Venema, G. Lactococcal plasmid pWV01 as an integration vector for lactococci. Appl. Environ. Microbiol. 1991, 57, 2562–2567. [Google Scholar] [CrossRef]
- Maguin, E.; Prévost, H.; Ehrlich, S.D.; Gruss, A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 1996, 178, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.C.; Klaenhammer, T.R. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J. Bacteriol. 1994, 176, 5330–5340. [Google Scholar] [CrossRef]
- Martinussen, J.; Hammer, K. Cloning and characterization of upp, a gene encoding uracil phosphoribosyltransferase from Lactococcus lactis. J. Bacteriol. 1994, 176, 6457–6463. [Google Scholar] [CrossRef] [PubMed]
- Solem, C.; Defoor, E.; Jensen, P.R.; Martinussen, J. Plasmid pCS1966, a New Selection/Counterselection Tool for Lactic Acid Bacterium Strain Construction Based on the oroP Gene, Encoding an Orotate Transporter from Lactococcus lactis. Appl. Environ. Microbiol. 2008, 74, 4772–4775. [Google Scholar] [CrossRef]
- Xin, Y.; Guo, T.; Mu, Y.; Kong, J. Development of a counterselectable seamless mutagenesis system in lactic acid bacteria. Microb. Cell Fact. 2017, 16, 116. [Google Scholar] [CrossRef]
- O’sullivan, T.F.; Fitzgerald, G.F. Electrotransformation of industrial strains of Streptococcus thermophilus. J. Appl. Microbiol. 1999, 86, 275–283. [Google Scholar] [CrossRef]
- Ito, M.; Kim, Y.-G.; Tsuji, H.; Takahashi, T.; Kiwaki, M.; Nomoto, K.; Danbara, H.; Okada, N. Transposon Mutagenesis of Probiotic Lactobacillus casei Identifies asnH, an Asparagine Synthetase Gene Involved in Its Immune-Activating Capacity. PLoS ONE 2014, 9, e83876. [Google Scholar] [CrossRef]
- Perpetuini, G.; Scornec, H.; Tofalo, R.; Serror, P.; Schirone, M.; Suzzi, G.; Corsetti, A.; Cavin, J.F.; Licandro-Seraut, H. Identification of Critical Genes for Growth in Olive Brine by Transposon Mutagenesis of Lactobacillus pentosus C11. Appl. Environ. Microbiol. 2013, 79, 4568–4575. [Google Scholar] [CrossRef][Green Version]
- Palud, A.; Scornec, H.; Cavin, J.-F.; Licandro, H. New Genes Involved in Mild Stress Response Identified by Transposon Mutagenesis in Lactobacillus paracasei. Front. Microbiol. 2018, 9, 535. [Google Scholar] [CrossRef] [PubMed]
- Court, D.L.; Sawitzke, J.A.; Thomason, L.C. Genetic Engineering Using Homologous Recombination. Annu. Rev. Genet. 2002, 36, 361–388. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.C. Use of Bacteriophage λ Recombination Functions To Promote Gene Replacement in Escherichia coli. J. Bacteriol. 1998, 180, 2063–2071. [Google Scholar] [CrossRef]
- Yu, D.; Ellis, H.M.; Lee, E.-C.; Jenkins, N.A.; Copeland, N.G.; Court, D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5978–5983. [Google Scholar] [CrossRef]
- Yang, P.; Wang, J.; Qi, Q. Prophage recombinases-mediated genome engineering in Lactobacillus plantarum. Microb. Cell Fact. 2015, 14, 154. [Google Scholar] [CrossRef]
- Xin, Y.; Guo, T.; Mu, Y.; Kong, J. Identification and functional analysis of potential prophage-derived recombinases for genome editing in Lactobacillus casei. FEMS Microbiol. Lett. 2017, 364, fnx243. [Google Scholar] [CrossRef]
- Sawitzke, J.A.; Costantino, N.; Li, X.; Thomason, L.C.; Bubunenko, M.; Court, C.; Court, D.L. Probing Cellular Processes with Oligo-Mediated Recombination and Using the Knowledge Gained to Optimize Recombineering. J. Mol. Biol. 2011, 407, 45–59. [Google Scholar] [CrossRef]
- Ellis, H.M.; Yu, D.; DiTizio, T.; Court, D.L. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 2001, 98, 6742–6746. [Google Scholar] [CrossRef]
- van Pijkeren, J.-P.; Britton, R.A. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012, 40, e76. [Google Scholar] [CrossRef]
- Dong, H.; Wang, H.; Fu, S.; Zhang, D. CRISPR/Cas tools for enhancing the biopreservation ability of lactic acid bacteria in aquatic products. Front. Bioeng. Biotechnol. 2022, 10, 1114588. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Zhou, D.; Jiang, Z.; Pang, Q.; Zhu, Y.; Wang, Q.; Qi, Q. CRISPR/Cas9-Assisted Seamless Genome Editing in Lactobacillus plantarum and Its Application in N-Acetylglucosamine Production. Appl. Environ. Microbiol. 2019, 85, e01367-19. [Google Scholar] [CrossRef] [PubMed]
- Van Zyl, W.F.; Dicks, L.M.T.; Deane, S.M. Development of a novel selection/counter-selection system for chromosomal gene integrations and deletions in lactic acid bacteria. BMC Mol. Biol. 2019, 20, 10. [Google Scholar] [CrossRef]
- Zhu, D.; Zhao, K.; Xu, H.; Zhang, X.; Bai, Y.; Saris, P.E.J.; Qiao, M. Construction of thyA deficient Lactococcus lactis using the Cre-loxP recombination system. Ann. Microbiol. 2015, 65, 1659–1665. [Google Scholar] [CrossRef]
- Ronda, C.; Pedersen, L.E.; Sommer, M.O.A.; Nielsen, A.T. CRMAGE: CRISPR Optimized MAGE Recombineering. Sci. Rep. 2016, 6, 19452. [Google Scholar] [CrossRef] [PubMed]
- Kababji, A.M.; Butt, H.; Mahfouz, M. Synthetic directed evolution for targeted engineering of plant traits. Front. Plant Sci. 2024, 15, 1449579. [Google Scholar] [CrossRef]
- Rozanov, A.S.; Shlyahtun, V.N.; Tekutieva, L.A.; Son, O.M.; Sizova, S.V.; Peltek, S.E. Methods of yeast genome editing. Vavilov J. Genet. Breed. 2017, 21, 969–978. [Google Scholar] [CrossRef][Green Version]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Cui, Y.; Qu, X. CRISPR-Cas systems of lactic acid bacteria and applications in food science. Biotechnol. Adv. 2024, 71, 108323. [Google Scholar] [CrossRef]
- Xie, Z.; McAuliffe, O.; Jin, Y.-S.; Miller, M.J. Invited review: Genomic modifications of lactic acid bacteria and their applications in dairy fermentation. J. Dairy Sci. 2024, 107, 8749–8764. [Google Scholar] [CrossRef]
- Mu, Y.; Zhang, C.; Li, T.; Jin, F.-J.; Sung, Y.-J.; Oh, H.-M.; Lee, H.-G.; Jin, L. Development and Applications of CRISPR/Cas9-Based Genome Editing in Lactobacillus. Int. J. Mol. Sci. 2022, 23, 12852. [Google Scholar] [CrossRef] [PubMed]
- Leenhouts, K.J.; Kok, J.; Venema, G. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. Lactis. Appl. Environ. Microbiol. 1989, 55, 394–400. [Google Scholar] [CrossRef]
- Lemay, M.-L.; Tremblay, D.M.; Moineau, S. Genome Engineering of Virulent Lactococcal Phages Using CRISPR-Cas9. ACS Synth. Biol. 2017, 6, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.-H.; van Pijkeren, J.-P. CRISPR–Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014, 42, e131. [Google Scholar] [CrossRef]
- Guo, T.; Xin, Y.; Zhang, Y.; Gu, X.; Kong, J. A rapid and versatile tool for genomic engineering in Lactococcus lactis. Microb. Cell Fact. 2019, 18, 22. [Google Scholar] [CrossRef]
- Leenay, R.T.; Vento, J.M.; Shah, M.; Martino, M.E.; Leulier, F.; Beisel, C.L. Genome Editing with CRISPR-Cas9 in Lactobacillus plantarum Revealed That Editing Outcomes Can Vary Across Strains and Between Methods. Biotechnol. J. 2019, 14, 1700583. [Google Scholar] [CrossRef]
- Song, X.; Huang, H.; Xiong, Z.; Ai, L.; Yang, S. CRISPR-Cas9 D10A Nickase-Assisted Genome Editing in Lactobacillus casei. Appl. Environ. Microbiol. 2017, 83, e01259-17. [Google Scholar] [CrossRef]
- Goh, Y.J.; Barrangou, R. Portable CRISPR-Cas9 N System for Flexible Genome Engineering in Lactobacillus acidophilus, Lactobacillus gasseri, and Lactobacillus paracasei. Appl. Environ. Microbiol. 2021, 87, e02669-20. [Google Scholar] [CrossRef]
- Tian, K.; Hong, X.; Guo, M.; Li, Y.; Wu, H.; Caiyin, Q.; Qiao, J. Development of Base Editors for Simultaneously Editing Multiple Loci in Lactococcus lactis. ACS Synth. Biol. 2022, 11, 3644–3656. [Google Scholar] [CrossRef]
- Caso, F.; Davies, B. Base editing and prime editing in laboratory animals. Lab. Anim. 2022, 56, 35–49. [Google Scholar] [CrossRef]
- Mitsunobu, H.; Kita, Y.; Nambu-Nishida, Y.; Miyazaki, S.; Nakajima, K.; Taoka, K.; Kondo, A.; Nishida, K. Development of a highly efficient base editing system for Lactobacilli to improve probiotics and dissect essential functions. Appl. Microbiol. Biotechnol. 2025, 109, 96. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yu, H.; Feng, X.; Tang, H.; Xiong, Z.; Xia, Y.; Ai, L.; Song, X. Specific bile salt hydrolase genes in Lactobacillus plantarum AR113 and relationship with bile salt resistance. LWT 2021, 145, 111208. [Google Scholar] [CrossRef]
- Wiull, K.; Haugen, L.K.; Eijsink, V.G.H.; Mathiesen, G. CRISPR/Cas9-mediated genomic insertion of functional genes into Lactiplantibacillus plantarum WCFS1. Microbiol. Spectr. 2025, 13, e02025-24. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Liu, X.; Zhang, Y.; Chen, Y.; Hang, H.; Chu, J.; Zhuang, Y. Metabolic engineering coupled with adaptive evolution strategies for the efficient production of high-quality L-lactic acid by Lactobacillus paracasei. Bioresour. Technol. 2021, 323, 124549. [Google Scholar] [CrossRef]
- Pechenov, P.Y.; Garagulya, D.A.; Stanovov, D.S.; Letarov, A.V. New Effective Method of Lactococcus Genome Editing Using Guide RNA-Directed Transposition. Int. J. Mol. Sci. 2022, 23, 13978. [Google Scholar] [CrossRef]
- Lecomte, X.; Gagnaire, V.; Lortal, S.; Dary, A.; Genay, M. Streptococcus thermophilus, an emerging and promising tool for heterologous expression: Advantages and future trends. Food Microbiol. 2016, 53, 2–9. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Myrbråten, I.S.; Wiull, K.; Salehian, Z.; Håvarstein, L.S.; Straume, D.; Mathiesen, G.; Kjos, M. CRISPR Interference for Rapid Knockdown of Essential Cell Cycle Genes in Lactobacillus plantarum. MSphere 2019, 4, 10-1128. [Google Scholar] [CrossRef]
- Xiong, Z.-Q.; Wei, Y.-Y.; Kong, L.-H.; Song, X.; Yi, H.-X.; Ai, L.-Z. Short communication: An inducible CRISPR/dCas9 gene repression system in Lactococcus lactis. J. Dairy Sci. 2020, 103, 161–165. [Google Scholar] [CrossRef]
- Kong, L.; Xiong, Z.; Song, X.; Xia, Y.; Ai, L. CRISPR/dCas9-based metabolic pathway engineering for the systematic optimization of exopolysaccharide biosynthesis in Streptococcus thermophilus. J. Dairy Sci. 2022, 105, 6499–6512. [Google Scholar] [CrossRef]
- Zheng, Y.; Su, T.; Qi, Q. Microbial CRISPRi and CRISPRa Systems for Metabolic Engineering. Biotechnol. Bioprocess Eng. 2019, 24, 579–591. [Google Scholar] [CrossRef]
- Liu, L.; Yang, D.; Zhang, Z.; Liu, T.; Hu, G.; He, M.; Zhao, S.; Peng, N. High-Efficiency Genome Editing Based on Endogenous CRISPR-Cas System Enhances Cell Growth and Lactic Acid Production in Pediococcus acidilactici. Appl. Environ. Microbiol. 2021, 87, e00948-21. [Google Scholar] [CrossRef]
- Ma, S.; Wang, F.; Zhang, X.; Qiao, L.; Guo, X.; Lu, X.; Qi, Q. Repurposing endogenous type II CRISPR-Cas9 system for genome editing in Streptococcus thermophilus. Biotechnol. Bioeng. 2024, 121, 749–756. [Google Scholar] [CrossRef]
- Gu, S.; Zhang, J.; Li, L.; Zhong, J. Repurposing the Endogenous CRISPR-Cas9 System for High-Efficiency Genome Editing in Lacticaseibacillus paracasei. ACS Synth. Biol. 2022, 11, 4031–4042. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; Goh, Y.J.; Pan, M.; Sanozky-Dawes, R.; Barrangou, R. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc. Natl. Acad. Sci. USA 2019, 116, 15774–15783. [Google Scholar] [CrossRef] [PubMed]

| Genome-Editing Tool/Approach | Nuclease(s) Employed | Editing Efficiencies Achieved | Validated Host Strains | Key Citation(s) |
|---|---|---|---|---|
| RecA-mediated double-crossover (classical homologous recombination) | None (RecA-dependent HR only) | Very low (~10−6–10−5 per cell without selection). Marker selection or counter-selection is usually required to recover mutants. | L. lactis, L. plantarum, L. casei (broadly all LAB, but extremely low efficiency without selection) | [2] |
| Phage RecT/RecE recombineering (λ-Red analogs from LAB prophages) | None (phage-encoded recombinases, e.g., RecT/Beta and RecE/Exo) | High for single-gene edits: often >50% of colonies correct; up to ~100% in optimized cases (e.g., 167 bp deletion or GFP insertion). Larger edits (~5 kb insertions) feasible with selection | L. plantarum WCFS1, L. casei BL23, L. lactis (with heterologous RecT). Primarily species-specific systems (phage origin) but functional across some LAB genus | [15,16] |
| CRISPR–Cas9 (DSB with HDR template) (Cas9-mediated counter-selection) | SpCas9 (wild-type, DSB-forming) | Point mutations and small deletions: ~90–100% efficiency (near-complete editing in L. reuteri with oligo donors). Small insertions (≤100 bp): ~50–75% efficiency in one step. Larger inserts (~1–2 kb) achieved at high frequency with optimized two-plasmid systems (e.g., ~80–90% for ~1 kb insert) | L. reuteri ATCC 6475, L. lactis NZ9000, L. plantarum WJL, L. paracasei, L. plantarum WCFS1, others (many LAB with species-specific optimization) | [34,35] |
| CRISPR–nCas9 (nickase-mediated HDR) (Cas9-D10A single-strand nick) | SpCas9 D10A (nickase variant) | 25–62% efficiency for precise deletions/insertions in L. casei. Up to ~60% efficiency in various Lactobacillus species using a portable nCas9 system. Significantly improved cell survival compared to DSB-causing Cas9 | L. casei BL23, L. acidophilus, L. gasseri, L. paracasei (demonstrated across multiple probiotic Lactobacillus) | [37,38] |
| CRISPR base editors (dCas9/nCas9 fused to deaminase for C→T or A→G editing) | Cas9 nickase or dCas9 fused with cytidine deaminase (CBE) or adenine deaminase (ABE) | High efficiency single-nucleotide conversions without DSB or donor DNA. For example, ~80–100% of cells acquired target C→T or A→G mutations in L. lactis, and similarly high efficiencies were achieved at multiple loci simultaneously. In L. plantarum, a multiplexable base-editing (Target-AID) system showed efficient point mutations with no survival penalty | L. lactis NZ9000, L. plantarum WCFS1, other Lactobacillus spp. (feasibility shown in multiple strains) | [39,41] |
| CRISPR-guided transposase (CAST; CRISPR-associated transposon integration) | Type I-F Cascade (Cas6/7/8 complex, no cutting) + TnsABC transposase (Tn7-like) | Allows large DNA insertions without HDR. ~2 × 10−4 efficiency for ~1 kb insertions, and on the order of 10−5 for ~10 kb payloads in L. lactis. Notably, inserts up to 10 kb were stably integrated in one step. Currently low absolute efficiency (10−4–10−5), but a major advance for payload size | L. lactis MG1363 (first demonstrated in LAB). Species-specific (system requires retooling for each host) | [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaposhnikov, L.A.; Rozanov, A.S.; Sazonov, A.E. Genome-Editing Tools for Lactic Acid Bacteria: Past Achievements, Current Platforms, and Future Directions. Int. J. Mol. Sci. 2025, 26, 7483. https://doi.org/10.3390/ijms26157483
Shaposhnikov LA, Rozanov AS, Sazonov AE. Genome-Editing Tools for Lactic Acid Bacteria: Past Achievements, Current Platforms, and Future Directions. International Journal of Molecular Sciences. 2025; 26(15):7483. https://doi.org/10.3390/ijms26157483
Chicago/Turabian StyleShaposhnikov, Leonid A., Aleksei S. Rozanov, and Alexey E. Sazonov. 2025. "Genome-Editing Tools for Lactic Acid Bacteria: Past Achievements, Current Platforms, and Future Directions" International Journal of Molecular Sciences 26, no. 15: 7483. https://doi.org/10.3390/ijms26157483
APA StyleShaposhnikov, L. A., Rozanov, A. S., & Sazonov, A. E. (2025). Genome-Editing Tools for Lactic Acid Bacteria: Past Achievements, Current Platforms, and Future Directions. International Journal of Molecular Sciences, 26(15), 7483. https://doi.org/10.3390/ijms26157483






