Genome-Wide Analyses of the XTH Gene Family in Brachypodium distachyon and Functional Analyses of the Role of BdXTH27 in Root Elongation
Abstract
1. Background
2. Results
2.1. Identification of the BdXTH Genes in Brachypodium distachyon
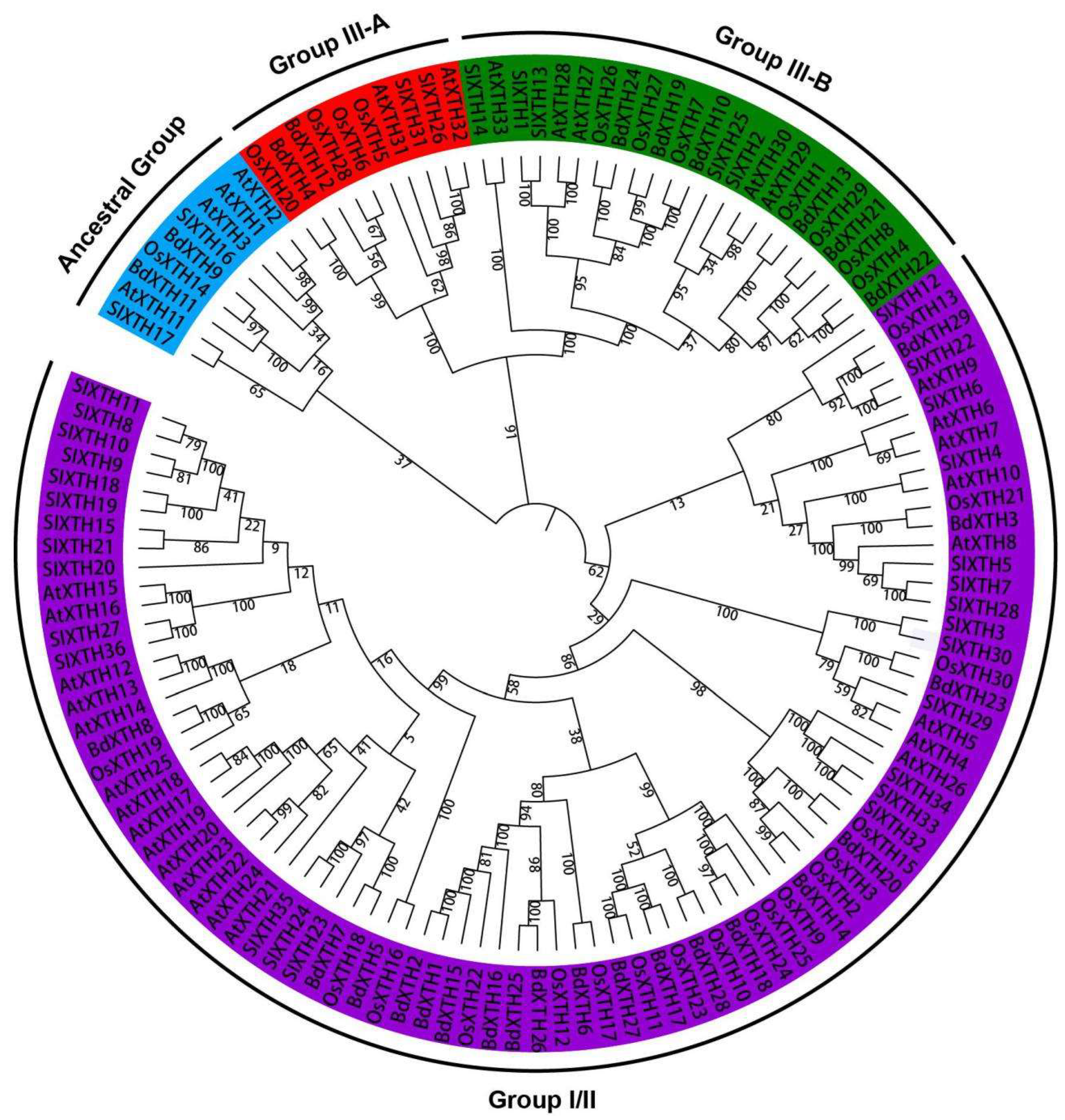
2.2. Evolutionary Analysis of BdXTH Proteins
2.3. Structural and Conserved Motif Analyses of the BdXTH Genes
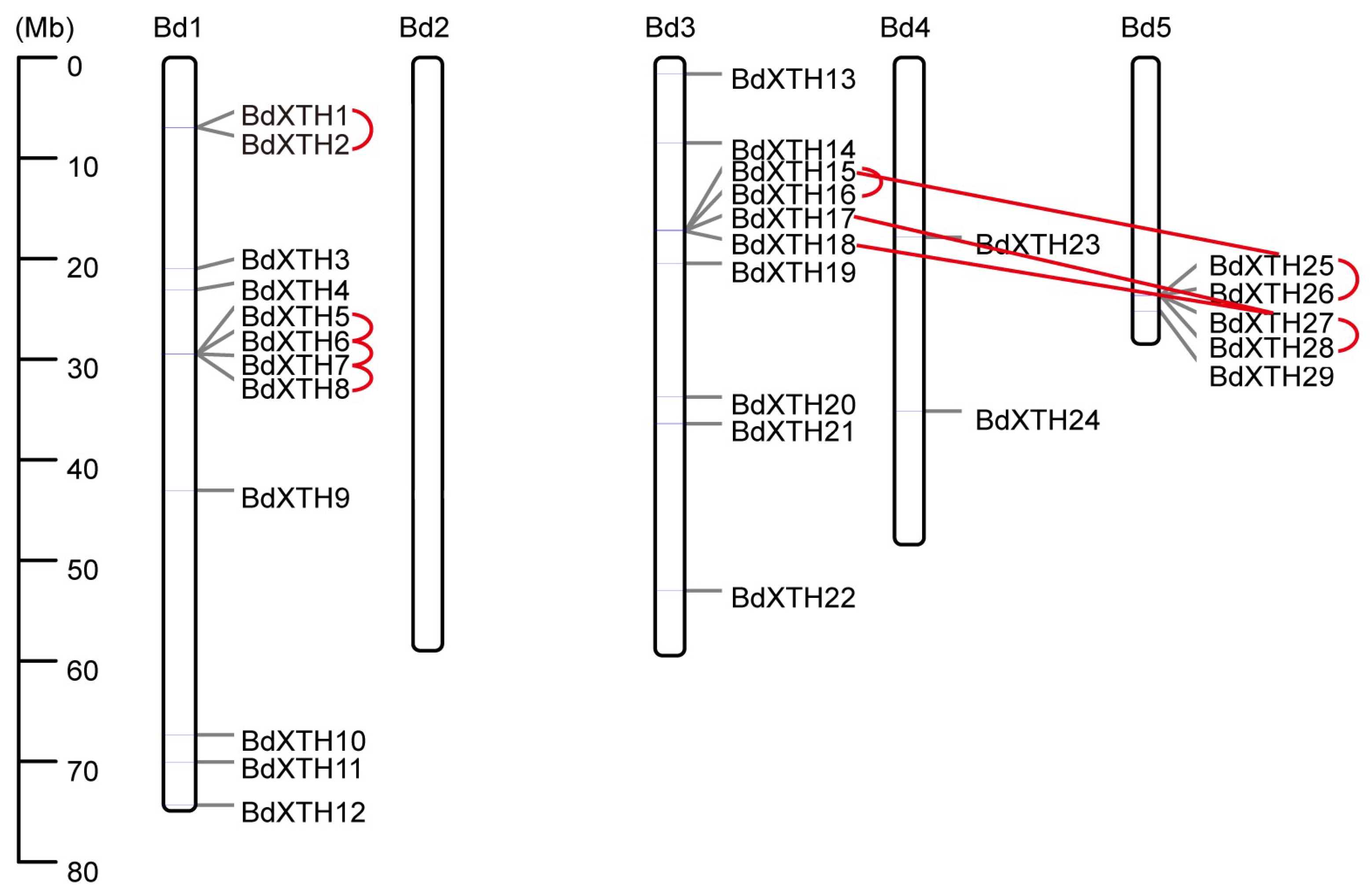
2.4. Chromosomal Location and Synteny Analysis of the BdXTH Genes
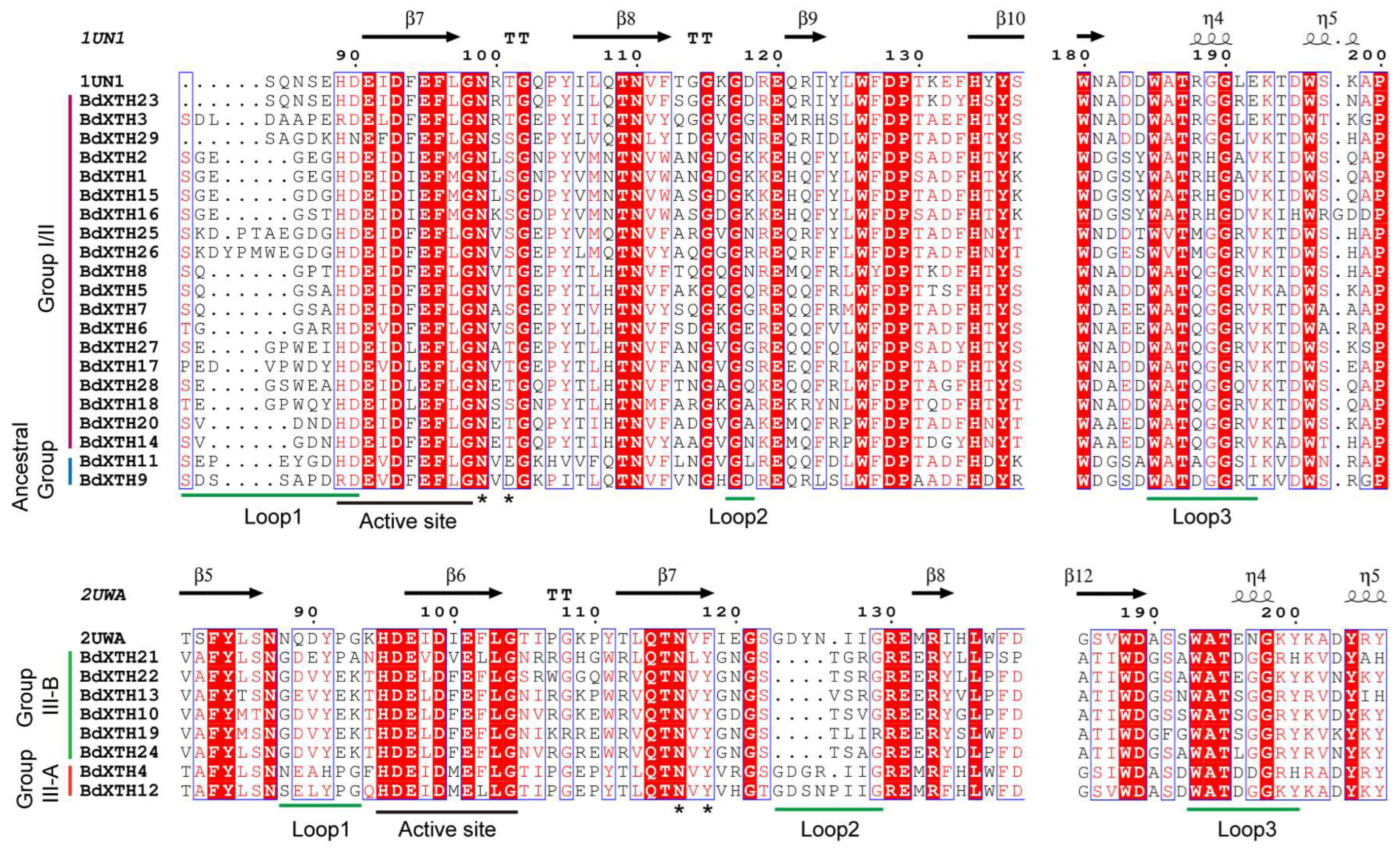
2.5. Structure-Based Sequence Alignment
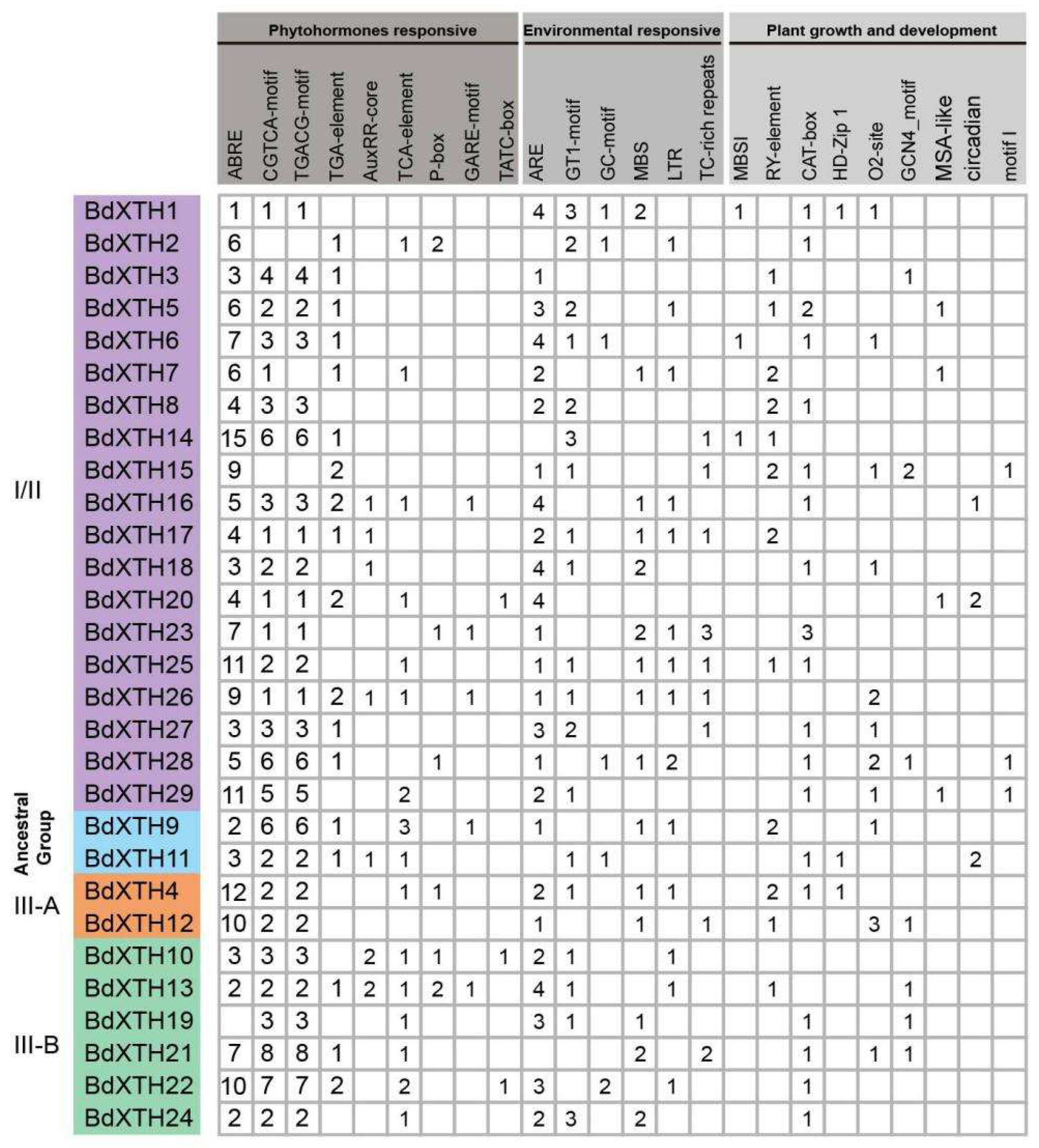
2.6. Cis-Element Analysis of the BdXTH Gene Promoter Regions
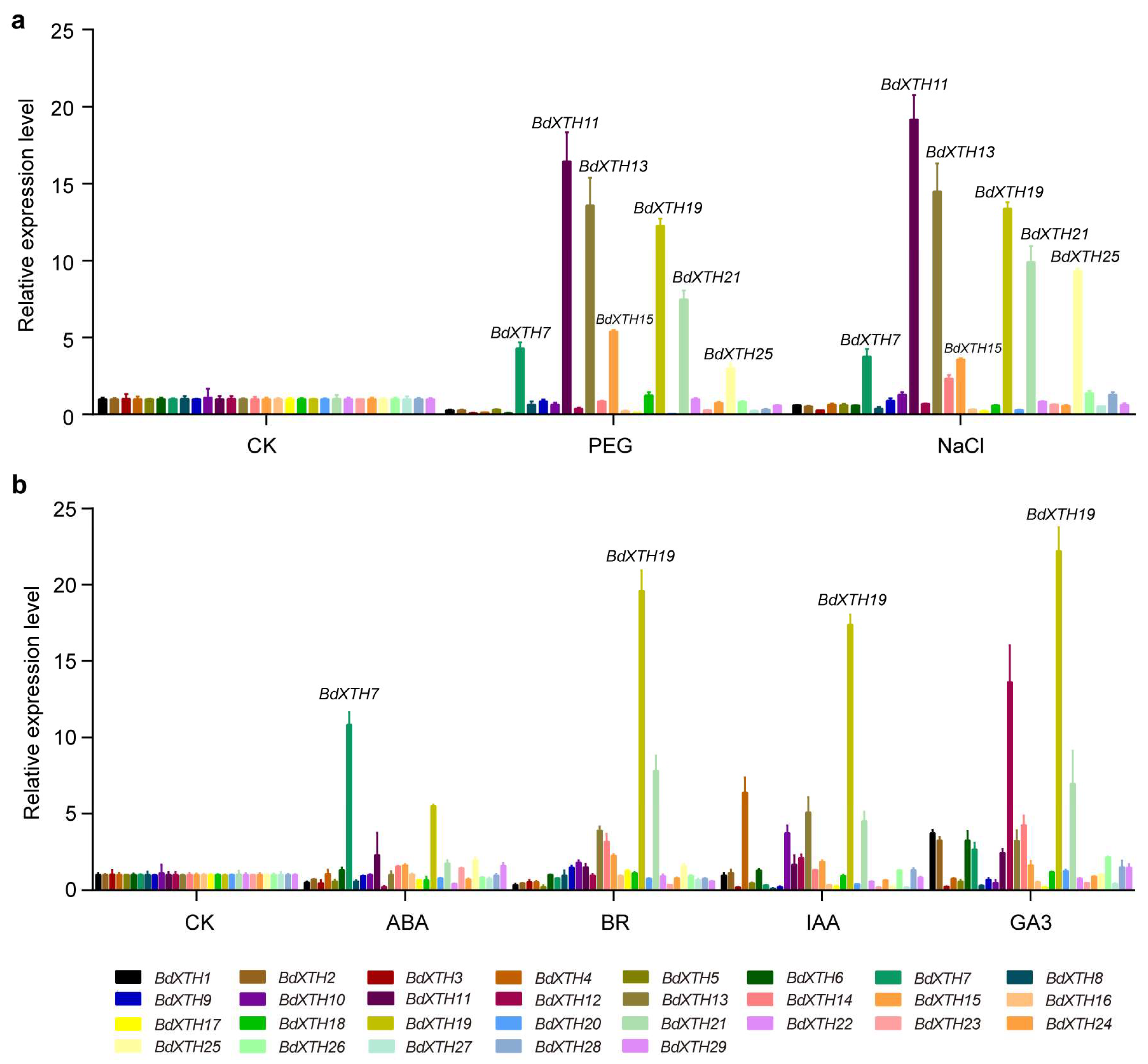
2.7. Expression Profiling of BdXTH Genes in Response to Abiotic Stress and Phytohormone Treatments Using qRT-PCR
2.8. Expression Patterns of the BdXTH Genes
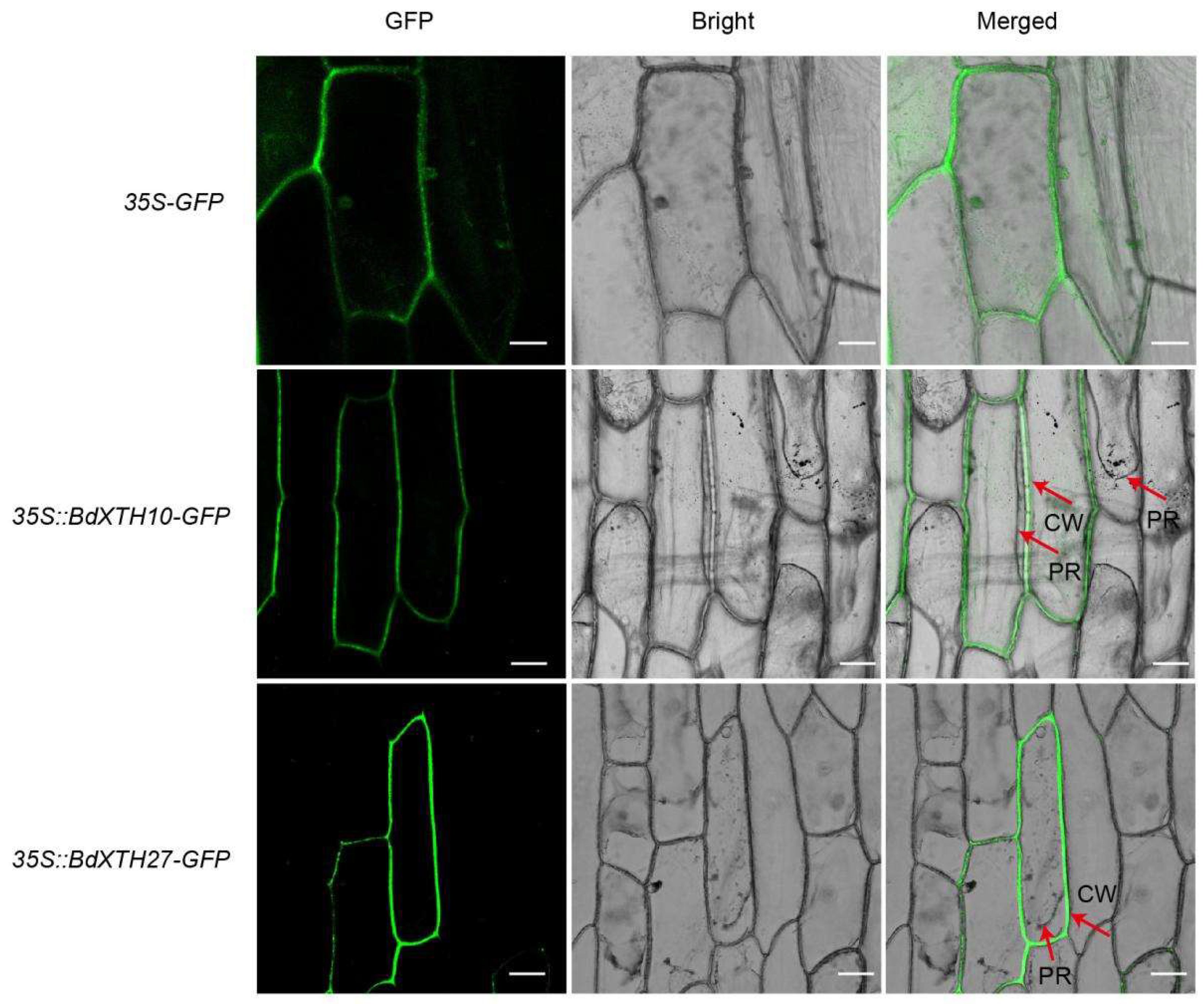
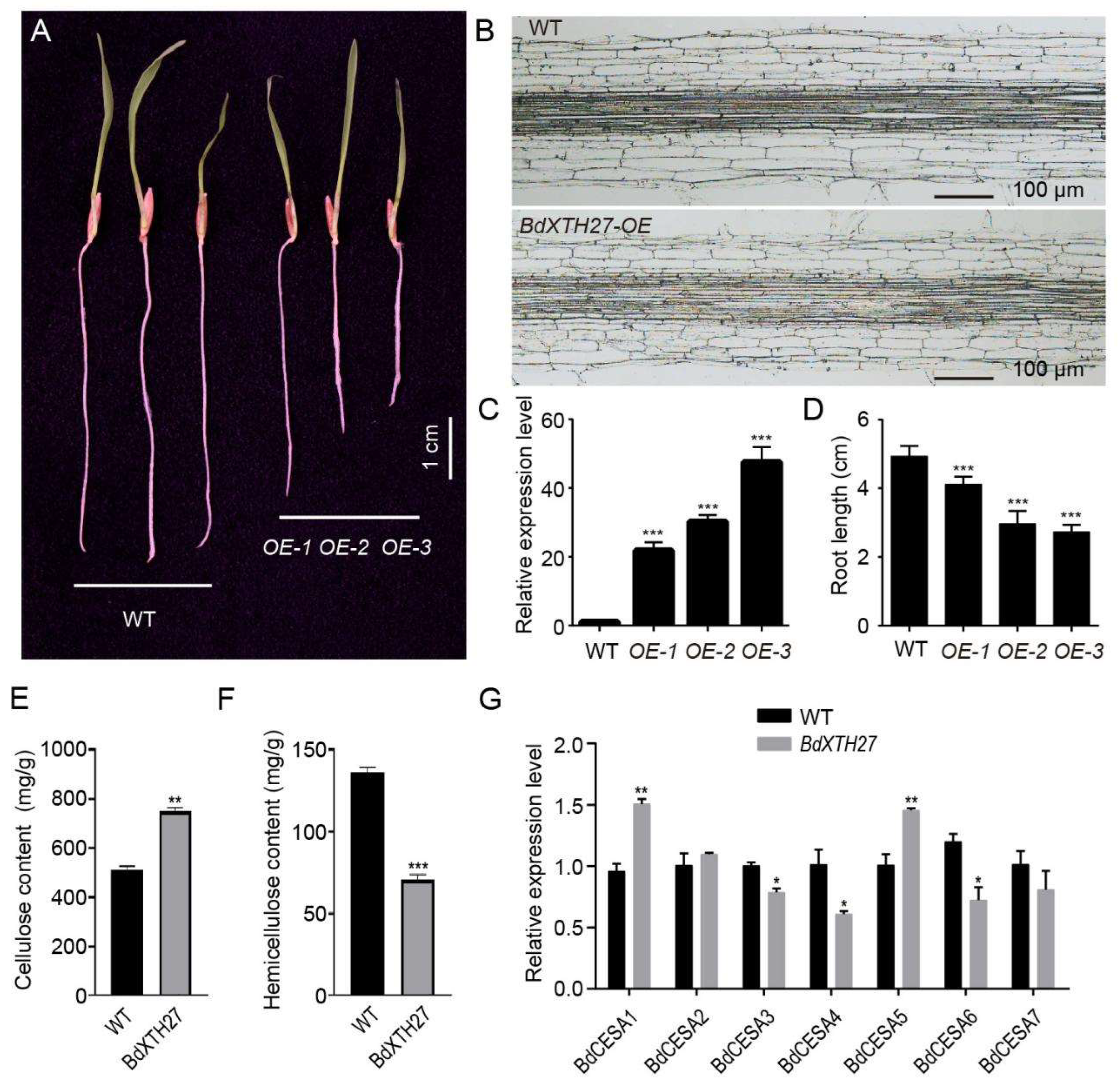
2.9. Functional Analysis of the BdXTH27 Gene in Brachypodium
3. Discussion
4. Methods
4.1. Identification of the XTH Family Genes in B. distachyon
4.2. Evolutionary Tree Construction
4.3. Gene Structures, Conserved Protein Motifs, and Cis-Acting Regulatory Element Analysis
4.4. Chromosomal Location and Gene Duplication
4.5. Structure-Based Sequence Alignment Analysis
4.6. Plant Materials and Treatments
4.7. Expression Pattern Analysis of BdXTH Genes Using the Public RNA-Seq Data
4.8. RNA Isolation and qRT-PCR Gene Expression Analysis
4.9. Identification of the BdCESA Genes in Brachypodium
4.10. Analysis of the BdXTH27-OE Transgenic Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Van Sandt, V.S.; Suslov, D.; Verbelen, J.P.; Vissenberg, K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 2007, 100, 1467–1473. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.; Braam, J. Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999, 4, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Nishitani, K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001, 42, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Rose, J.K.; Nishitani, K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 2004, 134, 1088–1099. [Google Scholar]
- Saladié, M.; Rose, J.K.; Cosgrove, D.J.; Catalá, C. Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. Plant J. 2006, 47, 282–295. [Google Scholar] [CrossRef]
- Bi, H.; Liu, Z.; Liu, S.; Qiao, W.; Zhang, K.; Zhao, M.; Wang, D. Genome-wide analysis of wheat xyloglucan endotransglucosylase/hydrolase (XTH) gene family revealed TaXTH17 involved in abiotic stress responses. BMC Plant Biol. 2024, 24, 640. [Google Scholar] [CrossRef]
- Yang, Y.; Miao, Y.; Zhong, S.; Fang, Q.; Wang, Y.; Dong, B.; Zhao, H. Genome-wide identification and expression analysis of XTH gene family during flower-opening stages in Osmanthus fragrans. Plants 2022, 11, 1015. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, L.; Li, D.; Qi, D.; Fang, F.; Luo, Y.; Zhang, H.; Zhang, S. Genome-wide characterization of the xyloglucan endotransglucosylase/hydrolase family genes and their response to plant hormone in sugar beet. Plant Physiol. Biochem. 2024, 206, 108239. [Google Scholar] [CrossRef]
- Refaiy, M.; Tahir, M.; Jiao, L.; Zhang, X.; Zhang, H.; Chen, Y.; Xu, Y.; Song, S.; Pang, X. Genome-wide identification of xyloglucan endotransglucosylase/hydrolase multigene family in Chinese Jujube (Ziziphus jujuba) and their expression patterns under different environmental stresses. Plants 2024, 13, 3503. [Google Scholar] [CrossRef]
- Yang, L.; Chen, Y.; Liu, X.; Zhang, S.; Han, Q. Genome-wide identification and expression analysis of xyloglucan endotransglucosylase/hydrolase genes family in Salicaceae during grafting. BMC Genom. 2023, 24, 676. [Google Scholar] [CrossRef]
- Ma, Y.S.; Jie, H.D.; Zhao, L.; Lv, X.Y.; Liu, X.C.; Tang, Y.Y.; Zhang, Y.; He, P.L.; Xing, H.C.; Jie, Y.C. Identification of the xyloglucan endotransglycosylase/hydrolase (XTH) gene family members expressed in Boehmeria nivea in response to cadmium stress. Int. J. Mol. Sci. 2022, 23, 16104. [Google Scholar] [CrossRef]
- Danso, B.; Ackah, M.; Jin, X.; Ayittey, D.M.; Amoako, F.K.; Zhao, W. Genome-wide analysis of the xyloglucan endotransglucosylase/hydrolase (XTH) gene family: Expression pattern during magnesium stress treatment in the mulberry plant (Morus alba L.) leaves. Plants 2024, 13, 902. [Google Scholar] [CrossRef]
- Tan, Y.; Zhan, H.; Chen, H.; Li, X.; Chen, C.; Liu, H.; Chen, Y.; Zhao, Z.; Xiao, Y.; Liu, J.; et al. Genome-wide identification of XTH gene family in Musa acuminata and response analyses of MaXTHs and xyloglucan to low temperature. Physiol. Plant 2024, 176, e14231. [Google Scholar] [CrossRef]
- Wu, D.; Liu, A.; Qu, X.; Liang, J.; Song, M. Genome-wide identification, and phylogenetic and expression profiling analyses, of XTH gene families in Brassica rapa L. and Brassica oleracea L. BMC Genom. 2020, 21, 782. [Google Scholar] [CrossRef] [PubMed]
- Baumann, M.J.; Eklöf, J.M.; Michel, G.; Kallas, A.M.; Teeri, T.T.; Czjzek, M.; Brumer, H., 3rd. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 2007, 19, 1947–1963. [Google Scholar] [CrossRef]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; Mao, C.Z.; et al. XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef] [PubMed]
- Kaewthai, N.; Gendre, D.; Eklöf, J.M.; Ibatullin, F.M.; Ezcurra, I.; Bhalerao, R.P.; Brumer, H. Group III-A XTH genes of Arabidopsis encode predominant xyloglucan endohydrolases that are dispensable for normal growth. Plant Physiol. 2013, 161, 440–454. [Google Scholar] [CrossRef]
- Han, Y.; Zhu, Q.; Zhang, Z.; Meng, K.; Hou, Y.; Ban, Q.; Suo, J.; Rao, J. Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes and diverse roles of isoenzymes during persimmon fruit development and postharvest softening. PLoS ONE 2015, 10, e0123668. [Google Scholar] [CrossRef] [PubMed]
- Witasari, L.D.; Huang, F.C.; Hoffmann, T.; Rozhon, W.; Fry, S.C.; Schwab, W. Higher expression of the strawberry xyloglucan endotransglucosylase/hydrolase genes FvXTH9 and FvXTH6 accelerates fruit ripening. Plant J. 2019, 100, 1237–1253. [Google Scholar] [CrossRef]
- Lee, Y.K.; Rhee, J.Y.; Lee, S.H.; Chung, G.C.; Park, S.J.; Segami, S.; Maeshima, M.; Choi, G. Functionally redundant LNG3 and LNG4 genes regulate turgor-driven polar cell elongation through activation of XTH17 and XTH24. Plant Mol. Biol. 2018, 97, 23–36. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, D. Association of allelic variation in PtoXET16A with growth and wood properties in Populus tomentosa. Int. J. Mol. Sci. 2014, 15, 16949–16974. [Google Scholar] [CrossRef]
- Zhu, X.F.; Wan, J.X.; Sun, Y.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Xyloglucan Endotransglucosylase-Hydrolase17 Interacts with Xyloglucan Endotransglucosylase-Hydrolase31 to Confer Xyloglucan Endotransglucosylase Action and Affect Aluminum Sensitivity in Arabidopsis. Plant Physiol. 2014, 165, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ban, Q.; Hou, Y.; Meng, K.; Suo, J.; Rao, J. Isolation and Characterization of Two Persimmon Xyloglucan Endotransglycosylase/Hydrolase (XTH) Genes That Have Divergent Functions in Cell Wall Modification and Fruit Postharvest Softening. Front. Plant Sci. 2016, 7, 624. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Han, S.; Ban, Q.; He, Y.; Jin, M.; Rao, J. Overexpression of persimmon DkXTH1 enhanced tolerance to abiotic stress and delayed fruit softening in transgenic plants. Plant Cell Rep. 2017, 36, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Girin, T.; David, L.C.; Chardin, C.; Sibout, R.; Krapp, A.; Ferrario-Méry, S.; Daniel-Vedele, F. Brachypodium: A promising hub between model species and cereals. J. Exp. Bot. 2014, 65, 5683–5696. [Google Scholar] [CrossRef]
- Ozdemir, B.S.; Hernandez, P.; Filiz, E.; Budak, H. Brachypodium genomics. Int. J. Plant Genomics 2008, 2008, 536104. [Google Scholar] [CrossRef]
- Brkljacic, J.; Grotewold, E.; Scholl, R.; Mockler, T.; Garvin, D.F.; Vain, P.; Brutnell, T.; Sibout, R.; Bevan, M.; Budak, H.; et al. Brachypodium as a model for the grasses: Today and the future. Plant Physiol. 2011, 157, 3–13. [Google Scholar] [CrossRef]
- Hsia, M.M.; O’Malley, R.; Cartwright, A.; Nieu, R.; Gordon, S.P.; Kelly, S.; Williams, T.G.; Wood, D.F.; Zhao, Y.; Bragg, J.; et al. Sequencing and functional validation of the JGI Brachypodium distachyon T-DNA collection. Plant J. 2017, 91, 361–370. [Google Scholar] [CrossRef]
- Wu, H.; Xue, X.; Qin, C.; Xu, Y.; Guo, Y.; Li, X.; Lv, W.; Li, Q.; Mao, C.; Li, L.; et al. An Efficient System for Ds Transposon Tagging in Brachypodium distachyon. Plant Physiol. 2019, 180, 56–65. [Google Scholar] [CrossRef]
- International Brachypodium, I. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Contreras-Moreira, B.; Woods, D.P.; Des Marais, D.L.; Burgess, D.; Shu, S.; Stritt, C.; Roulin, A.C.; Schackwitz, W.; Tyler, L.; et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 2017, 8, 2184. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.; Brumer, H., 3rd; Baumann, M.J.; Kallas, A.M.; Henriksson, H.; Denman, S.E.; Teeri, T.T.; Jones, T.A. Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 2004, 16, 874–886. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Rose, J.K.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.; Zhang, H.; Gao, H.; Guo, X.; Wang, D.; Zhang, X.; Zhang, A. The alpha- and beta-expansin and xyloglucan endotransglucosylase/hydrolase gene families of wheat: Molecular cloning, gene expression, and EST data mining. Genomics 2007, 90, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Fanutti, C.; Gidley, M.J.; Reid, J.S. Action of a pure xyloglucan endo-transglycosylase (formerly called xyloglucan-specific endo-(1-->4)-beta-D-glucanase) from the cotyledons of germinated nasturtium seeds. Plant J. 1993, 3, 691–700. [Google Scholar] [CrossRef]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools-an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Davidson, R.M.; Gowda, M.; Moghe, G.; Lin, H.; Vaillancourt, B.; Shiu, S.H.; Jiang, N.; Robin Buell, C. Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 2012, 71, 492–502. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xu, G.; Zhao, X.; Zhang, Z.; Wang, X.; Liu, X.; Xiao, W.; Fu, X.; Chen, X.; Gao, D.; et al. Transcription factor TCP20 regulates peach bud endodormancy by inhibiting DAM5/DAM6 and interacting with ABF2. J. Exp. Bot. 2020, 19, 1585–1597. [Google Scholar] [CrossRef]
- Zhao, Y.Q.; Song, X.Q.; Zhou, H.J.; Wei, K.L.; Jiang, C.; Wang, J.N.; Cao, Y.; Tang, F.; Zhao, S.; Lu, M.-Z. KNAT2/6b, a class I KNOX gene, impedes xylem differentiation by regulating NAC domain transcription factors in poplar. New Phytol. 2020, 225, 1531–1544. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Chromosome | Start | End | Length (aa) | MW (kDa) | PI | II | AI | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BdXTH1 | Bradi1g09690 | Bd1 | 6944672 | 6946132 | 284 | 31.55 | 5.67 | 36.99 | 63.87 | −0.27 | Cell wall |
| BdXTH2 | Bradi1g09700 | Bd1 | 6956014 | 6957576 | 284 | 31.54 | 5.67 | 36.12 | 63.87 | −0.26 | Cell wall |
| BdXTH3 | Bradi1g25847 | Bd1 | 20978556 | 20980916 | 307 | 34.16 | 5.09 | 39.79 | 63.03 | −0.34 | Cell wall |
| BdXTH4 | Bradi1g27867 | Bd1 | 23074430 | 23076645 | 302 | 34.11 | 8.73 | 40.95 | 58.91 | −0.46 | Cell wall |
| BdXTH5 | Bradi1g33810 | Bd1 | 29475206 | 29476739 | 290 | 32.39 | 8.78 | 30.99 | 58.24 | −0.42 | Cell wall |
| BdXTH6 | Bradi1g33817 | Bd1 | 29478854 | 29481513 | 300 | 34.28 | 6.71 | 43.09 | 71.2 | −0.36 | Cell wall |
| BdXTH7 | Bradi1g33827 | Bd1 | 29485795 | 29487172 | 284 | 31.45 | 5.73 | 38.54 | 62.64 | −0.34 | Cell wall |
| BdXTH8 | Bradi1g33840 | Bd1 | 29494938 | 29496678 | 293 | 32.59 | 6.22 | 31.26 | 69.66 | −0.28 | Cell wall/Cytoplasm |
| BdXTH9 | Bradi1g44777 | Bd1 | 43043568 | 43045106 | 301 | 33.45 | 5.05 | 43.37 | 68.37 | −0.33 | Cell wall |
| BdXTH10 | Bradi1g68590 | Bd1 | 67356916 | 67359656 | 328 | 36.46 | 6.02 | 52.51 | 67.65 | −0.39 | Cell wall |
| BdXTH11 | Bradi1g71937 | Bd1 | 70046381 | 70048307 | 284 | 31.51 | 6.44 | 36.9 | 78.27 | −0.12 | Cell wall |
| BdXTH12 | Bradi1g77990 | Bd1 | 74331199 | 74332831 | 318 | 35.24 | 7.00 | 46.49 | 61.07 | −0.41 | Cell wall |
| BdXTH13 | Bradi3g02700 | Bd3 | 1633396 | 1635150 | 336 | 37.66 | 8.57 | 51.44 | 76.10 | −0.22 | Cell wall |
| BdXTH14 | Bradi3g10290 | Bd3 | 8496465 | 8498025 | 288 | 32.10 | 4.91 | 32.13 | 66.32 | −0.33 | Cell wall |
| BdXTH15 | Bradi3g18590 | Bd3 | 17168440 | 17171803 | 280 | 31.02 | 4.91 | 38.97 | 59.21 | −0.38 | Cell wall |
| BdXTH16 | Bradi3g18600 | Bd3 | 17175111 | 17176766 | 301 | 33.71 | 5.29 | 28.49 | 60.66 | −0.52 | Cell wall |
| BdXTH17 | Bradi3g18607 | Bd3 | 17180627 | 17182323 | 291 | 33.41 | 4.85 | 42.45 | 63.68 | −0.51 | Cell wall |
| BdXTH18 | Bradi3g18690 | Bd3 | 17286604 | 17292191 | 289 | 32.83 | 6.89 | 32.97 | 71.87 | −0.41 | Cell wall |
| BdXTH19 | Bradi3g21337 | Bd3 | 20473847 | 20476548 | 354 | 39.32 | 8.75 | 48.62 | 75.06 | −0.25 | Cell wall |
| BdXTH20 | Bradi3g31767 | Bd3 | 20473847 | 20476548 | 289 | 32.32 | 5.78 | 26.27 | 69.24 | −0.26 | Cell wall |
| BdXTH21 | Bradi3g34227 | Bd3 | 36405786 | 36409262 | 330 | 35.58 | 7.83 | 44.98 | 79.88 | −0.18 | Cell wall |
| BdXTH22 | Bradi3g52307 | Bd3 | 53017512 | 53019765 | 345 | 37.56 | 8.83 | 33.85 | 64.09 | −0.23 | Cell wall |
| BdXTH23 | Bradi4g16990 | Bd4 | 17884860 | 17887177 | 294 | 33.37 | 6.25 | 37.64 | 64.76 | −0.49 | Cell wall/Cytoplasm |
| BdXTH24 | Bradi4g29707 | Bd4 | 35169644 | 35172434 | 338 | 37.31 | 6.41 | 52.96 | 70.74 | −0.29 | Cell wall |
| BdXTH25 | Bradi5g20718 | Bd5 | 23655071 | 23657848 | 316 | 34.23 | 4.67 | 44.62 | 44.62 | −0.19 | Cell wall |
| BdXTH26 | Bradi5g20726 | Bd5 | 23658483 | 23663566 | 372 | 40.94 | 5.79 | 43.33 | 67.96 | −0.25 | Cell wall |
| BdXTH27 | Bradi5g20734 | Bd5 | 23661776 | 23664848 | 295 | 33.66 | 4.83 | 38.07 | 76.41 | −0.29 | Cell wall |
| BdXTH28 | Bradi5g20742 | Bd5 | 23668827 | 23670387 | 279 | 30.54 | 5.19 | 40.05 | 67.89 | −0.26 | Cell wall |
| BdXTH29 | Bradi5g22907 | Bd5 | 25246820 | 25248701 | 314 | 34.27 | 5.37 | 31.8 | 73.34 | −0.25 | Cell wall |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, H.; Tan, Q.; Zhao, W.; Zhang, M.; Qin, C.; Liu, Z.; Wang, X.; An, S.; An, H.; Wu, H. Genome-Wide Analyses of the XTH Gene Family in Brachypodium distachyon and Functional Analyses of the Role of BdXTH27 in Root Elongation. Int. J. Mol. Sci. 2025, 26, 7457. https://doi.org/10.3390/ijms26157457
Shen H, Tan Q, Zhao W, Zhang M, Qin C, Liu Z, Wang X, An S, An H, Wu H. Genome-Wide Analyses of the XTH Gene Family in Brachypodium distachyon and Functional Analyses of the Role of BdXTH27 in Root Elongation. International Journal of Molecular Sciences. 2025; 26(15):7457. https://doi.org/10.3390/ijms26157457
Chicago/Turabian StyleShen, Hongyan, Qiuping Tan, Wenzhe Zhao, Mengdan Zhang, Cunhao Qin, Zhaobing Liu, Xinsheng Wang, Sendi An, Hailong An, and Hongyu Wu. 2025. "Genome-Wide Analyses of the XTH Gene Family in Brachypodium distachyon and Functional Analyses of the Role of BdXTH27 in Root Elongation" International Journal of Molecular Sciences 26, no. 15: 7457. https://doi.org/10.3390/ijms26157457
APA StyleShen, H., Tan, Q., Zhao, W., Zhang, M., Qin, C., Liu, Z., Wang, X., An, S., An, H., & Wu, H. (2025). Genome-Wide Analyses of the XTH Gene Family in Brachypodium distachyon and Functional Analyses of the Role of BdXTH27 in Root Elongation. International Journal of Molecular Sciences, 26(15), 7457. https://doi.org/10.3390/ijms26157457





