Inhibitory Effect of Allyl Isothiocyanate on Cariogenicity of Streptococcus mutans
Abstract
1. Introduction
2. Results
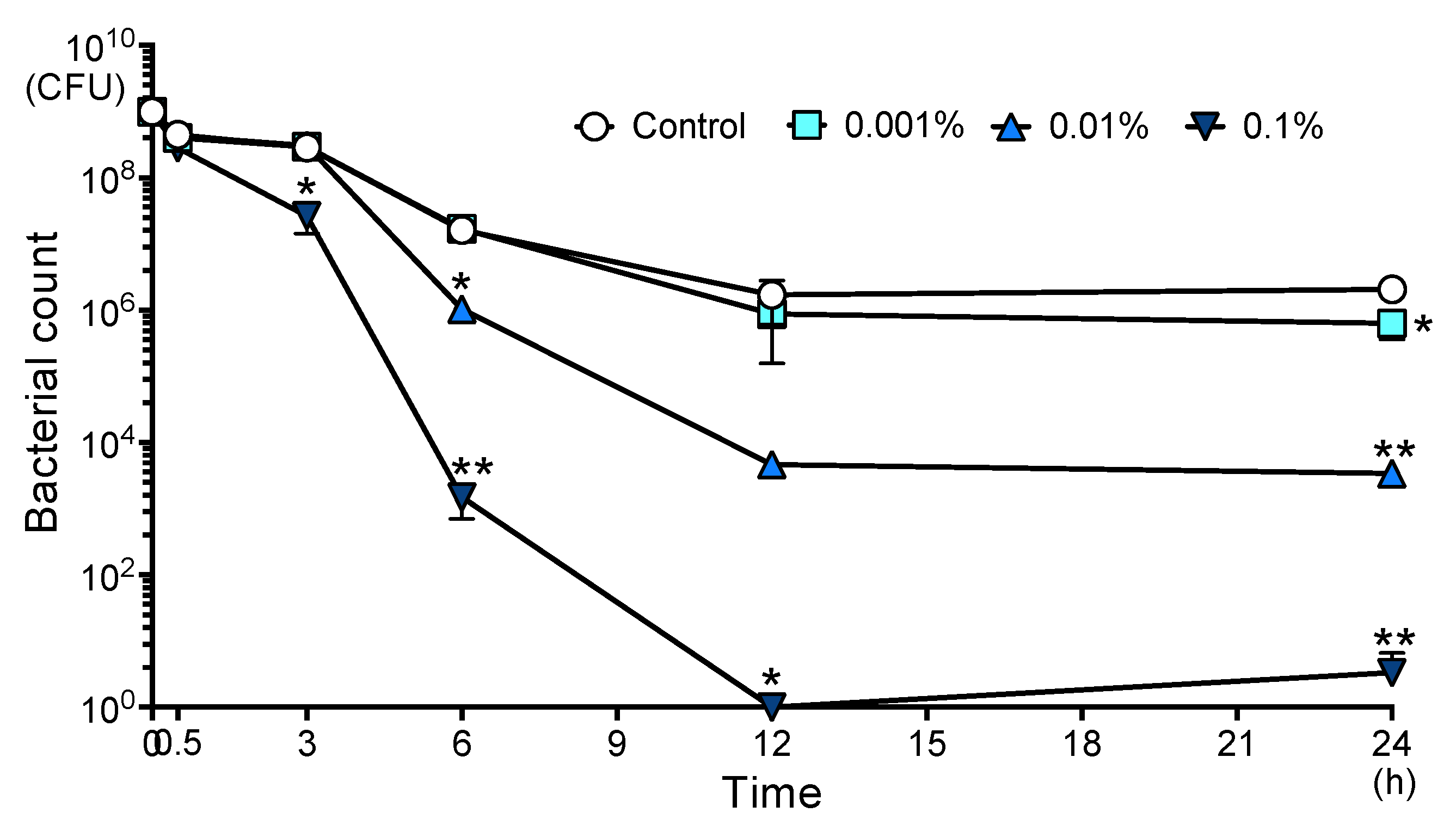
2.1. AITC Became Extinct S. mutans in a Concentration- and Time-Dependent Manner
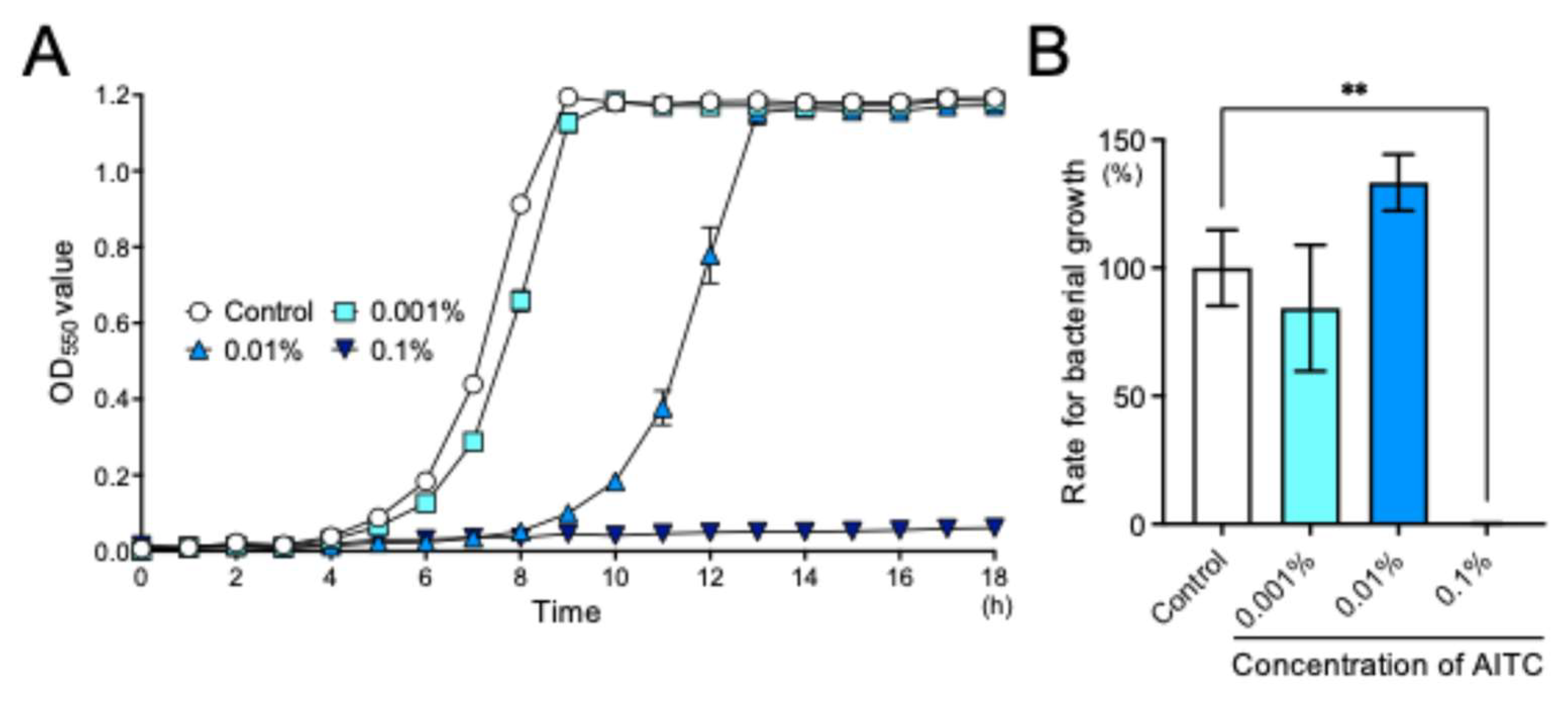
2.2. AITC Inhibited the Bacterial Growth
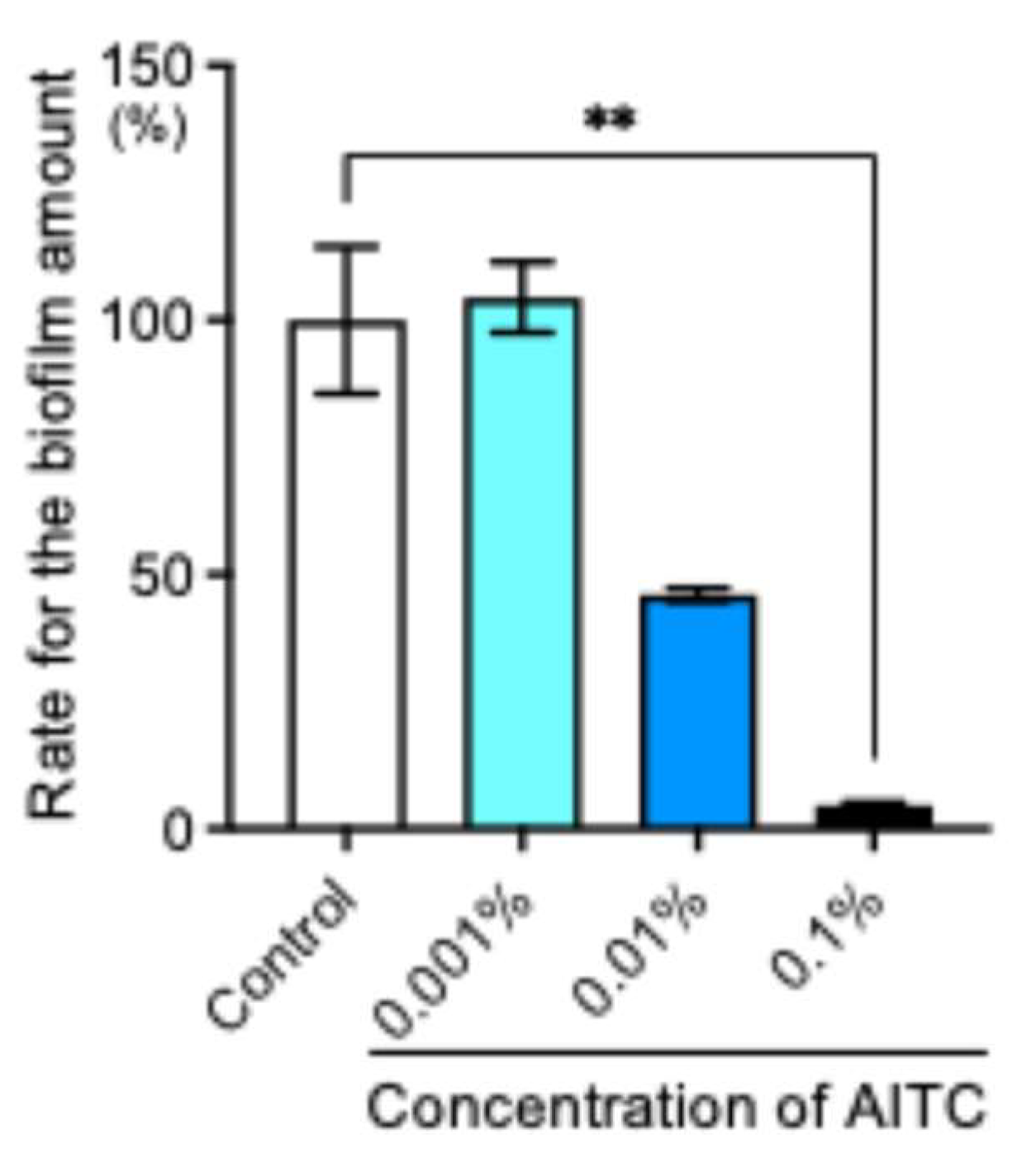
2.3. AITC Inhibited the Biofilm Formation
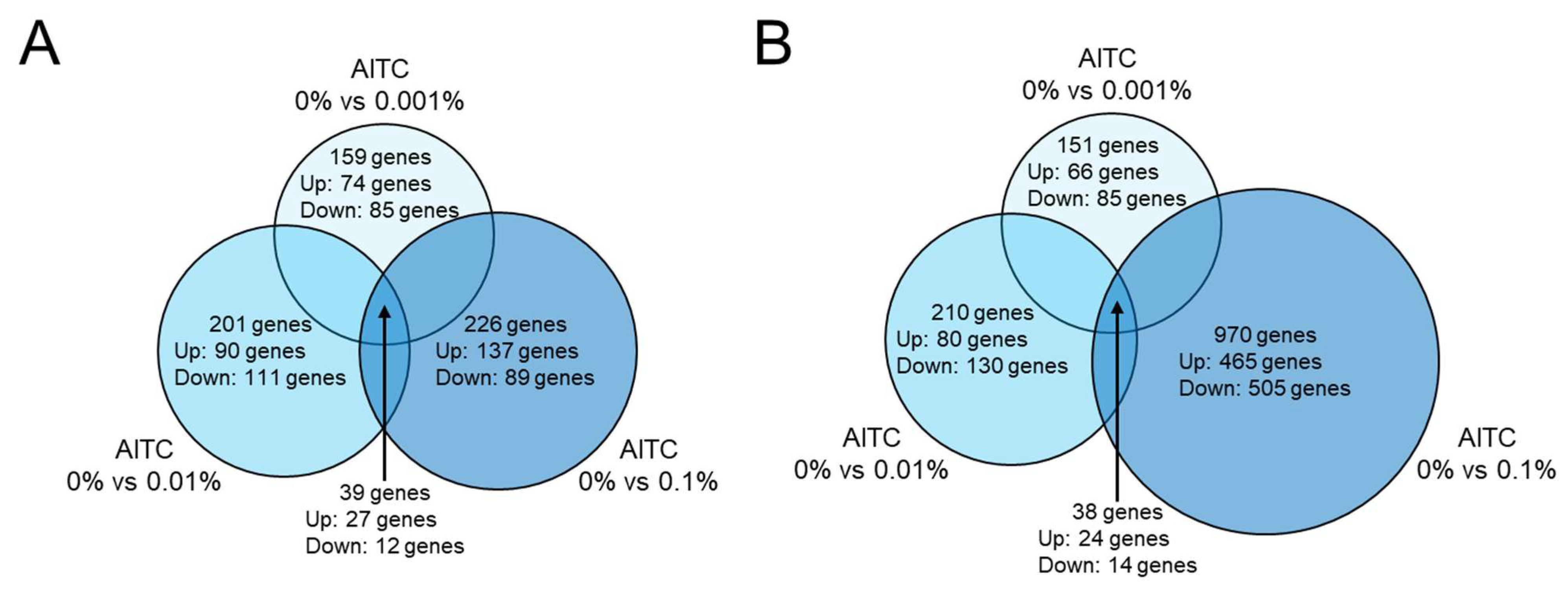
2.4. RNA Sequencing Analysis Revealed That Expression of 39 Genes Is Altered in Survival Assay and 38 Genes in Growth Assay in Three Conditions
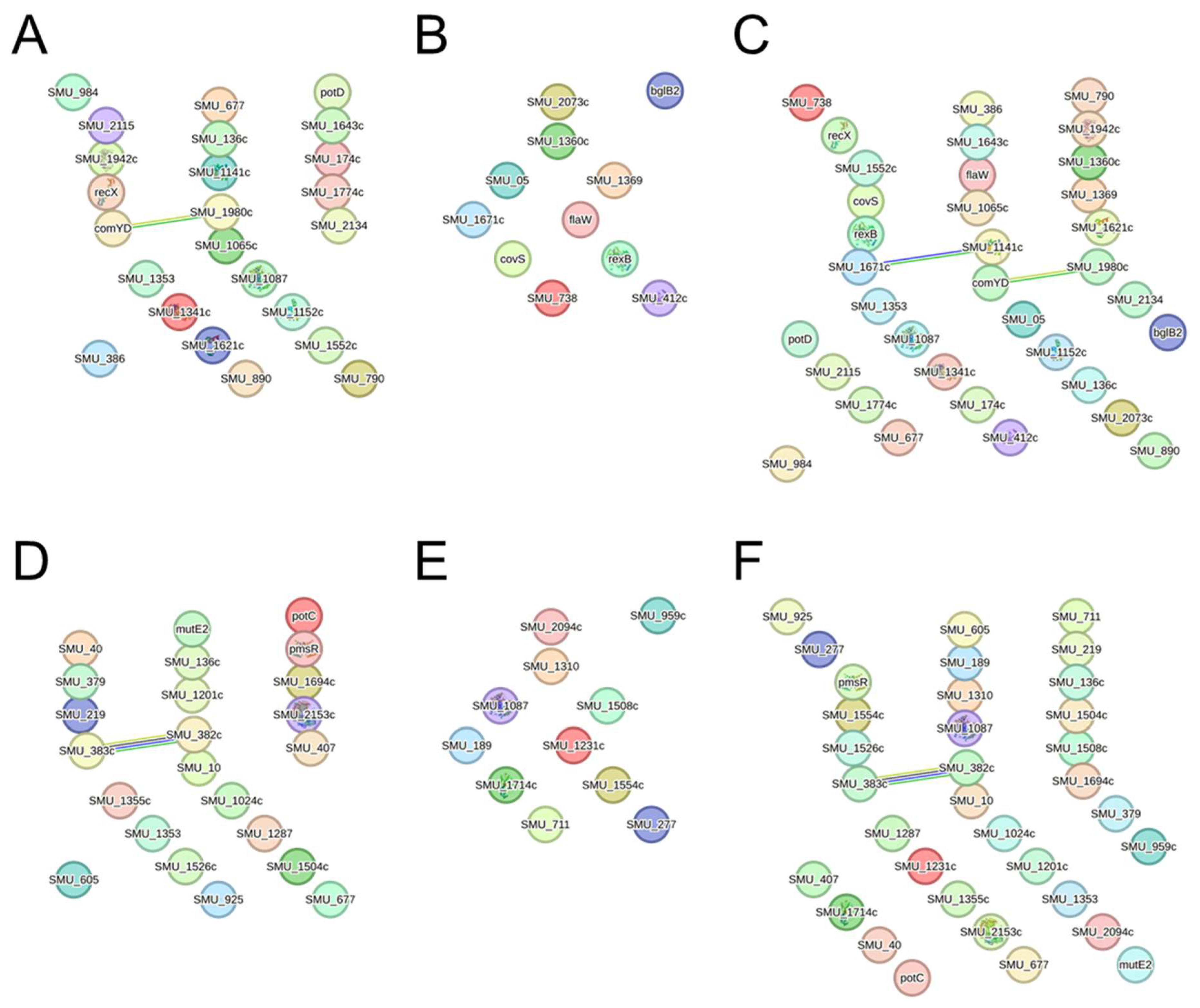
2.5. Protein–Protein Interaction Network with Genes Whose Expression Was Altered by AITC Was Constructed
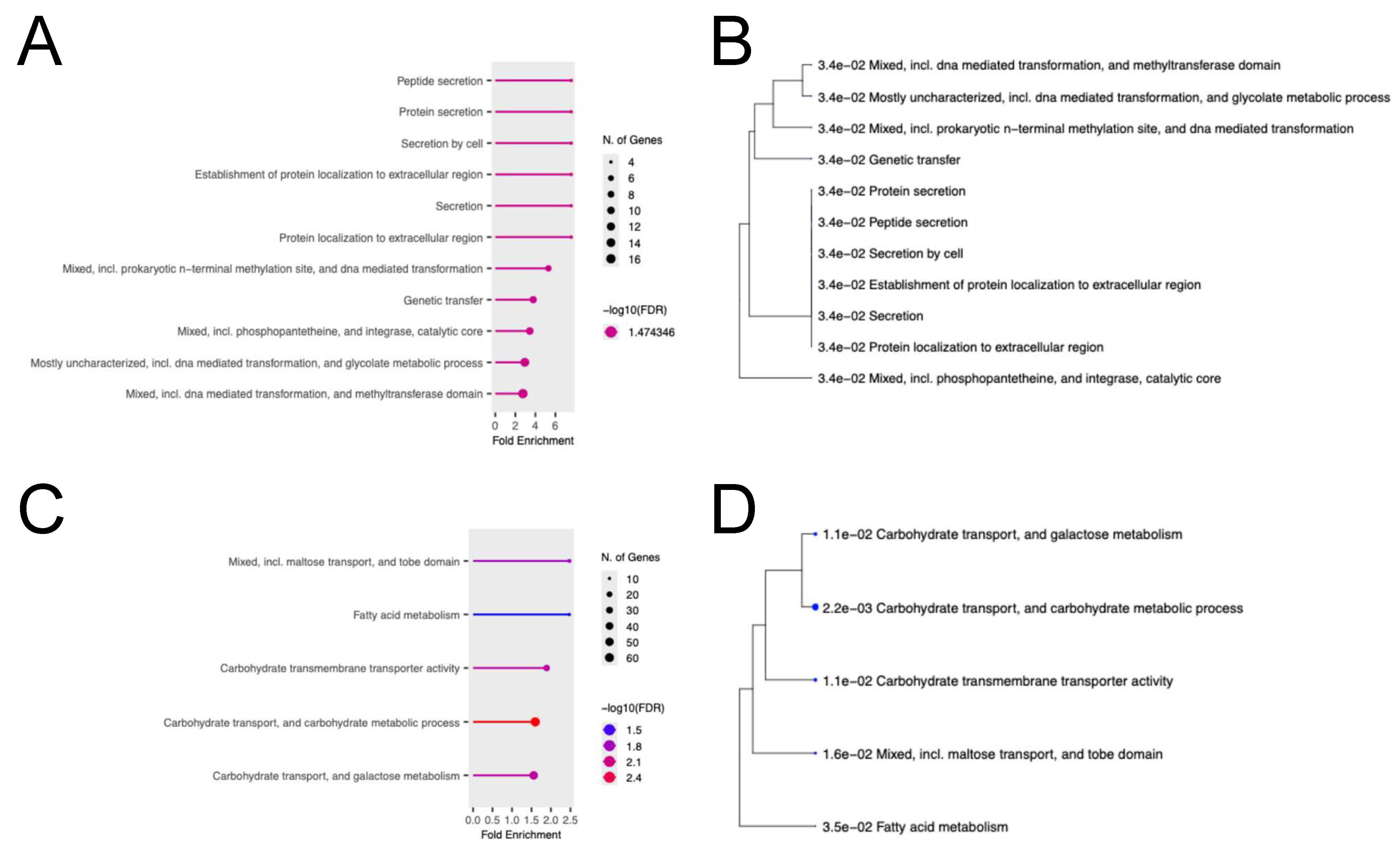
2.6. Gene Ontology (GO) Enrichment Analysis with Genes Whose Expression Was Altered by AITC Revealed Functional Interpretations
3. Discussion
3.1. Inhibitory Effect
3.2. Concentration
3.3. Analysis of Inhibitory Mechanism
3.4. Comparison with Existing Drugs
3.5. Current Limitations and Future Challenges for Clinical Application
4. Materials and Methods
4.1. S. mutans Strain and Culture Conditions
4.2. Materials
4.3. Bacterial Survival Assay
4.4. Bacterial Growth Assay
4.5. Biofilm Assay
4.6. RNA Sequencing Analysis
4.7. Bioinformatics Analyses
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, Y.; Chen, J.; Zhou, X.; Li, Y. Inhibition of Streptococcus mutans biofilm formation by strategies targeting the metabolism of exopolysaccharides. Crit. Rev. Microbiol. 2021, 47, 667–677. [Google Scholar] [CrossRef]
- Moynihan, P.; Petersen, P.E. Diet, nutrition and the prevention of dental diseases. Public Health Nutr. 2004, 7, 201–226. [Google Scholar] [CrossRef]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, K.D.; Na, H.S.; Jeong, S.Y.; Park, H.R.; Kim, S.; Chung, J. Tumor necrosis factor-α and interleukin-1β expression pathway induced by Streptococcus mutans in macrophage cell line RAW 264.7. Mol. Oral. Microbiol. 2012, 27, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Baik, J.E.; Kang, S.S.; Yun, C.H.; Seo, D.G.; Han, S.H. Lipoteichoic acid of Streptococcus mutans interacts with Toll-like receptor 2 through the lipid moiety for induction of inflammatory mediators in murine macrophages. Mol. Immunol. 2014, 57, 284–291. [Google Scholar] [CrossRef]
- Lucchese, A. Streptococcus mutans antigen I/II and autoimmunity in cardiovascular diseases. Autoimmun. Rev. 2017, 16, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Hong, Y.; Maishi, N.; Matsuda, A.Y.; Hida, Y.; Hasebe, A.; Kitagawa, Y.; Hida, K. Oral bacterium Streptococcus mutans promotes tumor metastasis through thrombosis formation. Cancer Sci. 2024, 115, 648–659. [Google Scholar] [CrossRef]
- Zhao, G.N.; Wong, H.M.; Wen, P.Y.F.; Wu, Y.; Zhong, Y.J.; Jiang, Y. Burden, Trends, and inequality of dental caries in the U.S., 1990–2019. Am. J. Prev. Med. 2023, 64, 788–796. [Google Scholar] [CrossRef]
- Chopra, B.; Dhingra, A.K. Natural products: A lead for drug discovery and development. Phytother. Res. 2021, 35, 4660–4702. [Google Scholar] [CrossRef]
- Naureen, Z.; Medori, M.C.; Dhuli, K.; Donato, K.; Connelly, S.T.; Bellinato, F.; Gisondi, P.; Bertelli, M. Polyphenols and Lactobacillus reuteri in oral health. J. Prev. Med. Hyg. 2022, 63 (Suppl. 3), E246–E254. [Google Scholar]
- Nomura, R.; Suehiro, Y.; Tojo, F.; Matayoshi, S.; Okawa, R.; Hamada, M.; Naka, S.; Matsumoto-Nakano, M.; Unesaki, R.; Koumoto, K.; et al. Inhibitory Effects of Shikonin Dispersion, an Extract of Lithospermum erythrorhizon Encapsulated in β-1,3-1,6 Glucan, on Streptococcus mutans and Non-Mutans Streptococci. Int. J. Mol. Sci. 2024, 25, 1075. [Google Scholar] [CrossRef]
- Usuda, M.; Kametani, M.; Hamada, M.; Suehiro, Y.; Matayoshi, S.; Okawa, R.; Naka, S.; Matsumoto-Nakano, M.; Akitomo, T.; Mitsuhata, C.; et al. Inhibitory effect of adsorption of Streptococcus mutans onto scallop-derived hydroxyapatite. Int. J. Mol. Sci. 2023, 24, 11371. [Google Scholar] [CrossRef]
- Abbas, M.; Gururani, M.A.; Ali, A.; Bajwa, S.; Hassan, R.; Batool, S.W.; Imam, M.; Wei, D. Antimicrobial properties and therapeutic potential of bioactive compounds in Nigella sativa: A Review. Molecules 2024, 29, 4914. [Google Scholar] [CrossRef]
- Mendes, P.M.; Gomes Fontoura, G.M.; Rodrigues, L.D.S.; Souza, A.S.; Viana, J.P.M.; Fernandes Pereira, A.L.; Dutra, R.P.; Nogueira Ferreira, A.G.; Neto, M.S.; Reis, A.S.; et al. Therapeutic potential of Punica granatum and isolated compounds: Evidence-based advances to treat bacterial infections. Int. J. Microbiol. 2023, 2023, 4026440. [Google Scholar] [CrossRef]
- Salimi, Y.; Tavahodi, N.; Taheri, H.; Masoudi, M.; Modaber, M.S.; Azimi, N.; Amin, M.N.; Bagharianlemraski, M.; Namadkolahi, R.; Khorami, M.; et al. Effect of Mangifera indica (Mango) on dental caries: A systematic review. Nutr. Metab. Insights 2023, 16, 11786388231204200. [Google Scholar] [CrossRef] [PubMed]
- Tarar, A.; Peng, S.; Cheema, S.; Peng, C.A. Anticancer activity, mechanism, and delivery of allyl isothiocyanate. Bioengineering 2022, 9, 470. [Google Scholar] [CrossRef]
- Duda-Chodak, A.; Tarko, T.; Petka-Poniatowska, K. Antimicrobial compounds in food packaging. Int. J. Mol. Sci. 2023, 24, 2457. [Google Scholar] [CrossRef]
- Joković, N.; Pešić, S.; Vitorović, J.; Bogdanović, A.; Sharifi-Rad, J.; Calina, D. Glucosinolates and their hydrolytic derivatives: Promising phytochemicals with anticancer potential. Phytother. Res. 2025, 39, 1035–1089. [Google Scholar] [CrossRef] [PubMed]
- Rajakumar, T.; Pugalendhi, P. Allyl isothiocyanate regulates oxidative stress, inflammation, cell proliferation, cell cycle arrest, apoptosis, angiogenesis, invasion and metastasis via interaction with multiple cell signaling pathways. Histochem. Cell Biol. 2024, 161, 211–221. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Zhang, Z.; Cao, Y.; Li, J.; Luo, L. Allyl isothiocyanate (AITC) triggered toxicity and FsYvc1 (a STRPC Family Member) responded sense in Fusarium solani. Front. Microbiol. 2020, 11, 870. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.M.; Preston, J.F., 3rd; Wei, C.I. Antibacterial mechanism of allyl isothiocyanate. J. Food Prot. 2000, 63, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Nomura, R.; Yoneyama, R.; Naka, S.; Otsugu, M.; Ogaya, Y.; Hatakeyama, R.; Morita, Y.; Maruo, J.; Matsumoto-Nakano, M.; Yamada, O.; et al. The in vivo inhibition of oral biofilm accumulation and Streptococcus mutans by ceramic water. Caries Res. 2017, 51, 58–67. [Google Scholar] [CrossRef]
- Scopelliti, F.; Dimartino, V.; Cattani, C.; Cavani, A. Functional TRPA1 channels regulate CD56dimCD16+NK cell cytotoxicity against tumor cells. Int. J. Mol. Sci. 2023, 24, 14736. [Google Scholar] [CrossRef]
- Hashimoto, T.; Nakamura, S.; Suzuki, T.; Hasegawa, Y.; Kanazawa, K. Cytotoxicity of allyl isothiocyanate and its metabolites in hepatocellular carcinoma HepG2 cells. Mutat. Res. 2025, 830, 111899. [Google Scholar] [CrossRef]
- Hashimoto, T.; Yoshioka, S.; Iwanaga, S.; Kanazawa, K. Anti-Malarial Activity of Allyl isothiocyanate and N-acetyl-S-(N-allylthiocarbamoyl)-l-Cysteine. Mol. Nutr. Food Res. 2023, 67, e2300185. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Liu, L.; Zhou, L.; Zhao, Z. Impacts of antibiotics on biofilm bacterial community and disinfection performance on simulated drinking water supply pipe wall. Environ. Pollut. 2021, 288, 117736. [Google Scholar] [CrossRef]
- Tavaddod, S.; Dawson, A.; Allen, R.J. Bacterial aggregation triggered by low-level antibiotic-mediated lysis. npj Biofilms Microbiomes 2024, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.O.; Kim, M.B.; Lim, S.B. Relationship between chemical structure and antimicrobial activities of isothiocyanates from cruciferous vegetables against oral pathogens. J. Microbiol. Biotechnol. 2016, 26, 2036–2042. [Google Scholar] [CrossRef]
- Ajdić, D.; McShan, W.M.; McLaughlin, R.E.; Savić, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.N.; Kim, D. Biofilm ecology associated with dental caries: Understanding of microbial interactions in oral communities leads to development of therapeutic strategies targeting cariogenic biofilms. Adv. Appl. Microbiol. 2023, 122, 27–75. [Google Scholar]
- Son, M.; Kaspar, J.; Ahn, S.J.; Burne, R.A.; Hagen, S.J. Threshold regulation and stochasticity from the MecA/ClpCP proteolytic system in Streptococcus mutans competence. Mol. Microbiol. 2018, 110, 914–930. [Google Scholar] [CrossRef]
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 2014, 12, 181–196. [Google Scholar] [CrossRef]
- Fontaine, L.; Wahl, A.; Fléchard, M.; Mignolet, J.; Hols, P. Regulation of competence for natural transformation in streptococci. Infect. Genet. Evol. 2015, 33, 343–360. [Google Scholar] [CrossRef]
- Merritt, J.; Qi, F.; Shi, W. A unique nine-gene comY operon in Streptococcus mutans. Microbiology 2005, 151, 157–166. [Google Scholar] [CrossRef]
- Son, M.; Shields, R.; Ahn, S.J.; Burne, R.A.; Hagen, S.J. Bidirectional signaling in the competence regulatory pathway of Streptococcus mutans. FEMS Microbiol. Lett. 2015, 362, fnv159. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Son, M.; Rosa-Alberty, A.E.; Zeng, L.; Ahn, S.; Hagen, S.J.; Burne, R.A. Effects of carbohydrate source on genetic competence in Streptococcus mutans. Appl. Environ. Microbiol. 2016, 82, 4821–4834. [Google Scholar] [CrossRef] [PubMed]
- De Furio, M.; Ahn, S.J.; Burne, R.A.; Hagen, S.J. Oxidative stressors modify the response of Streptococcus mutans to its competence signal peptides. Appl. Environ. Microbiol. 2017, 83, e01345-17. [Google Scholar] [CrossRef]
- Kametani, M.; Akitomo, T.; Hamada, M.; Usuda, M.; Kaneki, A.; Ogawa, M.; Ikeda, S.; Ito, Y.; Hamaguchi, S.; Kusaka, S.; et al. Inhibitory effects of surface pre-reacted glass ionomer filler eluate on Streptococcus mutans in the presence of sucrose. Int. J. Mol. Sci. 2024, 25, 9541. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.L.; Dixon, J.E. Bacterial and viral protein tyrosine phosphatases. Semin. Cell Biol. 1993, 4, 389–396. [Google Scholar] [CrossRef][Green Version]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. Biomed. Res. Int. 2013, 2013, 329121. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J.A. Biology of oral streptococci. Microbiol. Spectr. 2018, 6, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Bloch, S.; Hager-Mair, F.F.; Andrukhov, O.; Schäffer, C. Oral streptococci: Modulators of health and disease. Front. Cell Infect. Microbiol. 2024, 14, 1357631. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef]
- Finkel, S.E.; Kolter, R. DNA as a nutrient: Novel role for bacterial competence gene homologs. J. Bacteriol. 2001, 183, 6288–6293. [Google Scholar] [CrossRef]
- Jamali, H.; Akrami, F.; Layeghkhavidaki, H.; Bouakkaz, S. Bacterial protein secretion systems: Mechanisms, functions, and roles in virulence. Microb. Pathog. 2025, 206, 107790. [Google Scholar] [CrossRef]
- Carreón-Rodríguez, O.E.; Gosset, G.; Escalante, A.; Bolívar, F. Glucose transport in Escherichia coli: From basics to transport engineering. Microorganisms 2023, 11, 1588. [Google Scholar] [CrossRef]
- Kolmas, J.; Groszyk, E.; Kwiatkowska-Różycka, D. Substituted hydroxyapatites with antibacterial properties. Biomed. Res. Int. 2014, 2014, 178123. [Google Scholar] [CrossRef]
- Poppolo Deus, F.; Ouanounou, A. Chlorhexidine in dentistry: Pharmacology, uses, and adverse effects. Int. Dent. J. 2022, 72, 269–277. [Google Scholar] [CrossRef]
- Guedes, B.N.; Krambeck, K.; Durazzo, A.; Lucarini, M.; Santini, A.; Oliveira, M.B.P.P.; Fathi, F.; Souto, E.B. Natural antibiotics against antimicrobial resistance: Sources and bioinspired delivery systems. Braz. J. Microbiol. 2024, 55, 2753–2766. [Google Scholar] [CrossRef] [PubMed]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms 2021, 9, 2041. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.A.; Kim, W.Y.; Park, H.J. Effects of microencapsulated Allyl isothiocyanate (AITC) on the extension of the shelf-life of Kimchi. Int. J. Food Microbiol. 2012, 153, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Kobayashi, K.; Murata, E.; Enomoto, H. Effects of drying temperature and storage conditions on anthocyanins and isothiocyanates in red turnip (Brassica campestris L.). Bull. Fukui Agric. Exp. Stn. 2009, 46, 55–58. (In Japanese) [Google Scholar]
- Minato, Y.; Aoki-Nonaka, Y.; Lwin, H.Y.; Ando, D.; Warita, Y.; Matsugishi-Nasu, A.; Hiyoshi, T.; Takahashi, N.; Tabeta, K. Allyl isothiocyanate suppressed periodontal tissue destruction in mice via bacteriostatic and anti-inflammatory activities against Porphyromonas gingivalis. Arch. Oral Biol. 2025, 169, 106118. [Google Scholar] [CrossRef] [PubMed]

| Fold Change | ||||
|---|---|---|---|---|
| Gene Name | Product | AITC 0.001% | AITC 0.01% | AITC 0.1% |
| SMU_1341c | putative gramicidin S synthetase | 3.128 | 3.517 | 2.162 |
| SMU_677 | putative transcriptional regulator (MerR family) | 5.758 | 4.382 | 2.307 |
| SMU_790 | hypothetical protein | 2.879 | 2.191 | 2.401 |
| SMU_1942c | putative amino acid binding protein | 2.720 | 2.009 | 2.798 |
| SMU_1065c | putative transcriptional regulator (GntR family) | 2.506 | 3.104 | 3.335 |
| SMU_984 | hypothetical protein | 2.112 | 2.739 | 3.481 |
| SMU_1141c | hypothetical protein | 2.150 | 4.659 | 3.484 |
| SMU_386 | putative ribosomal-protein-alanine acetyltransferase | 4.072 | 2.858 | 3.868 |
| SMU_1621c | hypothetical protein | 3.272 | 3.706 | 4.242 |
| SMU_2115 | putative short-chain dehydrogenase | 4.799 | 6.035 | 4.802 |
| SMU_174c | hypothetical protein | 2.160 | 2.189 | 5.405 |
| SMU_1774c | hypothetical protein | 3.841 | 4.385 | 7.207 |
| SMU_1780 | hypothetical protein | 6.727 | 7.679 | 9.617 |
| SMU_890 | hypothetical protein | 8.656 | 7.685 | 12.031 |
| SMU_1983 (comYD) | putative competence protein ComYD | 5.710 | 3.037 | 12.204 |
| SMU_t36 | tRNA-Ser | 2.399 | 4.382 | 12.604 |
| SMU_1980c | hypothetical protein | 27.975 | 3.112 | 17.253 |
| SMU_2134 | putative transcriptional regulator | 33.375 | 29.623 | 18.550 |
| SMU_976 (potD) | putative ABC transporter, periplasmic spermidine/putrescine-binding protein | 37.522 | 57.046 | 24.312 |
| SMU_1552c | hypothetical protein | 3.626 | 27.443 | 30.070 |
| SMU_1643c | hypothetical protein | 22.405 | 42.631 | 37.369 |
| SMU_136c | putative transcriptional regulator | 25.906 | 9.852 | 64.797 |
| SMU_1087 | putative 4-oxalocrotonate tautomerase | 349.836 | 399.330 | 70.345 |
| SMU_1353 | putative transposase | 759.405 | 247.650 | 135.672 |
| SMU_1152c | hypothetical protein | 141.089 | 140.197 | 169.344 |
| SMU_t34 | tRNA-Ser | 3937.040 | 6291.730 | 7878.940 |
| SMU_t38 | tRNA-Gln | 13,594.000 | 12,069.100 | 18,892.200 |
| Fold Change | ||||
|---|---|---|---|---|
| Gene Name | Product | AITC 0.001% | AITC 0.01% | AITC 0.01% |
| SMU_t64 | tRNA-Glu | −2.084 | −3695.480 | −3695.480 |
| SMU_738 | hypothetical protein | −2.085 | −361.099 | −361.099 |
| SMU_1369 | hypothetical protein | −267.274 | −267.274 | −267.274 |
| SMU_2073c | hypothetical protein | −206.566 | −206.566 | −206.566 |
| SMU_1516 (covS) | putative histidine kinase CovS; VicK-like protein | −3.413 | −159.146 | −159.146 |
| SMU_1360c | hypothetical protein | −105.478 | −2.327 | −105.478 |
| SMU_1500 (rexB) | putative exonuclease RexB | −4.356 | −111.655 | −3.110 |
| SMU_05 | hypothetical protein | −9.701 | −489.950 | −2.518 |
| SMU_1671c | hypothetical protein | −2.747 | −5.295 | −2.416 |
| SMU_982 (bglB2) | putative BglB fragment | −2.214 | −3.104 | −2.361 |
| SMU_412c | putative Hit-like protein involved in cell-cycle regulation | −5.706 | −2.199 | −2.330 |
| SMU_1294 (flaW) | putative flavodoxin | −3.460 | −56.287 | −2.075 |
| Fold Change | ||||
|---|---|---|---|---|
| Gene Name | Product | AITC 0.001% | AITC 0.01% | AITC 0.1% |
| SMU_975 (potC) | putative spermidine/putrescine ABC transporter, permease protein | 2.542 | 2.850 | 2.155 |
| SMU_40 | conserved hypothetical protein | 4.581 | 5.199 | 2.534 |
| SMU_1694c | putative permease | 2.231 | 2.120 | 2.569 |
| SMU_10 | conserved hypothetical protein | 3.192 | 2.741 | 3.359 |
| SMU_1504c | hypothetical protein | 3.088 | 3.004 | 3.578 |
| SMU_677 | putative transcriptional regulator (MerR family) | 4.617 | 2.414 | 3.985 |
| SMU_605 | hypothetical protein | 7.036 | 6.022 | 4.183 |
| SMU_925 | hypothetical protein | 2.307 | 2.156 | 6.673 |
| SMU_219 | hypothetical protein | 4.394 | 3.506 | 8.167 |
| SMU_2153c | putative peptidase | 2.143 | 2.003 | 11.997 |
| SMU_1622 (pmsR) | putative peptide methionine sulfoxide reductase | 3.725 | 3.278 | 13.382 |
| SMU_1355c | putative transposase fregment | 2.649 | 3.007 | 17.162 |
| SMU_1287 | putative transcriptional regulator | 2.649 | 4.010 | 18.061 |
| SMU_407 | conserved hypothetical protein | 3.175 | 3.986 | 19.023 |
| SMU_382c | putative oxidoreductase | 2.037 | 5.078 | 24.988 |
| SMU_383c | conserved hypothetical protein; putative reductase | 2.171 | 4.262 | 25.245 |
| SMU_1201c | conserved hypothetical protein | 87.466 | 95.263 | 85.115 |
| SMU_136c | putative transcriptional regulator | 15.933 | 9.040 | 87.519 |
| SMU_1526c | conserved hypothetical protein | 106.669 | 40.362 | 132.068 |
| SMU_1024c | putative transposase fragment | 174.941 | 198.571 | 133.015 |
| SMU_656 (mutE2) | putative MutE | 33.486 | 33.501 | 151.823 |
| SMU_379 | hypothetical protein | 88.063 | 176.696 | 236.467 |
| SMU_1353 | putative transposase | 199.926 | 113.588 | 258.676 |
| SMU_t35 | tRNA-Leu | 2832.920 | 1071.800 | 7180.290 |
| Fold Change | ||||
|---|---|---|---|---|
| Gene Name | Product | AITC 0.001% | AITC 0.01% | AITC 0.1% |
| SMU_t25 | tRNA-Glu | −1855.060 | −1855.060 | −1855.060 |
| SMU_t38 | tRNA-Gln | −1580.070 | −1580.070 | −1580.070 |
| SMU_t33 | tRNA-Leu | −1023.210 | −1023.210 | −1023.210 |
| SMU_1231c | hypothetical protein | −672.496 | −672.496 | −672.496 |
| SMU_1310 | hypothetical protein | −176.481 | −176.481 | −176.481 |
| SMU_1554c | hypothetical protein | −98.370 | −98.370 | −98.370 |
| SMU_711 | conserved hypothetical protein | −2.645 | −3.496 | −51.212 |
| SMU_1714c | hypothetical protein | −27.984 | −27.984 | −27.984 |
| SMU_1508c | putative coenzyme PQQ synthesis protein | −2.455 | −22.668 | −22.668 |
| SMU_959c | hypothetical protein | −3.507 | −6.431 | −6.998 |
| SMU_189 | hypothetical protein | −42.282 | −42.282 | −6.815 |
| SMU_277 | hypothetical protein | −3.401 | −2.248 | −6.710 |
| SMU_1087 | putative 4-oxalocrotonate tautomerase | −3.401 | −5.994 | −5.751 |
| SMU_2094c | conserved hypothetical protein | −2.721 | −5.996 | −3.372 |
| Annotations | |||
|---|---|---|---|
| Assay | Gene Name | Molecular Function (Term) | Biological Process (Term) |
| Survival | SMU_1980c | - | - |
| Survival | SMU_1983 (comYD) | - | Response to stimulus (Establishment of competence for transformation) |
| Survival | SMU_1141c | - | - |
| Survival | SMU_1671c | - | - |
| Growth | SMU_382c | Catalytic activity (Oxidoreductase activity) | - |
| Growth | SMU_383c | Catalytic activity (Aldehyde dehydrogenase activity) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akitomo, T.; Kaneki, A.; Ogawa, M.; Ito, Y.; Hamaguchi, S.; Ikeda, S.; Kametani, M.; Usuda, M.; Kusaka, S.; Hamada, M.; et al. Inhibitory Effect of Allyl Isothiocyanate on Cariogenicity of Streptococcus mutans. Int. J. Mol. Sci. 2025, 26, 7443. https://doi.org/10.3390/ijms26157443
Akitomo T, Kaneki A, Ogawa M, Ito Y, Hamaguchi S, Ikeda S, Kametani M, Usuda M, Kusaka S, Hamada M, et al. Inhibitory Effect of Allyl Isothiocyanate on Cariogenicity of Streptococcus mutans. International Journal of Molecular Sciences. 2025; 26(15):7443. https://doi.org/10.3390/ijms26157443
Chicago/Turabian StyleAkitomo, Tatsuya, Ami Kaneki, Masashi Ogawa, Yuya Ito, Shuma Hamaguchi, Shunya Ikeda, Mariko Kametani, Momoko Usuda, Satoru Kusaka, Masakazu Hamada, and et al. 2025. "Inhibitory Effect of Allyl Isothiocyanate on Cariogenicity of Streptococcus mutans" International Journal of Molecular Sciences 26, no. 15: 7443. https://doi.org/10.3390/ijms26157443
APA StyleAkitomo, T., Kaneki, A., Ogawa, M., Ito, Y., Hamaguchi, S., Ikeda, S., Kametani, M., Usuda, M., Kusaka, S., Hamada, M., Mitsuhata, C., Kozai, K., & Nomura, R. (2025). Inhibitory Effect of Allyl Isothiocyanate on Cariogenicity of Streptococcus mutans. International Journal of Molecular Sciences, 26(15), 7443. https://doi.org/10.3390/ijms26157443





