Enhancing Oil Content in Oilseed Crops: Genetic Insights, Molecular Mechanisms, and Breeding Approaches
Abstract
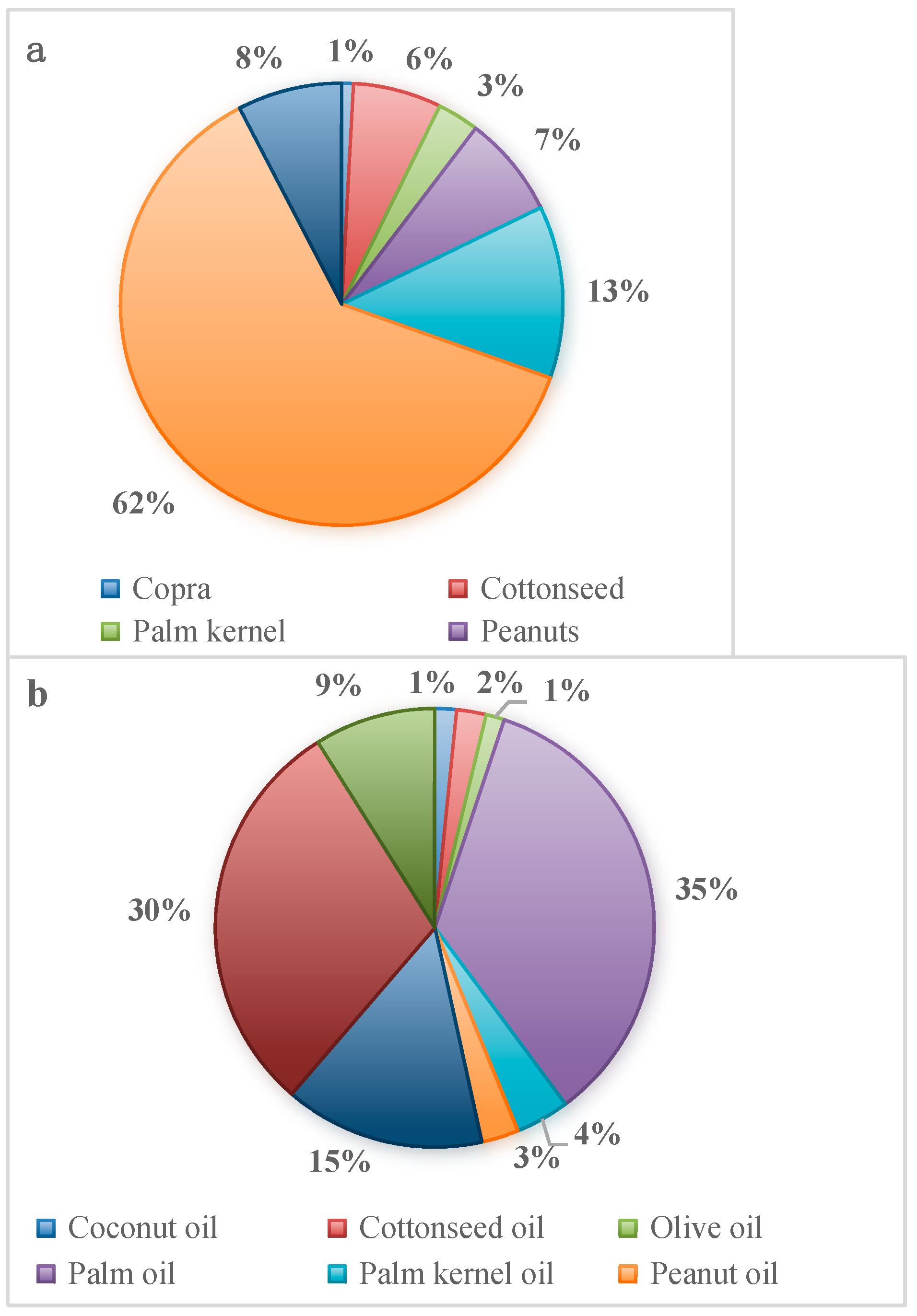
1. Introduction
2. Genetic and Environmental Regulation of Oil Content
2.1. Genetic Basis of Oil Content in Oilseed Crops
2.2. Environmental Factors Influencing Oil Content in Oilseed Crops
| Environmental Factor | Major Oilseeds | Impact on Oil Content | Reference |
|---|---|---|---|
| Temperature | Soybeans Rapeseed sunflower | ±2–5%: -Heat stress lowers rapeseed oil content by 3–5%; -cold stress reduces soybean oil by 2–4%. | [34,43] |
| Water Availability | Sunflower peanuts sesame | ±3–7%: -Drought reduces sunflower oil by 5–7%; -drought stress lowers peanut oil by 3–4%. | [42,44] |
| Soil Nutrients | Rapeseed Soybeans cottonseed | ±2–4%: -High N lowers rapeseed oil by 2–3%; -optimal P increases soybean oil by 1–2%. | [45,46] |
| Light Intensity/Duration | Sunflower sesame canola | ±3–6%: -Shading decreases sunflower oil by 4–6%; -long days increase canola oil by 2–3%. | [35] |
| Altitude | Rapeseed (high-altitude regions) peanuts | ±4–8%: -Rapeseed oil content drops by 5–8% at >2000 m compared to lowlands. | [36] |
3. Molecular Mechanisms of Oil Biosynthesis
3.1. High-Resolution QTL Mapping for Oil Content
3.2. Genome-Wide Mapping of Oil Content Loci
3.3. Key Enzymes and Pathways of Oil Biosynthesis
- Fatty acid (FA) synthesis: this stage occurs in plastids, where acetyl-CoA carboxylase (ACCase) and fatty acid synthase (FAS) catalyze the conversion of acetyl-CoA into palmitic acid (16:0) and stearic acids (18:0).
- Triacylglycerol (TAG) assembly via the Kennedy pathway: In the endoplasmic reticulum (ER), glycerol-3-phosphate is sequentially acylated by Glycerol-3-phosphate acyltransferase (GPAT), Lysophosphatidic acid acyltransferase (LPAT), and Diacylglycerol acyltransferase (DGAT, a critical rate-limiting enzyme) to form TAG. In oilseed, the Kennedy Pathway plays a pivotal role in oil biosynthesis by supplying phosphatidylcholine (PC), a key precursor for diacylglycerol (DAG), the direct substrate for TAG synthesis.
3.4. Transcriptional Regulation of Oil Biosynthesis
| Pathway | Gene | Gene Annotation | References |
|---|---|---|---|
| Fatty acid synthesis | ACCase | acetyl-CoA carboxylase | [73] |
| MCAT | fatty acid elongase | [74] | |
| KAS | β-ketoacyl-ACP synthase | [70] | |
| KAR | β-ketoacyl-ACP reductase | [71,72] | |
| HAD | β-hydroxylacyl-ACP dehydrase | [72,75] | |
| ENR | β-enoyl-ACP reductase | [71,72] | |
| SAD | stearoyl-acyl carrier protein desaturase | [61,76] | |
| FATA FATB | fatty acid thioesterase | [60,77] | |
| LACS | long-chain acyl-CoA synthase | [61] | |
| LPCAT | lysophosphatidyl choline acyltransferase | [61] | |
| FAD | fatty acid desaturase | [60,61] | |
| TAG synthesis | GPAT | glycerol-3-phosphate acyltransferase | [78] |
| LPAAT | lysophosphatidic acid acyltransferase | [79] | |
| PAP | phosphatidic acid phosphatase | [80] | |
| DGAT | diacylglycerol acyltransferase | [55] | |
| Transcription factors | WR11 | AP2/EREBP transcription factor | [59,68,69] |
| LEC | AP2/B3-like transcriptional factor | [71] | |
| ZF | Zinc finger transcription factor | [81] | |
| FUS3 | FUSCA3 | [71] | |
| ABI3 | ABSCISIC ACID INSENSITIVE 3 | [72] | |
| DOF | Dof zinc finger protein | [61] | |
| ZIP123 | bZIP transcription factor | [82] | |
| MYB | MYB transcription factor | [81] |
3.5. Post-Transcriptional and Epigenetic Regulation of Oil Biosynthesis
4. Biotechnological Strategies for Elevating Oil Content
4.1. Breeding Strategies for Enhancing Oil Content
4.2. Genetic Engineering for Enhancing Oil Content
| Strategies | Elevation of Seed Oil Content | Advantages | Disadvantages | Reference |
|---|---|---|---|---|
| Conventional Breeding (e.g., screening, hybridization) | - Soybean from 18–22% to 22–25% -sesame from 50–55% to 52–57% - Safflower from 28–32% to 30–35% - peanut from 45–50% to 48–55% | - Low cost - Expands genetic variation; - Ensures adoption in local agro-ecosystems | - Slow progress (5–8 generations) - Limited genetic gain - Heterosis effects vary across environments | [102,103,104,105] |
| Mutagenesis Breeding | - Sunflower from 38–42% to 45–48% - Linseed: 3–6% increase (from 35–40% to 36–42%) | - Creates novel variation - No regulatory barriers; - Suitable for orphan crops | - Requires large mutant populations, labor-intensive screening - Random mutations may introduce undesirable traits | [106,107] |
| Marker-Assisted Selection (MAS) | - Rapeseed from 40–45% to 47–50% - Sunflower from 40–45% to 42–48% | - Faster than conventional - Targets specific loci - Low cost; - Applicable to non-transgenic varieties; - Maintains genetic diversity | - Requires well-characterized genetic maps - Marker-trait - Linkage may break | [108,109] |
| Genomic Selection (GS) | - Peanut from 45–48% to 50–53% - Cottonseed from 18–22% to 19–24% | - Utilizes whole-genome markers - Higher prediction accuracy - Accelerates breeding cycles (3–4 generations faster than MAS) | - Requires large training population - Computationally intensive - Less effective for traits with low heritability | [96,110] |
| Transgenic Approach | - Rapeseed: 8–12% increase (from 40–45% to 43–50%) - Palm: 5–8% increase (from 45–50% to 47–54%) | - Direct genetic improvement - Stable heritability | - Regulatory challenges - Public acceptance issues | [73,111] |
| Gene editing | - Soybean: 10–15% increase (from 18–22% to 20–25%) - Camelina from 35–38% to 42–45% | - Precise editing - No foreign DNA | - Off-target effects - Regulatory restrictions in many regions - High technical cost for multiplex editing | [112,113] |
5. Future Prospects
5.1. Further Discovery and Exploitation of Functional Genes
5.2. Genome Editing as a Transformative Tool for Molecular Design Breeding
5.3. AI-Driven Polymerization Breeding
Author Contributions
Funding
Data Availability
Conflicts of Interest
References
- Zio, S.; Tarnagda, B.; Sankara, S.; Tapsoba, F.; Zongo, C.; Savadogo, A. Nutritional and therapeutic interest of most widely produced and consumed plant oils by human: A review. Appl. Food Res. 2025, 5, 101093. [Google Scholar] [CrossRef]
- Popoola, C.A.; Jibatswen, T.Y. Non-edible vegetable oils as bio-lubricant basestocks: A review. Open Access Res. J. Eng. Technol. 2024, 6, 080–086. [Google Scholar] [CrossRef]
- Vashist, A.; Kolishetti, N.; Arias, A.Y.; Raymond, A.D.; Vashist, A.; Bhunia, S.; Brooks, D.; Atluri, V.; Nair, M. Recent updates on the emerging role of vegetable oils in the development of hydrogel technology for biomedical applications. In Vegetable Oil-Based Polymers and Their Surface Applications; Elsevier: Amsterdam, The Netherlands, 2024; pp. 131–138. [Google Scholar] [CrossRef]
- Wang, W.; Hu, Z.; Lei, W. Research progress on vegetable oil-Based UV-curing resins. Polymers 2025, 17, 1890. [Google Scholar] [CrossRef]
- Maulana, S.; Wibowo, E.S.; Mardawati, E.; Iswanto, A.H.; Papadopoulos, A.; Lubis, M.A.R. Eco-friendly and high-performance bio-polyurethane adhesives from vegetable oils: A review. Polymers 2024, 16, 1613. [Google Scholar] [CrossRef]
- Feng, H.T.; Wang, H.Z. Security strategy for the nation’s edible vegetable oil supplies under the new circumstances. Chin. J. Oil Crop Sci. 2024, 46, 221–227. [Google Scholar] [CrossRef]
- Gao, Y.; Hu, J.; Su, X.; Li, Q.; Su, C.; Li, Y.; Ma, G.; Zhang, S.; Yu, X. Extraction, chemical components, bioactive functions and adulteration identification of walnut oils: A review. Grain Oil Sci. Technol. 2024, 7, 30–41. [Google Scholar] [CrossRef]
- Qi, W.; Lu, H.; Zhang, Y.; Cheng, J.; Huang, B.; Lu, X.; Sheteiwy, M.S.A.; Kuang, S.; Shao, H. Oil crop genetic modification for producing added value lipids. Crit. Rev. Biotechnol. 2020, 40, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Graham, I.A. Seed storage oil mobilization. Annu. Rev. Plant Biol. 2008, 59, 115–142. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.H.; Liu, N.; Hua, W. Research advances in the biosynthesis and regulation of lipid in oil crops. Chin. J. Oil Crop Sci. 2021, 43, 361–375. [Google Scholar] [CrossRef]
- Li, D.R.; Tian, J.H.; Chen, W.J.; Zhang, W.X.; Li, Y.H.; Wang, H. Breeding technologies and germplasm innovation on extra-high-oil content in Brassica napus. Acta Agric. Boreali-Occident. Sin. 2011, 20, 83–87. [Google Scholar] [CrossRef]
- Boem, F.; Lavado, R.S.; Porcelli, C.A. Note on the effects of winter and spring waterlogging on growth, chemical composition and yield of rapeseed. Field Crops Res. 1996, 47, 175–179. [Google Scholar] [CrossRef]
- Jensen, C.R.; Mogensen, V.O.; Mortensen, G.; Fieldsend, J.K.; Milford, G.F.J.; Andersen, M.N.; Thage, J.H. Seed glucosinolate, oil and protein contents of field-grown rape (Brassica napus L.) affected by soil drying and evaporative demand. Field Crops Res. 1996, 47, 93–105. [Google Scholar] [CrossRef]
- Si, P.; Mailer, R.; Galwey, N.; Turner, D. Influence of genotype and environment on oil and protein concentrations of canola (Brassica napus L.) grown across southern Australia. Crop Pasture Sci. 2003, 54, 397–407. [Google Scholar] [CrossRef]
- Zhao, J.; Becker, H.C.; Zhang, D.; Zhang, Y.; Ecke, W. Oil content in a European × Chinese rapeseed population: QTL with additive and epistatic effects and their genotype-environment interactions. Crop Sci. 2005, 45, 51–59. [Google Scholar] [CrossRef]
- Shi, J.; Li, R.; Qiu, D.; Jiang, C.; Long, Y.; Morgan, C.; Bancroft, I.; Zhao, J.; Meng, J. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 2009, 182, 851–861. [Google Scholar] [CrossRef]
- Li, X.; Chen, L.; Hong, M.; Zhang, Y.; Zu, F.; Wen, J.; Yi, B.; Ma, C.; Shen, J.; Tu, J.; et al. A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PLoS ONE 2012, 7, e44145. [Google Scholar] [CrossRef]
- Wu, J.G.; Shi, C.H.; Zhang, H.Z. Partitioning genetic effects due to embryo, cytoplasm and maternal parent for oil content in oilseed rape (Brassica napus L.). Genet. Mol. Biol. 2006, 29, 533–538. [Google Scholar] [CrossRef]
- Wang, X.F.; Liu, G.H.; Yang, Q.; Hua, W.; Liu, J.; Wang, H.Z. Genetic analysis on oil content in rapeseed (Brassica napus L.). Euphytica 2010, 173, 17–24. [Google Scholar] [CrossRef]
- Hua, W.; Liu, J.; Wang, H.Z. Molecular regulation and genetic improvement of seed oil content in Brassica napus L. Front. Agric. Sci. Eng. 2016, 3, 186–194. [Google Scholar] [CrossRef]
- Hua, W.; Li, R.J.; Zhan, G.M.; Liu, J.; Li, J.; Wang, X.F.; Liu, G.H.; Wang, H.Z. Maternal control of seed oil content in Brassica napus: The role of silique wall photosynthesis. Plant J. 2012, 69, 432–444. [Google Scholar] [CrossRef]
- Mu, D.L.; Han, X.M.; Li, X.Y.; Yao, D.; Yang, S.N.; Qu, Y.W.; Liang, J.N.; Zhang, J. Genetic analysis of major gene plus polygene on main quality traits in peanut. J. Peanut Sci. 2021, 50, 41–44. [Google Scholar] [CrossRef]
- Huang, B.Y.; Hu, J.Z.; Zhang, X.Y.; Miao, L.J.; Shi, L.; Lu, D.Y.; Chai, P.P.; Feng, S.P.; Liu, H.; Han, S.Y.; et al. Genetic analysis of direct and maternal effects of fat content in peanut seed. Chin. J. Oil Crop Sci. 2021, 43, 582–589. [Google Scholar] [CrossRef]
- Jin, X.N.; Li, J.; Wang, T.G.; Wu, X.Y.; Chen, S.L. Genetic analysis of major gene plus polygene of oil content of maize kernel. J. Maize Sci. 2019, 27, 47–51+57. [Google Scholar] [CrossRef]
- Gilsinger, J.J.; Burton, J.W.; Carter, T.E. Maternal effects on fatty acid composition of soybean seed oil. Crop Breed. Genet. 2010, 50, 1874–1881. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Lu, G.Y. Analysis of genetic characteristics of cotton oil content. Hubei Agric. Sci. 2019, 58, 14–16. [Google Scholar] [CrossRef]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, L.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef]
- Wang, M.L.; Tonnis, B.D.; Li, X.; Benke, R.L.; Huang, E.; Tallury, S.P.; Pupplala, N.; Peng, Z.; Wang, J. Genotype, environment, and their interaction effects on peanut seed protein, oil, and fatty acid content variability. Crop Sci. 2024, 116, 1440–1454. [Google Scholar] [CrossRef]
- Zeng, L.; Han, X.; Gou, X.; Pei, H.; Shao, Y.; Cao, Y.; Zhang, Z.; Li, X.; Yu, J.; Yan, J.; et al. Leveraging phenotypic plasticity in seed oil content for climate-adapted breeding and production. Plant Cell Environ. 2025, 48, 2856–2871. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010, 49, 235–249. [Google Scholar] [CrossRef]
- Singer, S.D.; Zou, J.; Weselake, R.J. Abiotic factors influence plant storage lipid accumulation and composition. Plant Sci. 2016, 243, 1–9. [Google Scholar] [CrossRef]
- Xiao, Z.C.; Zhang, C.; Tang, F.; Yang, B.; Zhang, L.Y.; Liu, J.S.; Huo, Q.; Wang, S.F.; Li, S.T.; Wei, L.J.; et al. Identification of candidate genes controlling oil content by combination of genome-wide association and transcriptome analysis in the oilseed crop Brassica napus. Biotechnol. Biofuels 2019, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- Balbino, S. Vegetable oil yield and composition influenced by environmental stress factors. In Oilseed Crops; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 80–101. [Google Scholar] [CrossRef]
- Zhu, L. Effect of Seed Dormancy on Oil Accumulation and the Genotypic Difference of Seed Vigor in Responding to Artificial Aging Treatment in Brassica napus. Ph.D. Dissertation, Zhejiang University, Hangzhou, China, 2022. [Google Scholar]
- Goffman, F.D.; Alonso, A.P.; Schwender, J.; Shachar-Hill, Y.; Ohirogge, B.O. Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiol. 2005, 138, 2269–2279. [Google Scholar] [CrossRef] [PubMed]
- Tang, L. Research on Accumulation Protential of Brassica napus L. Oil in Qing-Tiber High Plateau. Master’s Dissertation, Chinese Academy of Agricultural Sciences, Beijing, China, 2013. [Google Scholar]
- Zhang, K. Influence of Light Intensity on Photosynthetic Characteristics, Yield and Quality of Peanut and Its Growth Model. Ph.D. Dissertation, Shandong Agricultural University, Tai’an, China, 2009. [Google Scholar]
- Fu, S.X.; Wu, X.M.; Li, C.L. Effect of different geographical positions on oil content of Brassica napus L. Jiangsu J. Agric. Sci. 2009, 2, 247–252. [Google Scholar] [CrossRef]
- Chen, B.Y.; Xu, K.; Gao, G.Z.; Wu, X.M. Effect of planting density on yield and oil content of different rape varieties. Jiangsu Agric. Sci. 2018, 46, 83–89. [Google Scholar] [CrossRef]
- Qian, W.; Dai, C.M.; Zheng, C.Y.; Lin, X.X.; Liang, Q.X.; Li, F.; Wu, J.F.; Chen, H.J. Effects of different sowing dates on oil content and yield of winter rape (Brassica napus L.). South China Agric. 2018, 12, 23–24. [Google Scholar] [CrossRef]
- Hu, W.G.; Qiu, Q.S.; Li, Z.C.; Wu, L.R.; Dong, J. Studies of the effect factors on peanut qualities II. cultural factors. J. Peanut Sci. 2002, 31, 14–18. [Google Scholar] [CrossRef]
- Ai, X.; Wan, S.; Dai, R.; Ma, X.; Li, C.; Zhong, C.; Wang, J.; Liu, X.; Zhao, X.; Zhang, H.; et al. Dynamic changes of seed development, oil accumulation and fatty acid composition in peanut under soil water deficit. Plant Physiol Biochem 2025, 219, 109336. [Google Scholar] [CrossRef]
- Staniak, M.; Czopek, K.; Stępień-Warda, A.; Kocira, A.; Przybyś, M. Cold stress during flowering alters plant structure, yield and seed quality of different soybean genotypes. Agronomy 2021, 11, 2059. [Google Scholar] [CrossRef]
- Ghaffari, M.; Gholizadeh, A.; Rauf, S.; Shariati, F. Drought-stress induced changes of fatty acid composition affecting sunflower grain yield and oil quality. Food Sci. Nutr. 2023, 11, 7718–7731. [Google Scholar] [CrossRef]
- Brennan, R.F.; Bolland, M.D.A. Significant nitrogen by sulfur interactions occurred for canola grain production and oil concentration in grain on sandy soils in the mediterranean-type climate of southwestern Australia. J. Plant Nutr. 2008, 31, 1174–1187. [Google Scholar] [CrossRef]
- Ning, H.; Song, X.; Wang, X.; Cui, X.; Wang, J.; Li, W.; Hu, G.; Li, W. Effects of nitrogen, phosphorus and potassium fertilizers on oil content in soybean. Chin. J. Oil Crop Sci. 2007, 29, 302–307. [Google Scholar] [CrossRef]
- Delourme, R.; Falentin, C.; Huteau, V.; Clouet, V.; Horvais, R.; Gandon, B.; Specel, S.; Hanneton, L.; Dheu, J.E.; Deschamps, M.; et al. Genetic control of oil content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2006, 113, 1331–1345. [Google Scholar] [CrossRef]
- Qiu, D.; Morgan, C.; Shi, J.; Long, Y.; Liu, J.; Li, R.; Zhuang, X.; Wang, Y.; Dietrich, E.; Weihmann, C.; et al. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor. Appl. Genet. 2006, 114, 67–80. [Google Scholar] [CrossRef]
- Smooker, A.M.; Wells, R.; Morgan, C.; Beaudoin, F.; Cho, K.; Fraser, F.; Bancroft, J. The identification and mapping of candidate genes and QTL involved in the fatty acid desaturation pathway in Brassica napus. Theor. Appl. Genet. 2011, 122, 1075–1090. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.T.; Wang, M.; Fu, S.X.; Yang, W.C.; Qi, C.K.; Wang, X.J. Small RNA profiling in two Brassica napus cultivars identifies microRNAs with oil production and development-correlated expression and new small RNA classes. Plant Physiol. 2012, 158, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Jiang, C.C.; Cao, Z.Y.; Li, R.Y.; Long, Y.; Chen, S.; Meng, J.J. Association mapping of seed oil content in Brassica napus and comparison with quantitative trait loci identified from linkage mapping. Genome 2010, 53, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Li, H.N.; Hu, J.; Liu, Y.; Huang, Q.Y.; Li, X.; Wang, P.F.; Zhou, X.M.; Yang, G.S. A stable quantitative trait locus on chromosome A10 improves the oil content of a backbone parent in Brassica napus L. Ind. Crops Prod. 2023, 202, 117054. [Google Scholar] [CrossRef]
- Li, B.; Peng, J.Y.; Wu, Y.Y.; Hu, Q.; Huang, W.X.; Yuan, Z.H.; Tang, X.F.; Cao, D.; Xue, Y.G.; Luan, X.Y.; et al. Identification of an important QTL for seed oil content in soybean. Mol. Breed. 2023, 43, 43. [Google Scholar] [CrossRef]
- Zhang, Y.H.; He, J.B.; Wang, H.W.; Meng, S.; Xing, G.N.; Li, Y.; Yang, S.P.; Zhao, Y.H.; Zhao, T.J.; Gai, J.Y. Detecting the QTL-allele system of seed oil traits using multi-locus genome wide association analysis for population characterization and optimal cross prediction in soybean. Front. Plant Sci. 2018, 9, 1793. [Google Scholar] [CrossRef]
- Liu, M.Y. Enhanced Arabidopsis Seed Oil Content by Overexpressing Several Genes Related to Triacylglyceride Synthesis. Master’s Dissertation, Huazhong Agricultural University, Wuhan, China, 2020. [Google Scholar]
- Yang, X.H.; Guo, Y.Q.; Yan, J.B.; Zhang, J.; Song, T.M.; Rochford, T.; Li, J.S. Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor. Appl. Genet. 2010, 120, 665–678. [Google Scholar] [CrossRef]
- Tang, S.; Zhao, H.; Lu, S.; Yu, L.; Zhang, G.; Zhang, Y.; Yang, Q.Y.; Zhou, Y.; Wang, X.; Ma, W.; et al. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol. Plant 2021, 14, 470–487. [Google Scholar] [CrossRef]
- Hwang, E.Y.; Song, Q.; Jia, G.; Specht, J.E.; Hytem, D.; Costa, J.; Cregan, P.B. A genome-wide association study of seed protein and oil content in soybean. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Song, J.M.; Zhang, Y.T.; Zhou, Z.W.; Lu, S.P.; Ma, W.; Lu, C.F.; Chen, L.L.; Guo, L.G. Oil plant genomes: Current state of the science. J. Exp. Bot. 2021, 3, 2859–2874. [Google Scholar] [CrossRef] [PubMed]
- Sagun, J.V.; Yadav, U.P.; Alonso, A.P. Progress in understanding and improving oil content and quality in seeds. Front. Plant Sci. 2023, 14, 1116894. [Google Scholar] [CrossRef] [PubMed]
- Weselake, R.J.; Taylor, D.C.; Rahman, M.H.; Shah, S.; Laroche, A.; McVetty, P.B.E.; Harwood, J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009, 27, 866–878. [Google Scholar] [CrossRef]
- Jolivet, P.; Acevedo, F.; Boulard, C. Crop seed oil bodies: From challenges in protein identification to an emerging picture of the oil body proteome. Proteomics 2013, 13, 1836–1849. [Google Scholar] [CrossRef]
- Siloto, R.M.; Findlay, K.; Lopez-Villalobos, A.; Yeung, E.; Nykiforuk, C.; Moloney, M. The accumulation of oleosins determines the size of seed oil-bodies in Arabidopsis. Plant Cell 2006, 18, 1961–1974. [Google Scholar] [CrossRef]
- Liao, P.; Lechon, T.; Romsdahl, T.; Woodfield, H.; Fenyk, S.; Fawcett, T.; Wallington, E.; Bates, R.E.; Chye, M.L.; Chapman, K.D.; et al. Transgenic manipulation of triacylglycerol biosynthetic enzymes in B. napus alters lipid-associated gene expression and lipid metabolism. Sci. Rep. 2022, 12, 3352. [Google Scholar] [CrossRef]
- Xia, H.; Wang, X.J.; Li, M.J.; Xiao, H. Improving fatty acid composition and increasing triacylglycerol content in plants by gene engineering: A review. Shengwu Gongcheng Xuebao 2010, 26, 735–743. [Google Scholar] [CrossRef]
- Min, W.L.; Cao, X.T.; Ji, G.S.; Zhang, G.Z. Researches of plant transcription factors involving in fatty acid synthesis. Bull. Ferment. Sci. Technol. 2017, 46, 107–112. [Google Scholar] [CrossRef]
- Shen, B.; Allen, W.B.; Zheng, P.Z.; Li, C.J.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; Anna, E.T.; Pushkar, S.; Zhou, X.R.; Surinder, P.S.; James, R.P. Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. Febs Lett. 2013, 587, 364–369. [Google Scholar] [CrossRef]
- Yadav, R.; Kalia, S.; Rangan, P.; Pradheep, K.; Rao, G.P.; Kaur, V.; Pandey, R.; Rai, V.; Vasimalla, C.C.; Langyan, S.; et al. Current research trends and prospects for yield and quality improvement in sesame, an important oilseed crop. Front. Plant Sci. 2022, 13, 863521. [Google Scholar] [CrossRef] [PubMed]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef]
- Liu, L.Y.; Ruan, C.J.; Wang, L.; Zhang, W.C.; Wang, H.M.; Wu, B.; Yan, R. Coordinated regulation of multigenes formed by fatty acids in kernel of Xanthoceras sorbifolium. Mol. Plant Breed. 2019, 17, 1834–1842. [Google Scholar] [CrossRef]
- Wang, F.L.; Wu, G.T.; Lang, C.X.; Liu, R.H. Influence on Brassica seed oil content by transformation with heteromeric Acetyl–CoA Carboxylase (ACCase) gene. Mol. Plant Breed. 2017, 15, 920–927. [Google Scholar] [CrossRef]
- Jung, S.H.; Kim, R.J.; Kim, K.J.; Lee, D.H.; Suh, M.C. Plastidial and mitochondrial malonyl CoA-ACP malonyltransferase is essential for cell division and its overexpression increases storage oil content. Plant Cell Physiol. 2019, 60, 1239–1249. [Google Scholar] [CrossRef]
- Liu, Z.W.; Wang, Z.Y.; Gu, H.; You, J.; Hu, M.M.; Zhang, Y.J.; Zhu, Z.; Wang, Y.H.; Liu, S.J.; Chen, L.M.; et al. Identification and phenotypic characterization of ZEBRA LEAF16 encoding a β-Hydroxyacyl-ACP dehydratase in rice. Front. Plant Sci. 2018, 9, 782. [Google Scholar] [CrossRef]
- Liu, Q.; Singh, S.P.; Green, A.G. High-stearic and higholeic cottonseed oils produced by hairpin RNA-mediated post transcriptional gene silencing. Plant Physiol. 2002, 129, 1732–1743. [Google Scholar] [CrossRef]
- Sánchez-García, A.; Moreno-Pérez, A.J.; Muro-Pastor, A.M.; Salas, J.J.; Garces, R.; Martínez-Force, E. Acyl-ACP thioesterases from Castor (Ricinus communis L.): An enzymatic system appropriate for high rates of oil synthesis and accumulation. Phytochemistry 2010, 71, 860–869. [Google Scholar] [CrossRef]
- Hao, J.F. The Functions of Three Genes of Glycerol-3-Phosphate Acyltransferase (GPAT 6, 7, 9) in Regulation of Seed-Oil Content and Salt Tolerance in Arabidopsis. Master’s Dissertation, Nanjing Agricultural University, Nanjing, China, 2013. [Google Scholar]
- Woodfield, H.K.; Fenyk, S.; Wallington, E.; Bates, R.E.; Brown, A.; Guschina, I.A.; Marillia, E.F.; Taylor, D.C.; Fell, D.; Harwood, J.L.; et al. Increase in lysophosphatidate acyltransferase activity in oilseed rape (Brassica napus) increases seed triacylglycerol content despite its low intrinsic flux control coefficient. New Phytol. 2019, 224, 700–711. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Liu, Y.F.; Li, Q.T.; Lu, X.; Song, Q.X.; Lam, S.M.; Zhang, W.K.; Ma, B.; Lin, Q.; Man, W.Q.; Du, W.G.; et al. Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol. 2014, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.X.; Li, Q.T.; Liu, Y.F.; Zhang, F.X.; Ma, B.; Zhang, W.K.; Man, W.Q.; Du, W.G.; Wang, G.D.; Chen, S.Y.; et al. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J. Exp. Bot. 2013, 64, 4329–4341. [Google Scholar] [CrossRef]
- Kumar, N.; Chaudhary, A.; Singh, D.; Teotia, S. Transcriptional regulation of seed oil accumulation in Arabidopsis thaliana: Role of transcription factors and chromatin remodelers. J. Plant Biochem. Biotechnol. 2020, 29, 754–768. [Google Scholar] [CrossRef]
- Yadav, P.; Priyam, P.; Yadav, G.; Yadav, A.; Jain, R.; Sunderam, S.; Sharma, M.K.; Kaur, I.; Dhaka, N. Identification of lncRNAs regulating seed traits in Brassica juncea and development of a comprehensive seed omics database. Funct. Integr. Genom. 2024, 24, 189. [Google Scholar] [CrossRef]
- Kong, Q.; Yang, Y.; Low, P.M.; Guo, L.; Yuan, L.; Ma, W. The function of the WRI1-TCP4 regulatory module in lipid biosynthesis. Plant Signal. Behav. 2020, 15, 1812878. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Huang, Y.; Wang, Y.; Cui, Y.; Liu, Z.; Hua, J. RNA interference of GhPEPC2 enhanced seed oil accumulation and salt tolerance in Upland cotton. Plant Sci. 2018, 271, 52–61. [Google Scholar] [CrossRef]
- Lu, L.; Wei, W.; Li, Q.T.; Bian, X.H.; Lu, X.; Hu, Y.; Cheng, T.; Wang, Z.Y.; Jin, M.; Tao, J.J.; et al. A transcriptional regulatory module controls lipid accumulation in soybean. New Phytol. 2021, 231, 661–678. [Google Scholar] [CrossRef]
- Liao, B.; Hao, Y.; Lu, J.; Bai, H.; Guan, L.; Zhang, T. Transcriptomic analysis of Perilla frutescens seed to insight into the biosynthesis and metabolic of unsaturated fatty acids. BMC Genom. 2018, 19, 213. [Google Scholar] [CrossRef] [PubMed]
- Zafar, S.; Li, Y.L.; Li, N.N.; Zhu, K.M.; Tan, X.L. Recent advances in enhancement of oil content in oilseed crops. J. Biotechnol. 2019, 301, 35–44. [Google Scholar] [CrossRef]
- Marchive, C.; Nikovics, K.; To, A.; Lepiniec, L.; Baud, S. Transcriptional regulation of fatty acid production in higher plants: Molecular bases and biotechnological outcomes. Eur. J. Lipid Sci. Technol. 2014, 116, 1332–1343. [Google Scholar] [CrossRef]
- Wilmer, J.A.; Helsper, J.P.F.G.; van der Plas, L.H.W. Regulation of oil composition in oilseed rape (B. napus L.): Effects of abscisic acid and temperature. In Physiology, Biochemistry and Molecular Biology of Plant Lipids; Springer: Dordrecht, The Netherlands, 1997; pp. 301–303. [Google Scholar] [CrossRef]
- Amoah, S.; Kurup, S.; Rodriguez Lopez, C.M.; Welham, S.J.; Powers, S.J.; Hopkins, C.J.; Wilkinson, M.J.; King, G.J. A hypomethylated population of Brassica rapa for forward and reverse epi-genetics. BMC Plant Biol. 2012, 12, 193. [Google Scholar] [CrossRef]
- Yang, Y.; Kong, Q.; Lim, A.R.Q.; Lu, S.; Zhao, H.; Guo, L.; Yuan, L.; Ma, W. Transcriptional regulation of oil biosynthesis in seed plants: Current understanding, applications, and perspectives. Plant Commun. 2022, 3, 100328. [Google Scholar] [CrossRef]
- Cai, W.J.; Liang, X.F.; Yuan, X.C.; Li, A.X.; He, S. Changes of DNA methylation pattern in metabolic pathways induced by high-carbohydrate diet contribute to hyperglycemia and fat deposition in grass carp (Ctenopharyngodon idellus). Front. Endocrinol. 2020, 11, 398. [Google Scholar] [CrossRef]
- Lim, A.R.Q.; Kong, Q.; Singh, S.K.; Guo, L.; Yuan, L.; Ma, W. Sunflower WRINKLED1 plays a key role in transcriptional regulation of oil biosynthesis. Int. J. Mol. Sci. 2022, 23, 3054. [Google Scholar] [CrossRef]
- Janila, P.; Pandey, M.K.; Shasidhar, Y.; Variath, M.T.; Sriswathi, M.; Khera, P.; Manohar, S.S.; Nagesh, P.; Vishwakarma, M.K.; Mishra, G.P.; et al. Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 2016, 242, 203–213. [Google Scholar] [CrossRef]
- Zhao, G.L.; Chen, J.Q.; Yi, A.P.; Hu, Z.H.; Qian, X.Y.; Guo, D.Q. Transgenic soybean lines harbouring anti-PEP gene express super-high oil content. Mol. Plant Breed. 2005, 6, 792–796. [Google Scholar] [CrossRef]
- Lu, S.P.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. 2018, 94, 915–932. [Google Scholar] [CrossRef]
- Wang, Y.F.; Su, W.Y.; Cao, S.Y.; Zhang, L.; Lu, X.; Zhang, Y.H.; Xu, J.Q. Development of novel gene editing technologies and its applications in plant breeding. Acta Agric. Boreali-Occident. Sin. 2018, 27, 617–625. [Google Scholar] [CrossRef]
- Zhang, K.; Nie, L.L.; Cheng, Q.Q.; Yin, Y.T.; Chen, K.; Qi, F.Y.; Zou, D.S.; Zhao, W.G.; Wang, B.S.; Li, M.T. Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR-Cas9 system. Biotechnol. Biofuels 2019, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Lee, K.R.; Go, Y.S.; Jung, J.H.; Suh, M.C.; Kim, J.B. Endoplasmic reticulum-located PDAT1-2 from castor bean enhances hydroxy fatty acid accumulation in transgenic plants. Plant Cell Physiol. 2011, 52, 983–993. [Google Scholar] [CrossRef]
- Rodrigues, J.I.d.S.; Arruda, K.M.A.; Piovesan, N.D.; Barros, E.G.d.; Moreira, M.A. Selection of progenitors for increase in oil content in soybean. Rev. Ceres 2016, 63, 661–667. [Google Scholar] [CrossRef][Green Version]
- Uzun, B.; Arslan, Ç.; Furat, Ş. Variation in fatty acid compositions oil content oil yield in a germplasm collection of sesame (Sesamum indicum L.). J. Am. Oil Chem. Soc. 2008, 85, 1135–1142. [Google Scholar] [CrossRef]
- Kurt, C.; Altaf, M.T.; Liaqat, W.; Nadeem, M.A.; Çil, A.N.; Baloch, F.S. Oil content and fatty acid composition of safflower (Carthamus tinctorius L.) germplasm. Foods 2025, 14, 264. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Yan, L.; Chen, Y.; Kang, Y.; Huai, D.; Wang, X.; Liu, K.; Jiang, H.; Lei, Y.; et al. Correlation and variability analysis of yield and quality related traits in different peanut varieties across various ecological zones of China. Oil Crop Sci. 2023, 8, 236–242. [Google Scholar] [CrossRef]
- Skorić, D.; Jocić, S.; Sakac, Z.; Lecić, N. Genetic possibilities for altering sunflower oil quality to obtain novel oils. Can. J. Physiol. Pharmacol. 2008, 86, 215–221. [Google Scholar] [CrossRef]
- Green, A.G.; Marshall, D.R. Isolation of induced mutants in linseed (Linum usitatissimum) having reduced linolenic acid content. Euphytica 1984, 33, 321–328. [Google Scholar] [CrossRef]
- Spasibionek, S.; Mikołajczyk, K.; Ćwiek-Kupczyńska, H.; Piętka, T.; Krótka, K.; Matuszczak, M.; Nowakowska, J.; Michalski, K.; Bartkowiak-Broda, I. Marker assisted selection of new high oleic and low linolenic winter oilseed rape (Brassica napus L.) inbred lines revealing good agricultural value. PLoS ONE 2020, 15, e0233959. [Google Scholar] [CrossRef]
- Dimitrijevic, A.; Horn, R. Sunflower hybrid breeding: From markers to genomic selection. Front. Plant Sci. 2018, 8, 2238. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, J.; Pei, W.; Ma, Q.; Wang, N.; Zhang, X.; Cui, Y.; Li, D.; Liu, G.; Wu, M.; et al. Genome-wide association study of the oil content in upland cotton (Gossypium hirsutum L.) and identification of GhPRXR1, a candidate gene for a stable QTLqOC-Dt5-1. Plant Sci. 2019, 286, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bai, B.; Lee, M.; Alfiko, Y.; Suwanto, A.; Yue, G.H. Cloning and characterization of EgGDSL, a gene associated with oil content in oil palm. Sci. Rep. 2018, 8, 11406. [Google Scholar] [CrossRef]
- Liao, W.; Guo, R.; Li, J.; Liu, N.; Jiang, L.; Whelan, J.; Shou, H. CRISPR/Cas9-mediated mutagenesis of SEED FATTY ACID REDUCER genes significantly increased seed oil content in soybean. Plant Cell Physiol. 2025, 66, 273–284. [Google Scholar] [CrossRef]
- Cai, Y.; Liang, Y.; Shi, H.; Cui, J.; Prakash, S.; Zhang, J.; Anaokar, S.; Chai, J.; Schwender, J.; Lu, C.; et al. Creating yellow seed Camelina sativa with enhanced oil accumulation by CRISPR-mediated disruption of transparent testa 8. Plant Biotechnol. J. 2024, 22, 2773–2784. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhang, L.; Tang, M.; Liu, J.; Liu, H.; Yang, H.; Fan, S.; Terzaghi, W.; Wang, H.; Hua, W. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 2020, 18, 644–654. [Google Scholar] [CrossRef]
- Cvejić, S.; Hrnjaković, O.; Jocković, M.; Kupusinac, A.; Doroslovački, K.; Gvozdenac, S.; Jocić, S.; Miladinović, D. Oil yield prediction for sunflower hybrid selection using different machine learning algorithms. Sci. Rep. 2023, 13, 17611. [Google Scholar] [CrossRef]
- Zhu, H.; Liang, S.; Lin, C.; He, Y.; Xu, J.L. Using multi-sensor data fusion techniques and machine learning algorithms for improving UAV-based yield prediction of oilseed rape. Drones 2024, 8, 642. [Google Scholar] [CrossRef]

| Crop | Oil Content Range (%) | Crop | Oil Content Range (%) |
|---|---|---|---|
| Soybean | 18–28 | Cottonseed | 15–25 |
| Palm kernel | 45–55 | Sesame | 45–70 |
| Rapeseed | 40–65 | Linseed | 35–45 |
| Sunflower | 40–50 | Castor | 40–60 |
| Peanut | 45–62 | Safflower | 35–50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, G.; Zhang, L.; Tong, P.; Yan, G.; Wu, X. Enhancing Oil Content in Oilseed Crops: Genetic Insights, Molecular Mechanisms, and Breeding Approaches. Int. J. Mol. Sci. 2025, 26, 7390. https://doi.org/10.3390/ijms26157390
Gao G, Zhang L, Tong P, Yan G, Wu X. Enhancing Oil Content in Oilseed Crops: Genetic Insights, Molecular Mechanisms, and Breeding Approaches. International Journal of Molecular Sciences. 2025; 26(15):7390. https://doi.org/10.3390/ijms26157390
Chicago/Turabian StyleGao, Guizhen, Lu Zhang, Panpan Tong, Guixin Yan, and Xiaoming Wu. 2025. "Enhancing Oil Content in Oilseed Crops: Genetic Insights, Molecular Mechanisms, and Breeding Approaches" International Journal of Molecular Sciences 26, no. 15: 7390. https://doi.org/10.3390/ijms26157390
APA StyleGao, G., Zhang, L., Tong, P., Yan, G., & Wu, X. (2025). Enhancing Oil Content in Oilseed Crops: Genetic Insights, Molecular Mechanisms, and Breeding Approaches. International Journal of Molecular Sciences, 26(15), 7390. https://doi.org/10.3390/ijms26157390






