The Lysine at Position 177 Is Essential to Limit the Inhibitory Capacities of Sprouty4 Protein in Normal and Cancer-Derived Cells
Abstract
1. Introduction
2. Results
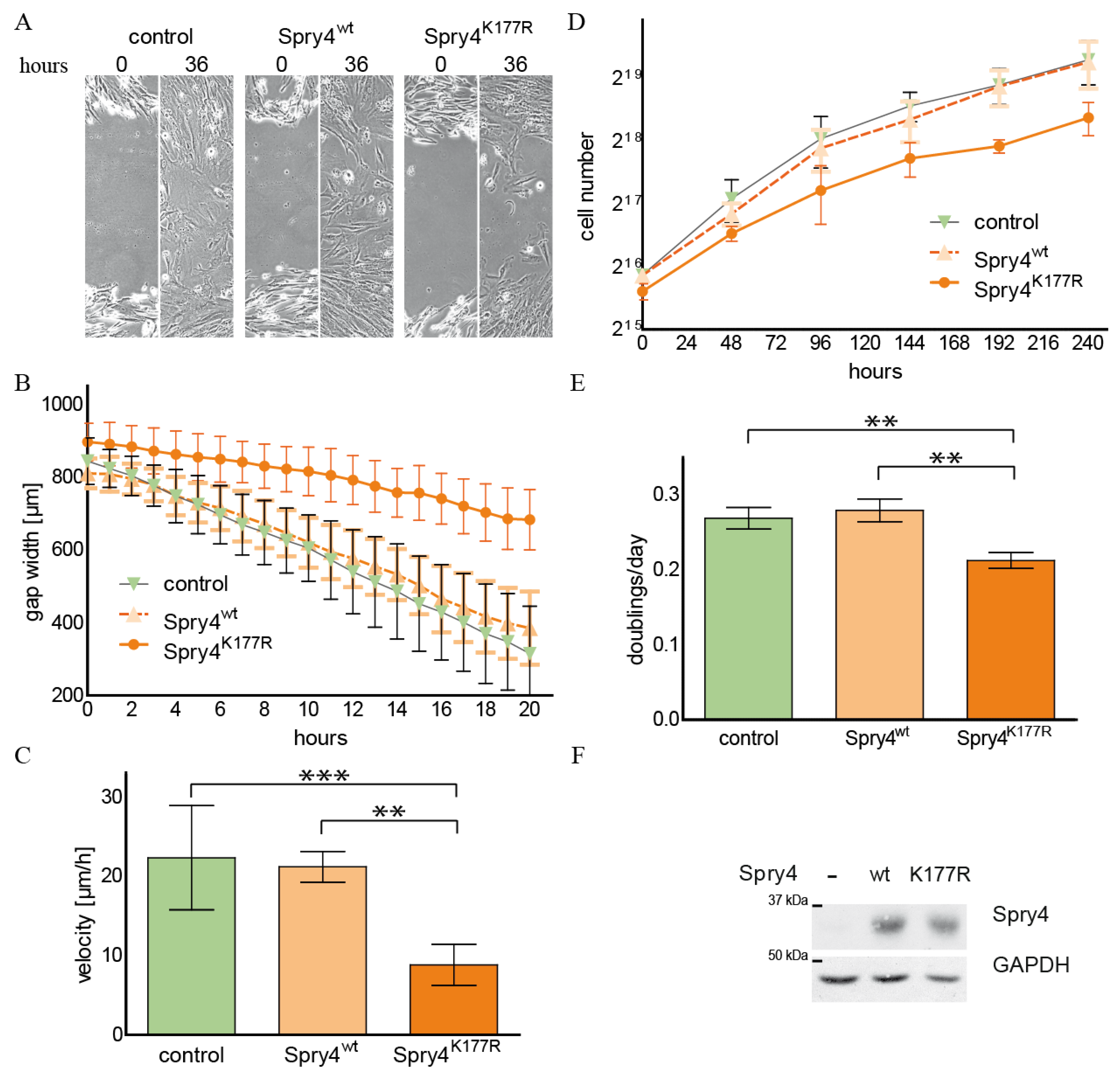
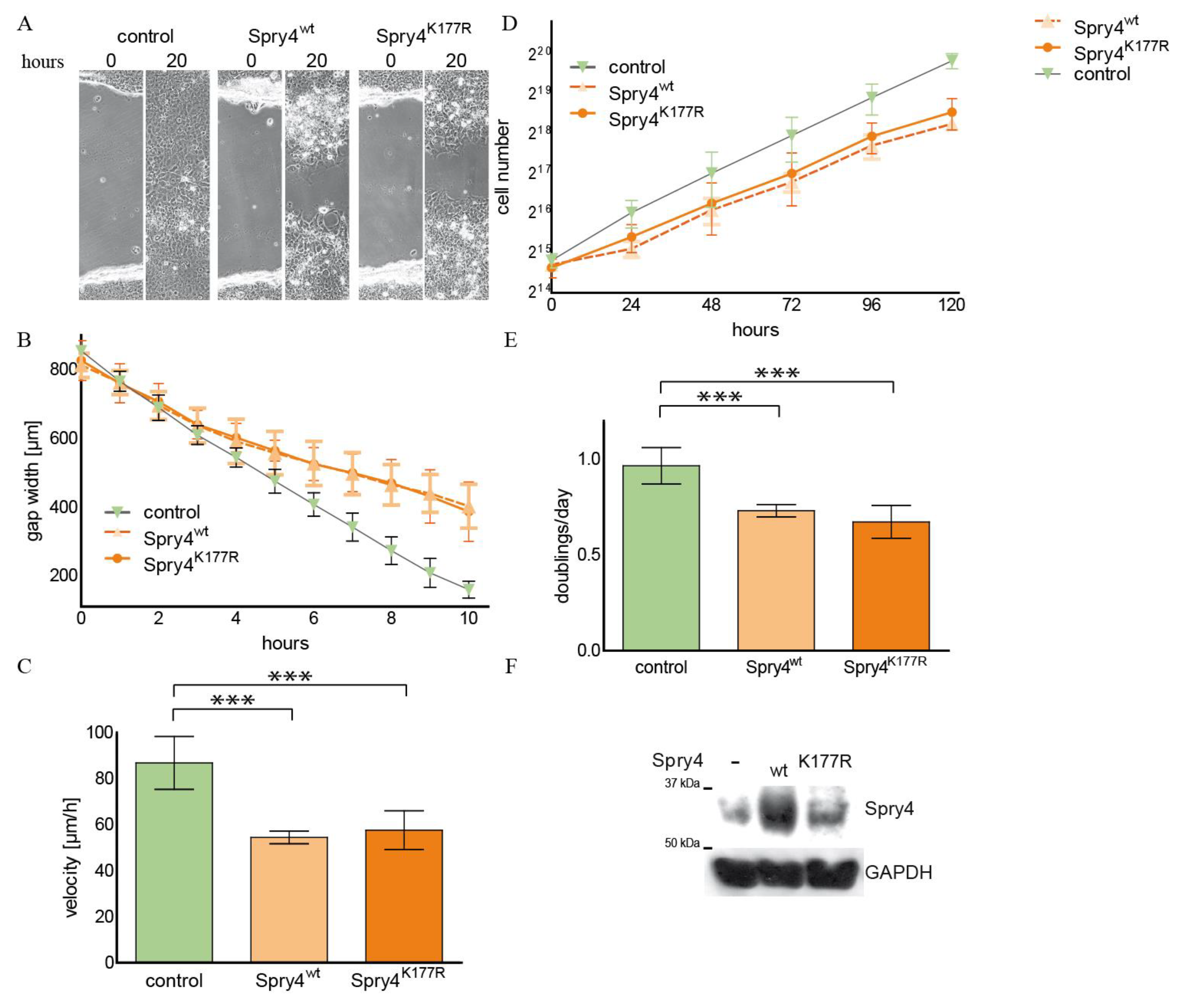
2.1. The Spry4 Variant Associated with Kallmann Syndrome Is Capable to Inhibit Migration and Proliferation of Normal Human Fibroblasts
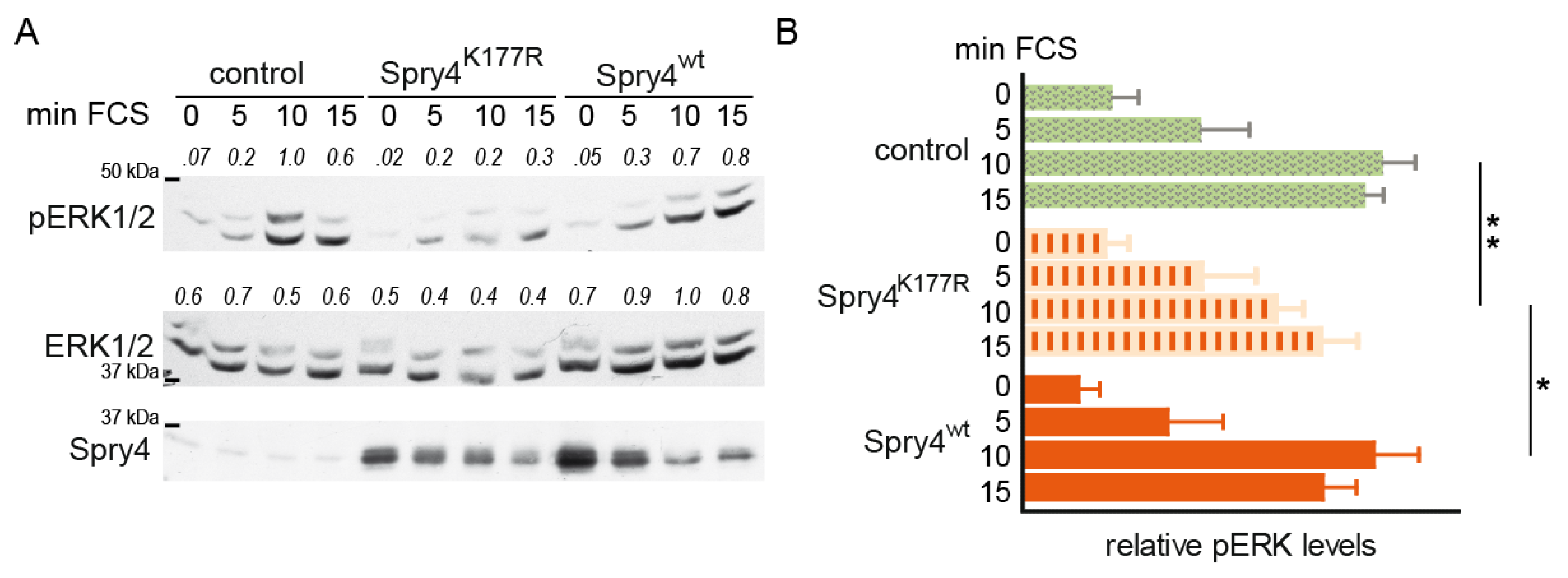
2.2. Replacement of the Lysine at the Position 177 by an Arginine Creates a Spry4 Protein Capable of Inhibiting Activation of MAPK Pathway in Normal Human Fibroblasts
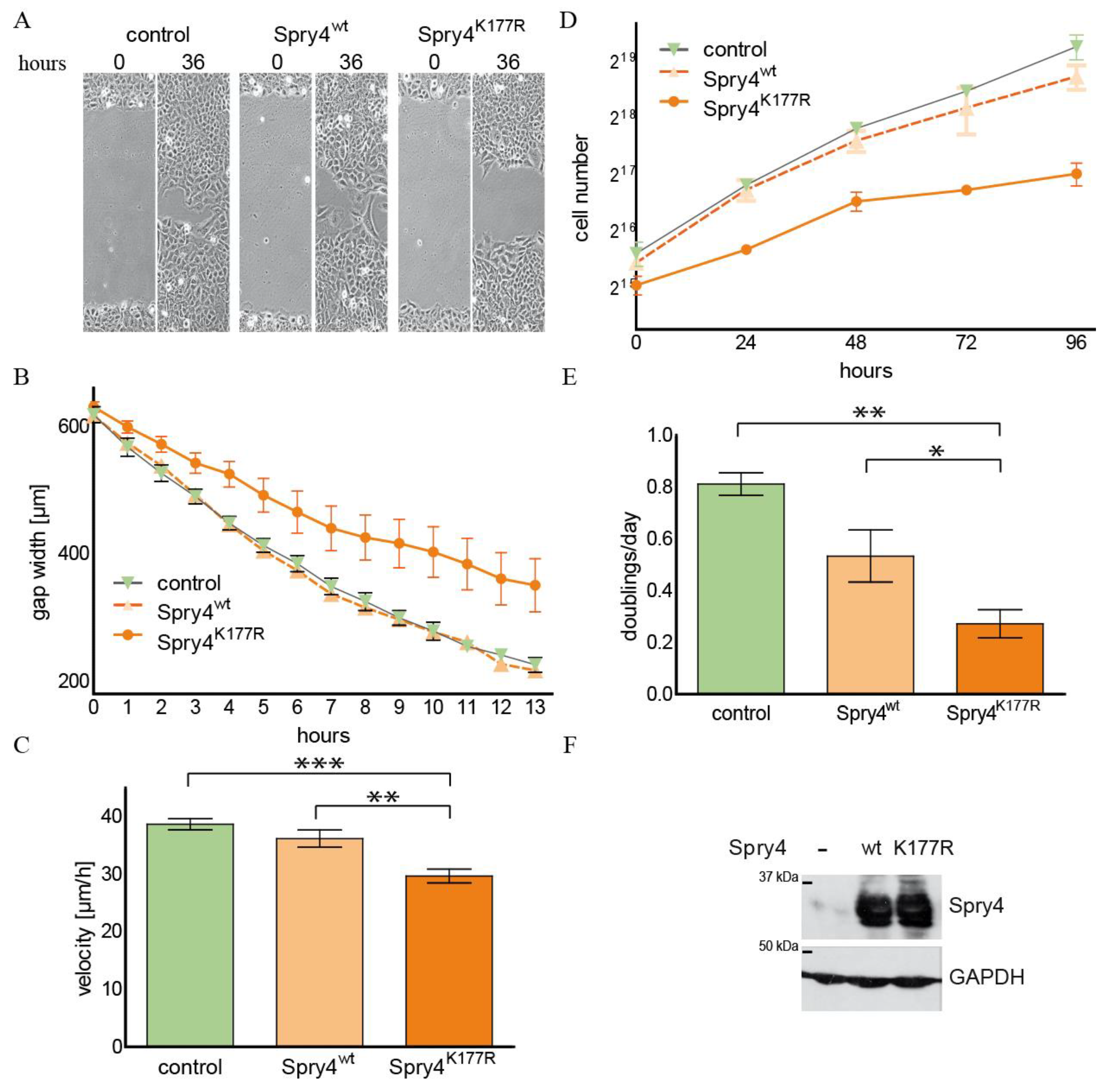
2.3. The Spry4K177R Variant Is Able to Inhibit Cell Migration and Proliferation in an Osteosarcoma-Derived Cell Line
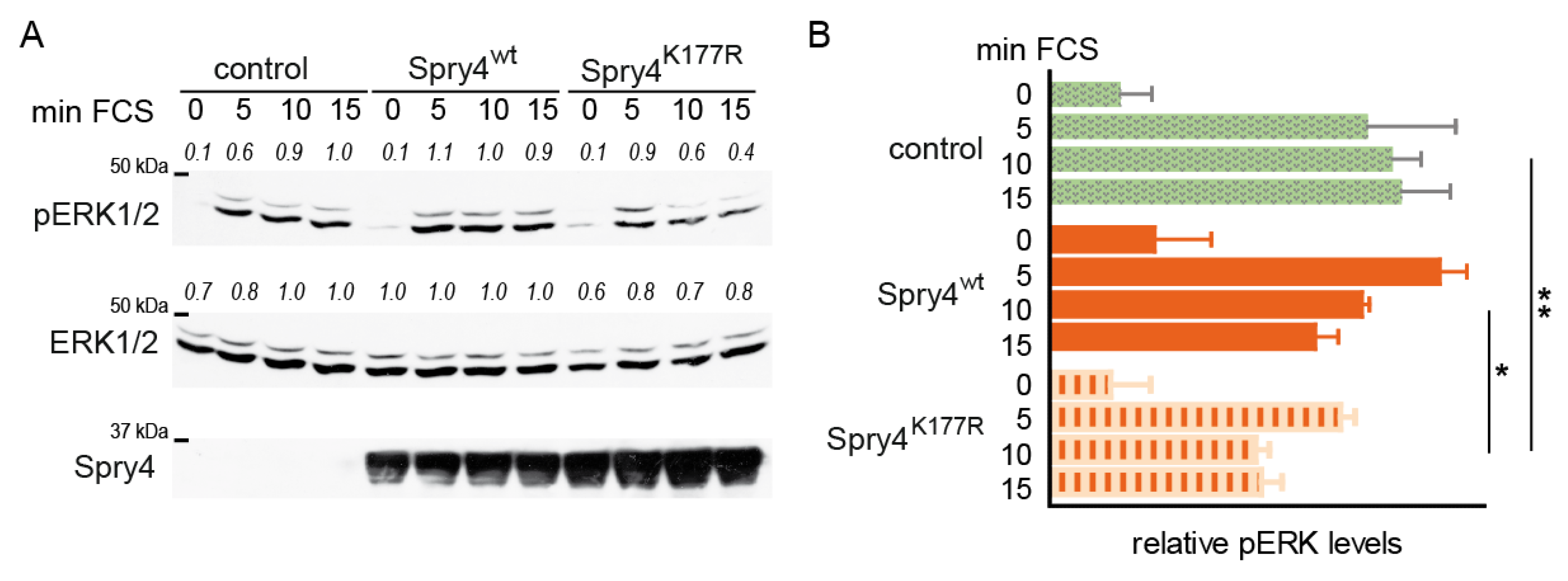
2.4. Like in Normal Fibroblasts, MAPK Activation Is Weakened in Presence of the Mutated Spry4 Version in Osteosarcoma-Derived Cells
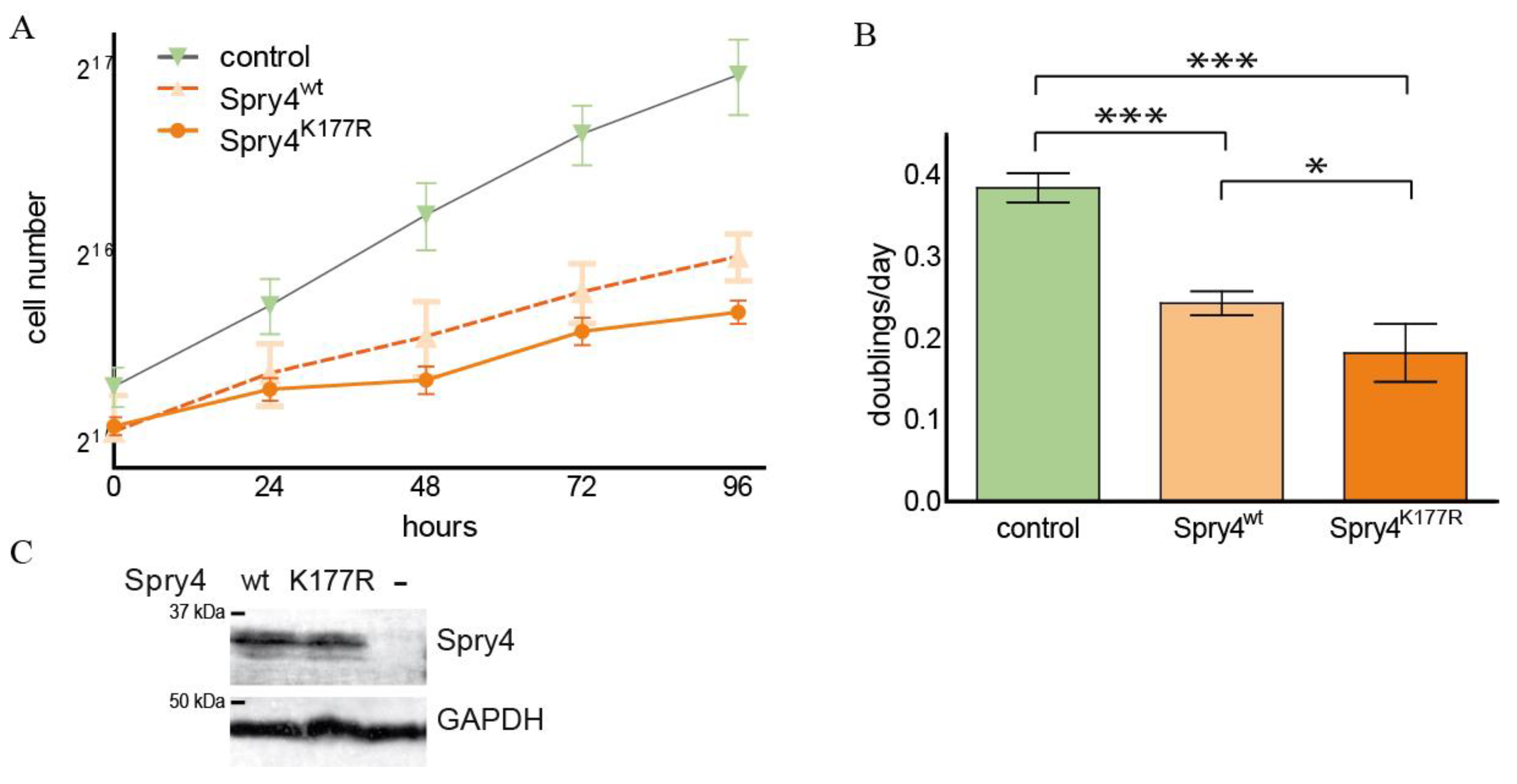
2.5. In a Lung Cancer-Derived Cell Line with an Activating EGFR Alteration, Both Spry4 Variants Are Equally Strong Inhibitors of Cell Proliferation and Migration
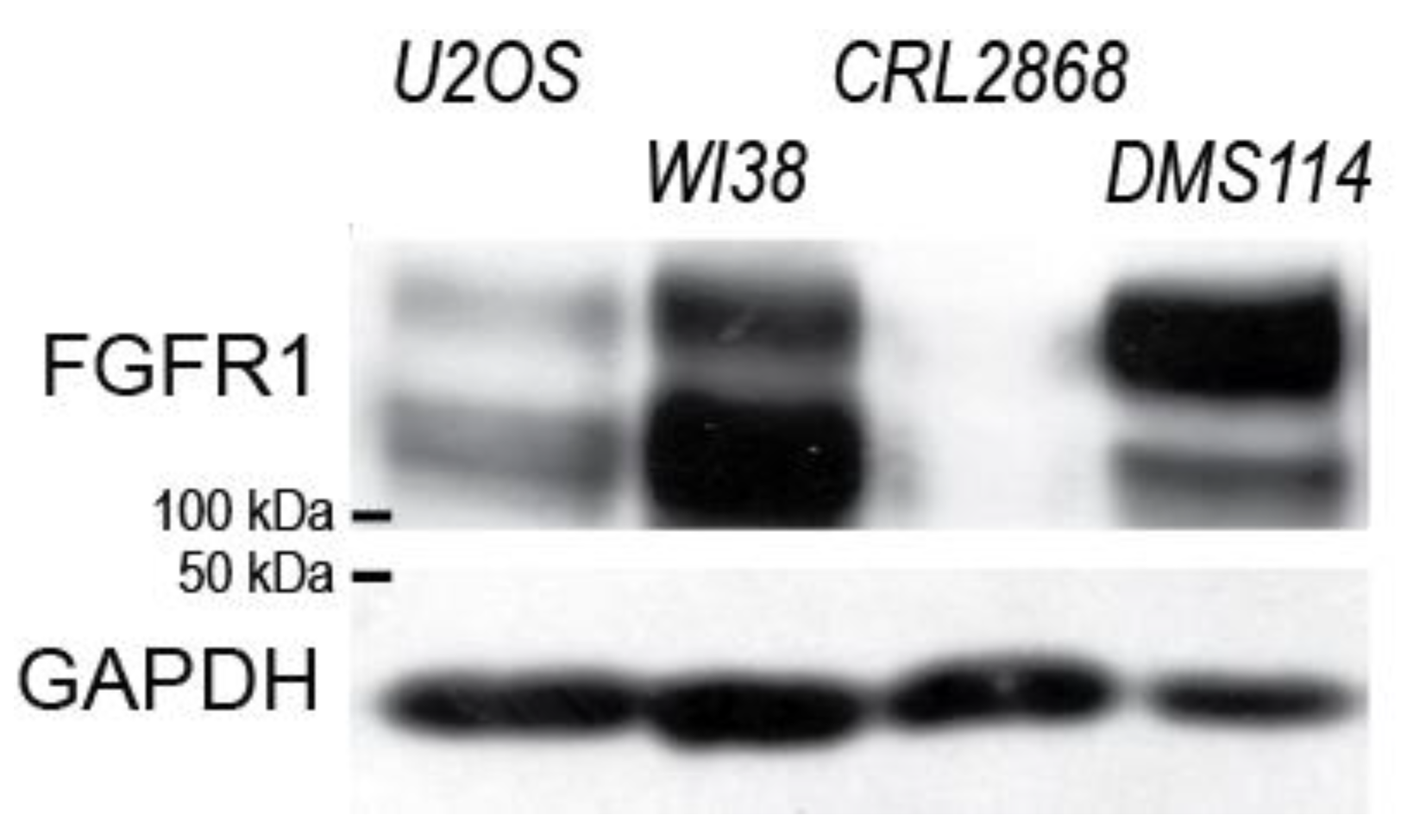
2.6. Spry4K177R Can Exacerbate the Inhibitory Effect of the Wt Variant in a Lung Cell Line Known to Be Dependent on FGFR1-Signalling
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Ectopic Expression of the Proteins Using the Adenoviral System
4.3. Calculation of Cell Doubling Time by Growth Curve Analysis
4.4. Migration Velocity Determined by Scratch Assay
4.5. Cell Signalling Assay
4.6. Immunoblotting
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Spry | Sprouty |
| FGFR | fibroblast growth factor receptor |
| pERK | phospho-extracellular-signal-regulated kinases |
| RTK | receptor tyrosine kinases |
| MAPK | mitogen-activated protein kinase |
| PLCγ | Phospholipase C gamma |
| PIK3 | Phosphoinositide 3-kinase |
| CFP | Cyan florescence protein |
| RAF | rapidly accelerated fibrosarcoma |
| MDM2 | Murine double minute 2 |
| FCS | fetal calf serum |
| ANOVA | analysis of variance |
| wt | wild-type |
| SD | standard deviation |
| SEM | standard error of the mean |
| DMEM | Dulbeccos Modified Eagle Medium |
| ATCC | American-type cell culture |
References
- Itoh, N.; Ornitz, D.M. Fibroblast growth factors: From molecular evolution to roles in development, metabolism and disease. J. Biochem. 2011, 149, 121–130. [Google Scholar] [CrossRef]
- Goetz, R.; Mohammadi, M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Huang, J.; Yan, W.; Liu, Z.; Liu, S.; Fang, W. FGFR families: Biological functions and therapeutic interventions in tumors. MedComm 2023, 4, e367. [Google Scholar] [CrossRef]
- Krook, M.A.; Reeser, J.W.; Ernst, G.; Barker, H.; Wilberding, M.; Li, G.; Chen, H.Z.; Roychowdhury, S. Fibroblast growth factor receptors in cancer: Genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br. J. Cancer 2021, 124, 880–892. [Google Scholar] [CrossRef]
- Domenichini, M.; Ravelli, C.; Corsini, M.; Codenotti, S.; Moreschi, E.; Gogna, A.; Capoferri, D.; Zizioli, D.; Bresciani, R.; Grillo, E.; et al. The D647N mutation of FGFR1 induces ligand-independent receptor activation. Biochim. Biophys. Acta Gen. Subj. 2023, 1867, 130470. [Google Scholar] [CrossRef]
- Holzmann, K.; Grunt, T.; Heinzle, C.; Sampl, S.; Steinhoff, H.; Reichmann, N.; Kleiter, M.; Hauck, M.; Marian, B. Alternative Splicing of Fibroblast Growth Factor Receptor IgIII Loops in Cancer. J. Nucleic Acids 2012, 2012, 950508. [Google Scholar] [CrossRef]
- De Luca, A.; Esposito Abate, R.; Rachiglio, A.M.; Maiello, M.R.; Esposito, C.; Schettino, C.; Izzo, F.; Nasti, G.; Normanno, N. FGFR Fusions in Cancer: From Diagnostic Approaches to Therapeutic Intervention. Int. J. Mol. Sci. 2020, 21, 6856. [Google Scholar] [CrossRef] [PubMed]
- Helsten, T.; Schwaederle, M.; Kurzrock, R. Fibroblast growth factor receptor signaling in hereditary and neoplastic disease: Biologic and clinical implications. Cancer Metastasis Rev. 2015, 34, 479–496. [Google Scholar] [CrossRef]
- Cadman, S.M.; Kim, S.H.; Hu, Y.; Gonzalez-Martinez, D.; Bouloux, P.M. Molecular pathogenesis of Kallmann’s syndrome. Horm. Res. 2007, 67, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Dode, C.; Hardelin, J.P. Kallmann syndrome: Fibroblast growth factor signaling insufficiency? J. Mol. Med. 2004, 82, 725–734. [Google Scholar] [CrossRef]
- Shim, K.; Minowada, G.; Coling, D.E.; Martin, G.R. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev. Cell 2005, 8, 553–564. [Google Scholar] [CrossRef]
- Taketomi, T.; Onimura, T.; Yoshiga, D.; Muratsu, D.; Sanui, T.; Fukuda, T.; Kusukawa, J.; Nakamura, S. Sprouty2 is involved in the control of osteoblast proliferation and differentiation through the FGF and BMP signaling pathways. Cell Biol. Int. 2018, 42, 1106–1114. [Google Scholar] [CrossRef]
- Taketomi, T.; Yoshiga, D.; Taniguchi, K.; Kobayashi, T.; Nonami, A.; Kato, R.; Sasaki, M.; Sasaki, A.; Ishibashi, H.; Moriyama, M.; et al. Loss of mammalian Sprouty2 leads to enteric neuronal hyperplasia and esophageal achalasia. Nat. Neurosci. 2005, 8, 855–857. [Google Scholar] [CrossRef]
- Taniguchi, K.; Ishizaki, T.; Ayada, T.; Sugiyama, Y.; Wakabayashi, Y.; Sekiya, T.; Nakagawa, R.; Yoshimura, A. Sprouty4 deficiency potentiates Ras-independent angiogenic signals and tumor growth. Cancer Sci. 2009, 100, 1648–1654. [Google Scholar] [CrossRef]
- Yu, T.; Yaguchi, Y.; Echevarria, D.; Martinez, S.; Basson, M.A. Sprouty genes prevent excessive FGF signalling in multiple cell types throughout development of the cerebellum. Development 2011, 138, 2957–2968. [Google Scholar] [CrossRef]
- Kawazoe, T.; Taniguchi, K. The Sprouty/Spred family as tumor suppressors: Coming of age. Cancer Sci. 2019, 110, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Holgren, C.; Dougherty, U.; Edwin, F.; Cerasi, D.; Taylor, I.; Fichera, A.; Joseph, L.; Bissonnette, M.; Khare, S. Sprouty-2 controls c-Met expression and metastatic potential of colon cancer cells: Sprouty/c-Met upregulation in human colonic adenocarcinomas. Oncogene 2010, 29, 5241–5253. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, G.; Hamdi, M.; Zwijnenburg, D.; Lakeman, A.; Geerts, D.; Versteeg, R.; Kool, M. Silencing of SPRY1 triggers complete regression of rhabdomyosarcoma tumors carrying a mutated RAS gene. Cancer Res. 2010, 70, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Celik-Selvi, B.E.; Stutz, A.; Mayer, C.E.; Salhi, J.; Siegwart, G.; Sutterluty, H. Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells. Cells 2019, 8, 808. [Google Scholar] [CrossRef]
- Park, J.W.; Wollmann, G.; Urbiola, C.; Fogli, B.; Florio, T.; Geley, S.; Klimaschewski, L. Sprouty2 enhances the tumorigenic potential of glioblastoma cells. Neuro Oncol. 2018, 20, 1044–1054. [Google Scholar] [CrossRef]
- Tennis, M.A.; Van Scoyk, M.M.; Freeman, S.V.; Vandervest, K.M.; Nemenoff, R.A.; Winn, R.A. Sprouty-4 inhibits transformed cell growth, migration and invasion, and epithelial-mesenchymal transition, and is regulated by Wnt7A through PPARgamma in non-small cell lung cancer. Mol. Cancer Res. MCR 2010, 8, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Vanas, V.; Muhlbacher, E.; Kral, R.; Sutterluty-Fall, H. Sprouty4 interferes with cell proliferation and migration of breast cancer-derived cell lines. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014, 35, 4447–4456. [Google Scholar] [CrossRef] [PubMed]
- Miraoui, H.; Dwyer, A.A.; Sykiotis, G.P.; Plummer, L.; Chung, W.; Feng, B.; Beenken, A.; Clarke, J.; Pers, T.H.; Dworzynski, P.; et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am. J. Hum. Genet. 2013, 92, 725–743. [Google Scholar] [CrossRef]
- Gach, A.; Pinkier, I.; Wysocka, U.; Salacinska, K.; Salachna, D.; Szarras-Czapnik, M.; Pietrzyk, A.; Sakowicz, A.; Nykel, A.; Rutkowska, L.; et al. New findings in oligogenic inheritance of congenital hypogonadotropic hypogonadism. Arch. Med. Sci. 2022, 18, 353–364. [Google Scholar] [CrossRef]
- Indirli, R.; Cangiano, B.; Profka, E.; Mantovani, G.; Persani, L.; Arosio, M.; Bonomi, M.; Ferrante, E. A Rare SPRY4 Gene Mutation Is Associated With Anosmia and Adult-Onset Isolated Hypogonadotropic Hypogonadism. Front. Endocrinol. 2019, 10, 781. [Google Scholar] [CrossRef]
- Amato, L.G.L.; Montenegro, L.R.; Lerario, A.M.; Jorge, A.A.L.; Guerra Junior, G.; Schnoll, C.; Renck, A.C.; Trarbach, E.B.; Costa, E.M.F.; Mendonca, B.B.; et al. New genetic findings in a large cohort of congenital hypogonadotropic hypogonadism. Eur. J. Endocrinol. 2019, 181, 103–119. [Google Scholar] [CrossRef]
- Stutz, A.; Kamptner, A.Z.M.; Sutterluty, H. A Sprouty4 Mutation Identified in Kallmann Syndrome Increases the Inhibitory Potency of the Protein towards FGF and Connected Processes. Int. J. Mol. Sci. 2021, 22, 2145. [Google Scholar] [CrossRef]
- Pan, H.; Xu, R.; Zhang, Y. Role of SPRY4 in health and disease. Front. Oncol. 2024, 14, 1376873. [Google Scholar] [CrossRef]
- de Maximy, A.A.; Nakatake, Y.; Moncada, S.; Itoh, N.; Thiery, J.P.; Bellusci, S. Cloning and expression pattern of a mouse homologue of drosophila sprouty in the mouse embryo. Mech. Dev. 1999, 81, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Klein, O.D.; Minowada, G.; Peterkova, R.; Kangas, A.; Yu, B.D.; Lesot, H.; Peterka, M.; Jernvall, J.; Martin, G.R. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev. Cell 2006, 11, 181–190. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, Y.; Itaranta, P.; Yagi, A.; Vainio, S. Expression of Sprouty genes 1, 2 and 4 during mouse organogenesis. Mech. Dev. 2001, 109, 367–370. [Google Scholar] [CrossRef]
- Masoumi-Moghaddam, S.; Amini, A.; Morris, D.L. The developing story of Sprouty and cancer. Cancer Metastasis Rev. 2014, 33, 695–720. [Google Scholar] [CrossRef]
- Guagnano, V.; Kauffmann, A.; Wohrle, S.; Stamm, C.; Ito, M.; Barys, L.; Pornon, A.; Yao, Y.; Li, F.; Zhang, Y.; et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012, 2, 1118–1133. [Google Scholar] [CrossRef]
- Rathmanner, N.; Haigl, B.; Vanas, V.; Doriguzzi, A.; Gsur, A.; Sutterluty-Fall, H. Sprouty2 but not Sprouty4 is a potent inhibitor of cell proliferation and migration of osteosarcoma cells. FEBS Lett. 2014, 587, 2597–2605. [Google Scholar] [CrossRef] [PubMed]
- Furugaki, K.; Mochizuki, M.; Kohno, M.; Shu, S.; Harada, N.; Yoshimura, Y. Expression of C-terminal ALK, RET, or ROS1 in lung cancer cells with or without fusion. BMC Cancer 2019, 19, 301. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.N.; Selinger, C.I.; Kohonen-Corish, M.R.J.; McCaughan, B.C.; Kennedy, C.W.; O’Toole, S.A.; Cooper, W.A. Fibroblast growth factor receptor 1 (FGFR1) copy number is an independent prognostic factor in non-small cell lung cancer. Lung Cancer 2013, 81, 462–467. [Google Scholar] [CrossRef]
- Malchers, F.; Ercanoglu, M.; Schutte, D.; Castiglione, R.; Tischler, V.; Michels, S.; Dahmen, I.; Bragelmann, J.; Menon, R.; Heuckmann, J.M.; et al. Mechanisms of Primary Drug Resistance in FGFR1-Amplified Lung Cancer. Clin. Cancer Res. 2017, 23, 5527–5536. [Google Scholar] [CrossRef]
- Sutterluty, H.; Mayer, C.E.; Setinek, U.; Attems, J.; Ovtcharov, S.; Mikula, M.; Mikulits, W.; Micksche, M.; Berger, W. Down-regulation of Sprouty2 in non-small cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and -independent mechanisms. Mol. Cancer Res. MCR 2007, 5, 509–520. [Google Scholar] [CrossRef]
- Jaggi, F.; Cabrita, M.A.; Perl, A.K.; Christofori, G. Modulation of endocrine pancreas development but not beta-cell carcinogenesis by Sprouty4. Mol. Cancer Res. MCR 2008, 6, 468–482. [Google Scholar] [CrossRef]
- Masoumi-Moghaddam, S.; Amini, A.; Wei, A.Q.; Robertson, G.; Morris, D.L. Sprouty 2 protein, but not Sprouty 4, is an independent prognostic biomarker for human epithelial ovarian cancer. Int. J. Cancer 2015, 137, 560–570. [Google Scholar] [CrossRef]
- Huang, Y.C.; Chen, W.C.; Yu, C.L.; Chang, T.K.; I-Chin Wei, A.; Chang, T.M.; Liu, J.F.; Wang, S.W. FGF2 drives osteosarcoma metastasis through activating FGFR1-4 receptor pathway-mediated ICAM-1 expression. Biochem. Pharmacol. 2023, 218, 115853. [Google Scholar] [CrossRef]
- Gong, Y.; Yang, X.; He, Q.; Gower, L.; Prudovsky, I.; Vary, C.P.; Brooks, P.C.; Friesel, R.E. Sprouty4 regulates endothelial cell migration via modulating integrin beta3 stability through c-Src. Angiogenesis 2013, 16, 861–875. [Google Scholar] [CrossRef]
- Zhou, X.; Xie, S.; Yuan, C.; Jiang, L.; Huang, X.; Li, L.; Chen, Y.; Luo, L.; Zhang, J.; Wang, D.; et al. Lower Expression of SPRY4 Predicts a Poor Prognosis and Regulates Cell Proliferation in Colorectal Cancer. Cell. Physiol. Biochem. 2016, 40, 1433–1442. [Google Scholar] [CrossRef]
- Paez, J.G.; Janne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef]
- Ozaki, K.; Miyazaki, S.; Tanimura, S.; Kohno, M. Efficient suppression of FGF-2-induced ERK activation by the cooperative interaction among mammalian Sprouty isoforms. J. Cell Sci. 2005, 118, 5861–5871. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Taketomi, T.; Kato, R.; Saeki, K.; Nonami, A.; Sasaki, M.; Kuriyama, M.; Saito, N.; Shibuya, M.; Yoshimura, A. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat. Cell Biol. 2003, 5, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Sos, M.L.; Seidel, D.; Peifer, M.; Zander, T.; Heuckmann, J.M.; Ullrich, R.T.; Menon, R.; Maier, S.; Soltermann, A.; et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl. Med. 2010, 2, 62ra93. [Google Scholar] [CrossRef]
- Hausott, B.; Pircher, L.; Kind, M.; Park, J.W.; Claus, P.; Obexer, P.; Klimaschewski, L. Sprouty2 Regulates Endocytosis and Degradation of Fibroblast Growth Factor Receptor 1 in Glioblastoma Cells. Cells 2024, 13, 1967. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, R.; Wang, X.; Lu, Y.; Chen, J.; Feng, D.; Zhou, J.; Xia, K.; Klein, O.; Xie, H.; et al. Sprouty genes regulate activated fibroblasts in mammary epithelial development and breast cancer. Cell Death Dis. 2024, 15, 256. [Google Scholar] [CrossRef]
- Azevedo, C.; Saiardi, A. Why always lysine? The ongoing tale of one of the most modified amino acids. Adv. Biol. Regul. 2016, 60, 144–150. [Google Scholar] [CrossRef]
- Guo, J.; Zhu, H.; Li, Q.; Dong, J.; Xiong, W.; Yu, K. SPRY4 suppresses proliferation and induces apoptosis of colorectal cancer cells by repressing oncogene EZH2. Aging 2021, 13, 11665–11677. [Google Scholar] [CrossRef]
- de Queiroz, R.M.; Efe, G.; Guzman, A.; Hashimoto, N.; Kawashima, Y.; Tanaka, T.; Rustgi, A.K.; Prives, C. Mdm2 requires Sprouty4 to regulate focal adhesion formation and metastasis independent of p53. Nat. Commun. 2024, 15, 7132. [Google Scholar] [CrossRef]
- Mayer, C.E.; Haigl, B.; Jantscher, F.; Siegwart, G.; Grusch, M.; Berger, W.; Sutterluty, H. Bimodal expression of Sprouty2 during the cell cycle is mediated by phase-specific Ras/MAPK and c-Cbl activities. Cell. Mol. Life Sci. 2010, 67, 3299–3311. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, J.; Stutz, A.; Vanas, V.; Salhi, J.; Reisecker, J.M.; Kral, R.M.; Sutterluty-Fall, H. Spatial signal repression as an additional role of Sprouty2 protein variants. Cell. Signal. 2019, 62, 109332. [Google Scholar] [CrossRef]
- Vanas, V.; Haigl, B.; Stockhammer, V.; Sutterluty-Fall, H. MicroRNA-21 Increases Proliferation and Cisplatin Sensitivity of Osteosarcoma-Derived Cells. PLoS ONE 2016, 11, e0161023. [Google Scholar] [CrossRef] [PubMed]
- Sutterluty, H.; Chatelain, E.; Marti, A.; Wirbelauer, C.; Senften, M.; Muller, U.; Krek, W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1999, 1, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Kral, R.M.; Mayer, C.E.; Vanas, V.; Gsur, A.; Sutterluty-Fall, H. In non-small cell lung cancer mitogenic signaling leaves Sprouty1 protein levels unaffected. Cell Biochem. Funct. 2014, 32, 96–100. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schiwek, M.; Ruhdorfer, K.; Pfurner, C.; Sutterlüty, H. The Lysine at Position 177 Is Essential to Limit the Inhibitory Capacities of Sprouty4 Protein in Normal and Cancer-Derived Cells. Int. J. Mol. Sci. 2025, 26, 7353. https://doi.org/10.3390/ijms26157353
Schiwek M, Ruhdorfer K, Pfurner C, Sutterlüty H. The Lysine at Position 177 Is Essential to Limit the Inhibitory Capacities of Sprouty4 Protein in Normal and Cancer-Derived Cells. International Journal of Molecular Sciences. 2025; 26(15):7353. https://doi.org/10.3390/ijms26157353
Chicago/Turabian StyleSchiwek, Maximilian, Kathrin Ruhdorfer, Christoph Pfurner, and Hedwig Sutterlüty. 2025. "The Lysine at Position 177 Is Essential to Limit the Inhibitory Capacities of Sprouty4 Protein in Normal and Cancer-Derived Cells" International Journal of Molecular Sciences 26, no. 15: 7353. https://doi.org/10.3390/ijms26157353
APA StyleSchiwek, M., Ruhdorfer, K., Pfurner, C., & Sutterlüty, H. (2025). The Lysine at Position 177 Is Essential to Limit the Inhibitory Capacities of Sprouty4 Protein in Normal and Cancer-Derived Cells. International Journal of Molecular Sciences, 26(15), 7353. https://doi.org/10.3390/ijms26157353






