Asynchrony Between Endometrial miRNA- and mRNA-Based Receptivity Stages Associated with Impaired Receptivity in Recurrent Implantation Failure
Abstract
1. Introduction
2. Results
2.1. Concordance Between mRNA (ERA) and miRNA (MIRA) Profiles
2.2. Staging Synchrony Between mRNA (ERA) and miRNA (MIRA) Profiles
2.3. Clinical Factors Associated with Lagging miRNA-Based Relative to mRNA-Based Receptivity Staging
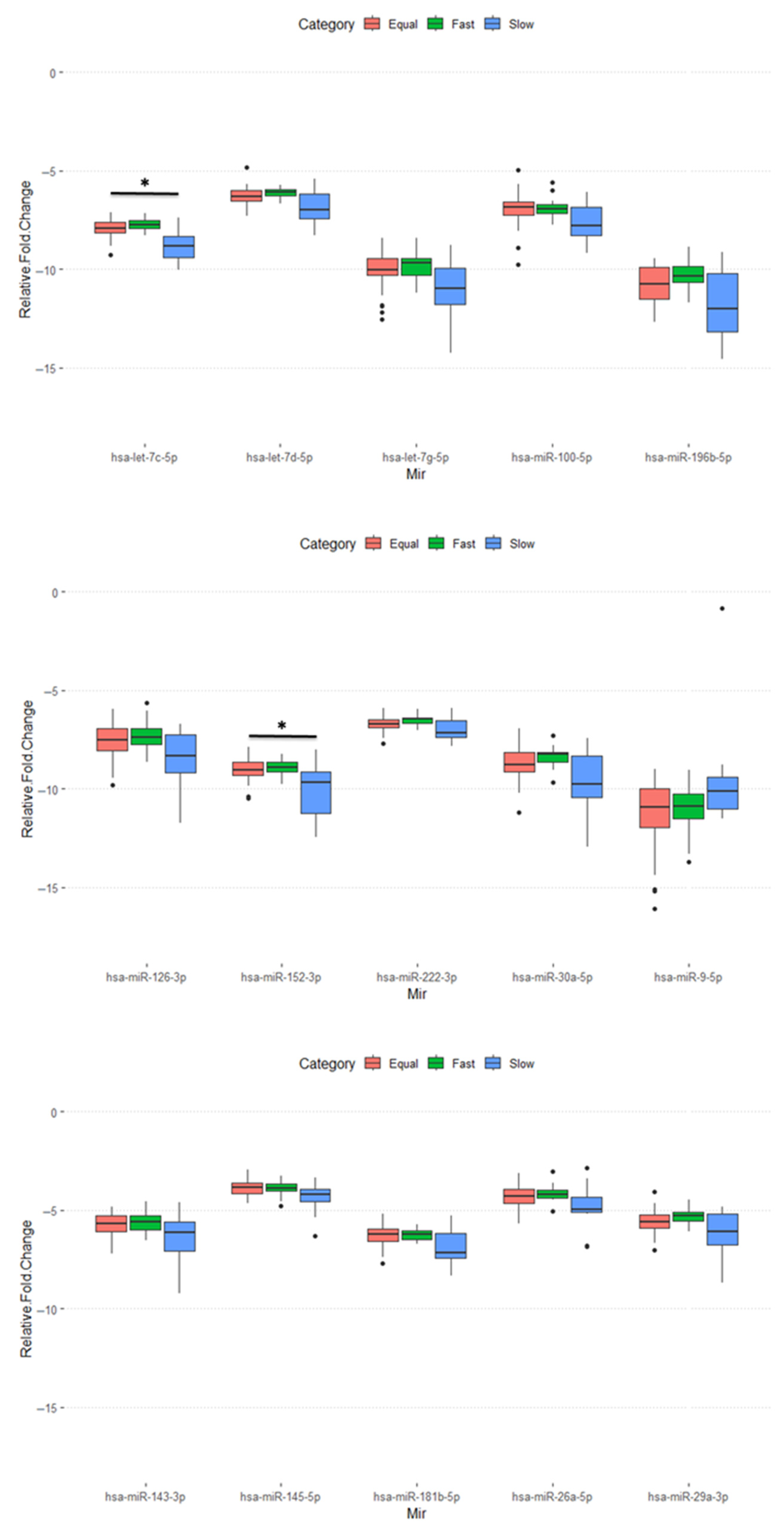
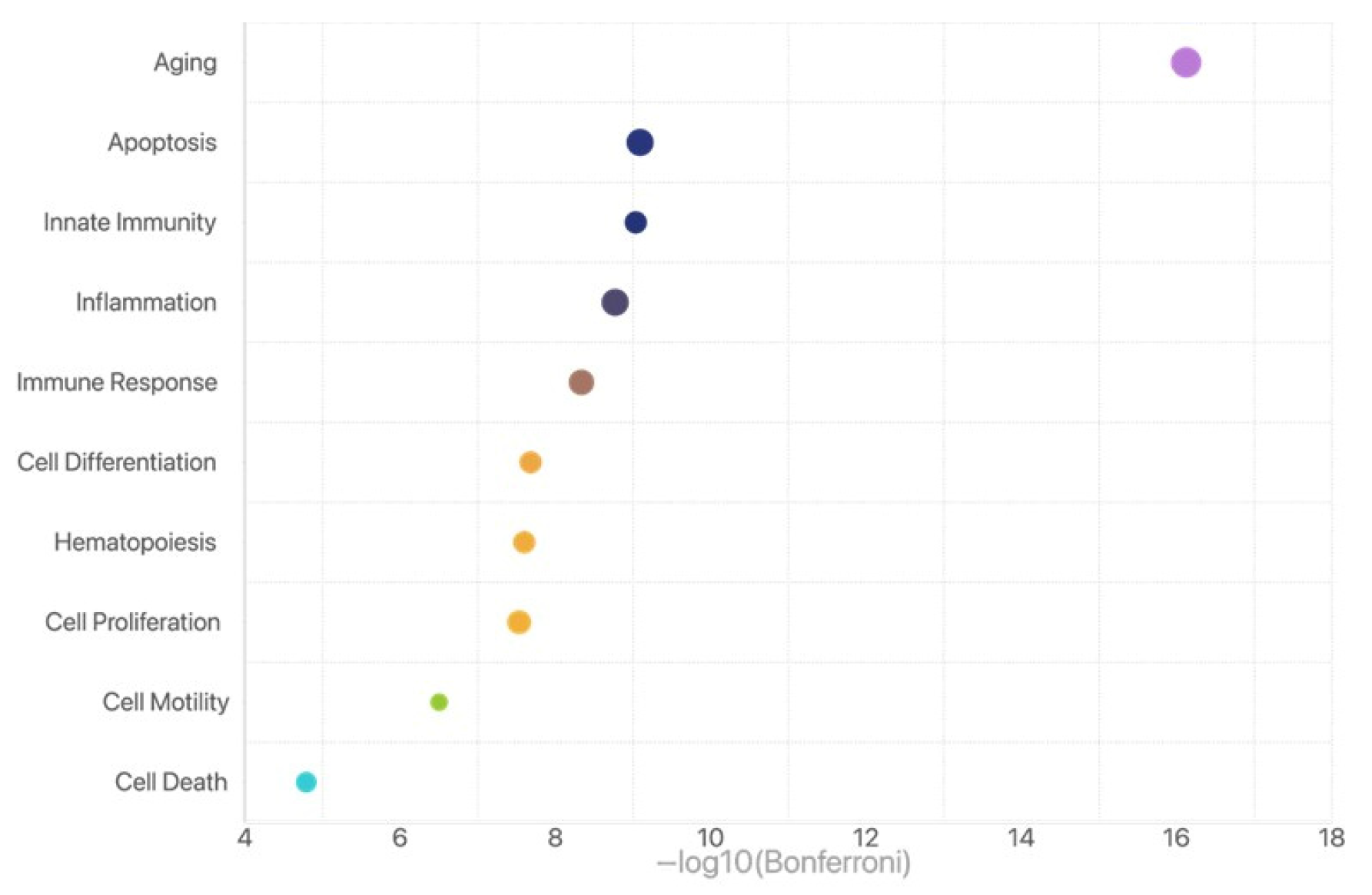
2.4. Function Enrichment Analysis for Those miRNAs with Differential Expression in the Slow Group
3. Discussion
4. Materials and Methods
4.1. Study Design and Patient Population
4.2. Endometrial Receptivity Array (ERA) Analysis
4.3. MicroRNA Receptivity Assay (MIRA) Analysis
4.4. Group Classification Based on miRNA and mRNA Timing
4.5. Personalized Embryo Transfer (ET) and Pregnancy Outcomes
4.6. Statistical Analysis
4.7. Functional Enrichment Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Altmae, S.; Koel, M.; Vosa, U.; Adler, P.; Suhorutsenko, M.; Laisk-Podar, T.; Kukushkina, V.; Saare, M.; Velthut-Meikas, A.; Krjutskov, K.; et al. Meta-signature of human endometrial receptivity: A meta-analysis and validation study of transcriptomic biomarkers. Sci. Rep. 2017, 7, 10077. [Google Scholar] [CrossRef]
- Cha, J.; Sun, X.; Dey, S.K. Mechanisms of implantation: Strategies for successful pregnancy. Nat. Med. 2012, 18, 1754–1767. [Google Scholar] [CrossRef]
- Lacconi, V.; Massimiani, M.; Carriero, I.; Bianco, C.; Ticconi, C.; Pavone, V.; Alteri, A.; Muzii, L.; Rago, R.; Pisaturo, V.; et al. When the Embryo Meets the Endometrium: Identifying the Features Required for Successful Embryo Implantation. Int. J. Mol. Sci. 2024, 25, 2834. [Google Scholar] [CrossRef]
- Van Vaerenbergh, I.; Fatemi, H.M.; Blockeel, C.; Van Lommel, L.; In’t Veld, P.; Schuit, F.; Kolibianakis, E.M.; Devroey, P.; Bourgain, C. Progesterone rise on HCG day in GnRH antagonist/rFSH stimulated cycles affects endometrial gene expression. Reprod. Biomed. Online 2011, 22, 263–271. [Google Scholar] [CrossRef]
- Evans, G.E.; Martínez-Conejero, J.A.; Phillipson, G.T.; Simón, C.; McNoe, L.A.; Sykes, P.H.; Horcajadas, J.A.; Lam, E.Y.; Print, C.G.; Sin, I.L.; et al. Gene and protein expression signature of endometrial glandular and stromal compartments during the window of implantation. Fertil. Steril. 2012, 97, 1365–1373.e1-2. [Google Scholar] [CrossRef] [PubMed]
- Rekker, K.; Altmäe, S.; Suhorutshenko, M.; Peters, M.; Martinez-Blanch, J.F.; Codoñer, F.M.; Vilella, F.; Simón, C.; Salumets, A.; Velthut-Meikas, A. A Two-Cohort RNA-seq Study Reveals Changes in Endometrial and Blood miRNome in Fertile and Infertile Women. Genes 2018, 9, 574. [Google Scholar] [CrossRef]
- Diaz-Gimeno, P.; Horcajadas, J.A.; Martinez-Conejero, J.A.; Esteban, F.J.; Alama, P.; Pellicer, A.; Simon, C. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil. Steril. 2011, 95, 50–60.e1-15. [Google Scholar] [CrossRef]
- Cimadomo, D.; de Los Santos, M.J.; Griesinger, G.; Lainas, G.; Le Clef, N.; McLernon, D.J.; Montjean, D.; Toth, B.; Vermeulen, N.; Macklon, N. ESHRE good practice recommendations on recurrent implantation failure. Hum. Reprod. Open 2023, 2023, hoad023. [Google Scholar] [CrossRef] [PubMed]
- Sha, A.G.; Liu, J.L.; Jiang, X.M.; Ren, J.Z.; Ma, C.H.; Lei, W.; Su, R.W.; Yang, Z.M. Genome-wide identification of micro-ribonucleic acids associated with human endometrial receptivity in natural and stimulated cycles by deep sequencing. Fertil. Steril. 2011, 96, 150–155.e5. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9. [Google Scholar] [CrossRef]
- Goharitaban, S.; Abedelahi, A.; Hamdi, K.; Khazaei, M.; Esmaeilivand, M.; Niknafs, B. Role of endometrial microRNAs in repeated implantation failure (mini-review). Front. Cell Dev. Biol. 2022, 10, 936173. [Google Scholar] [CrossRef] [PubMed]
- Zong, L.; Zheng, S.; Meng, Y.; Tang, W.; Li, D.; Wang, Z.; Tong, X.; Xu, B. Integrated Transcriptomic Analysis of the miRNA–mRNA Interaction Network in Thin Endometrium. Front. Genet. 2021, 12. [Google Scholar] [CrossRef]
- Rubin, S.C.; Abdulkadir, M.; Lewis, J.; Harutyunyan, A.; Hirani, R.; Grimes, C.L. Review of Endometrial Receptivity Array: A Personalized Approach to Embryo Transfer and Its Clinical Applications. J. Pers. Med. 2023, 13, 749. [Google Scholar] [CrossRef]
- Ruiz-Alonso, M.; Valbuena, D.; Gomez, C.; Cuzzi, J.; Simon, C. Endometrial Receptivity Analysis (ERA): Data versus opinions. Hum. Reprod. Open 2021, 2021, hoab011. [Google Scholar] [CrossRef] [PubMed]
- Gebremedhn, S.; Ali, A.; Hossain, M.; Hoelker, M.; Salilew-Wondim, D.; Anthony, R.V.; Tesfaye, D. MicroRNA-Mediated Gene Regulatory Mechanisms in Mammalian Female Reproductive Health. Int. J. Mol. Sci. 2021, 22, 938. [Google Scholar] [CrossRef]
- Revel, A.; Achache, H.; Stevens, J.; Smith, Y.; Reich, R. MicroRNAs are associated with human embryo implantation defects. Hum. Reprod. 2011, 26, 2830–2840. [Google Scholar] [CrossRef]
- Bidarimath, M.; Khalaj, K.; Wessels, J.M.; Tayade, C. MicroRNAs, immune cells and pregnancy. Cell. Mol. Immunol. 2014, 11, 538–547. [Google Scholar] [CrossRef]
- Hon, J.X.; Wahab, N.A.; Karim, A.K.A.; Mokhtar, N.M.; Mokhtar, M.H. MicroRNAs in Endometriosis: Insights into Inflammation and Progesterone Resistance. Int. J. Mol. Sci. 2023, 24, 15001. [Google Scholar] [CrossRef]
- Joshi, N.R.; Miyadahira, E.H.; Afshar, Y.; Jeong, J.W.; Young, S.L.; Lessey, B.A.; Serafini, P.C.; Fazleabas, A.T. Progesterone Resistance in Endometriosis Is Modulated by the Altered Expression of MicroRNA-29c and FKBP4. J. Clin. Endocrinol. Metab. 2017, 102, 141–149. [Google Scholar] [CrossRef]
- Simon, A.; Laufer, N. Assessment and treatment of repeated implantation failure (RIF). J. Assist. Reprod. Genet. 2012, 29, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Pirtea, P.; Cedars, M.I.; Devine, K.; Ata, B.; Franasiak, J.; Racowsky, C.; Toner, J.; Scott, R.T.; de Ziegler, D.; Barnhart, K.T. Recurrent implantation failure: Reality or a statistical mirage?: Consensus statement from the July 1, 2022 Lugano Workshop on recurrent implantation failure. Fertil. Steril. 2023, 120, 45–59. [Google Scholar] [CrossRef]
- Ma, J.; Gao, W.; Li, D. Recurrent implantation failure: A comprehensive summary from etiology to treatment. Front. Endocrinol. 2022, 13, 1061766. [Google Scholar] [CrossRef]
- Ruiz-Alonso, M.; Blesa, D.; Díaz-Gimeno, P.; Gómez, E.; Fernández-Sánchez, M.; Carranza, F.; Carrera, J.; Vilella, F.; Pellicer, A.; Simón, C. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertil. Steril. 2013, 100, 818–824. [Google Scholar] [CrossRef]
- Shekibi, M.; Heng, S.; Nie, G. MicroRNAs in the Regulation of Endometrial Receptivity for Embryo Implantation. Int. J. Mol. Sci. 2022, 23, 6210. [Google Scholar] [CrossRef] [PubMed]
- Segura-Benítez, M.; Bas-Rivas, A.; Juárez-Barber, E.; Carbajo-García, M.C.; Faus, A.; De Los Santos, M.J.; Pellicer, A.; Ferrero, H. Human blastocysts uptake extracellular vesicles secreted by endometrial cells containing miRNAs related to implantation. Hum. Reprod. 2023, 38, 1547–1559. [Google Scholar] [CrossRef]
- Kolanska, K.; Bendifallah, S.; Canlorbe, G.; Mekinian, A.; Touboul, C.; Aractingi, S.; Chabbert-Buffet, N.; Daraï, E. Role of miRNAs in Normal Endometrium and in Endometrial Disorders: Comprehensive Review. J. Clin. Med. 2021, 10, 3457. [Google Scholar] [CrossRef] [PubMed]
- Voros, C.; Varthaliti, A.; Athanasiou, D.; Mavrogianni, D.; Bananis, K.; Athanasiou, A.; Athanasiou, A.; Papahliou, A.-M.; Zografos, C.G.; Kondili, P.; et al. MicroRNA Signatures in Endometrial Receptivity—Unlocking Their Role in Embryo Implantation and IVF Success: A Systematic Review. Biomedicines 2025, 13, 1189. [Google Scholar] [CrossRef]
- Sadowska, A.; Molcan, T.; Wójtowicz, A.; Lukasik, K.; Pawlina-Tyszko, K.; Gurgul, A.; Ferreira-Dias, G.; Skarzynski, D.J.; Szóstek-Mioduchowska, A. Bioinformatic analysis of endometrial miRNA expression profile at day 26–28 of pregnancy in the mare. Sci. Rep. 2024, 14, 3900. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Kodithuwakku, S.P.; Pang, R.T.K.; Chiu, P.C.N.; Tang, M.H.Y.; Lee, K.-F. Role of microRNAs in embryo–endometrial interactions: Biological functions and clinical applications. Reprod. Dev. Med. 2023, 7, 238–251. [Google Scholar] [CrossRef]
- von Grothusen, C.; Frisendahl, C.; Modhukur, V.; Lalitkumar, P.G.; Peters, M.; Faridani, O.R.; Salumets, A.; Boggavarapu, N.R.; Gemzell-Danielsson, K. Uterine fluid microRNAs are dysregulated in women with recurrent implantation failure. Hum. Reprod. 2022, 37, 734–746. [Google Scholar] [CrossRef]
- Li, J.; Han, X.; Wan, Y.; Zhang, S.; Zhao, Y.; Fan, R.; Cui, Q.; Zhou, Y. TAM 2.0: Tool for MicroRNA set analysis. Nucleic Acids Res. 2018, 46, W180–W185. [Google Scholar] [CrossRef] [PubMed]
| ERA (mRNA Profiles) | MIRA (microRNA Profiles) | Total | ||
|---|---|---|---|---|
| Pre-Receptive (P + 4)mi | Receptive (P + 5)mi | Post-Receptive (P + 6)mi | ||
| Pre-receptive (P + 4)m | 13 | 9 | 0 | 22 (22.0%) |
| Receptive (P + 5)m | 6 | 51 | 8 | 65 (65.0%) |
| Post-receptive (P + 6)m | 1 | 4 | 8 | 13 (13.0%) |
| Total | 20 (20.0%) | 64 (64.0%) | 16 (16.0%) | 100 |
| Pregnancy Rate | Fast (n = 17) | Equal (n = 72) | Slow (n = 11) | p 1 |
|---|---|---|---|---|
| ERA pre-receptive | 8/9 (88.9%) | 10/13 (76.9%) | NA | 0.485 |
| ERA receptive | 8/8 (100%) | 42/51 (82.4%) | 4/6 (66.7%) | 0.247 |
| ERA post-receptive | NA | 7/8 (87.5%) | 2/5 (40.0%) | 0.083 |
| Total | 16/17 (94.1%) | 59/72 (81.9%) | 6/11 (54.5%) | 0.031 |
| Clinical Factors | Fast (n = 17) | Equal (n = 72) | Slow (n = 11) | p 1 |
|---|---|---|---|---|
| Age (Y) | 36.0 (34.0–42.0) | 39.0 (36.5–43.0) | 40.0 (35.5–45.5) | 0.260 |
| BMI (Kg/m2) | 20.7 (18.8–23.1) | 21.5 (20.3–23.4) | 21.1 (19.3–23.1) | 0.524 |
| Duration of infertility (Y) | 2.4 (1.7–4.1) | 3.3 (2.0–5.2) | 4.0 (1.5–6.8) | 0.633 |
| AMH (ng/mL) | 1.52 (0.22–3.21) | 2.07 (0.70–3.91) | 2.09 (0.88–7.03) | 0.334 |
| Embryo transfer times | 3.0 (2.0–3.3) | 3.0 (2.0–3.0) | 3.0 (2.0–3.8) | 0.875 |
| PCOS (%) | 2 (11.8%) | 9 (12.5%) | 4 (36.4%) | 0.109 |
| Endometriosis (%) | 5 (29.4%) | 17 (23.6%) | 3 (27.3%) | 0.869 |
| Endometrial polyps (%) | 10 (58.8%) | 23 (31.9%) | 2 (18.2%) | 0.019 |
| Clinical pregnancy (%) | 16 (94.1%) | 59 (81.9%) | 6 (54.5%) | 0.013 |
| Abortion (%) | 3 (17.6%) | 7 (9.7%) | 0 (0%) | 0.128 |
| Live birth (%) | 13 (76.5%) | 52 (72.2%) | 6 (54.5%) | 0.251 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-J.; Lee, C.-Y.; Cheng, E.-H.; Chen, W.-M.; Yang, P.E.; Lee, C.-I.; Lee, T.-H.; Lee, M.-S. Asynchrony Between Endometrial miRNA- and mRNA-Based Receptivity Stages Associated with Impaired Receptivity in Recurrent Implantation Failure. Int. J. Mol. Sci. 2025, 26, 7349. https://doi.org/10.3390/ijms26157349
Lee Y-J, Lee C-Y, Cheng E-H, Chen W-M, Yang PE, Lee C-I, Lee T-H, Lee M-S. Asynchrony Between Endometrial miRNA- and mRNA-Based Receptivity Stages Associated with Impaired Receptivity in Recurrent Implantation Failure. International Journal of Molecular Sciences. 2025; 26(15):7349. https://doi.org/10.3390/ijms26157349
Chicago/Turabian StyleLee, Yu-Jen, Chi-Ying Lee, En-Hui Cheng, Wei-Ming Chen, Pok Eric Yang, Chun-I Lee, Tsung-Hsien Lee, and Maw-Sheng Lee. 2025. "Asynchrony Between Endometrial miRNA- and mRNA-Based Receptivity Stages Associated with Impaired Receptivity in Recurrent Implantation Failure" International Journal of Molecular Sciences 26, no. 15: 7349. https://doi.org/10.3390/ijms26157349
APA StyleLee, Y.-J., Lee, C.-Y., Cheng, E.-H., Chen, W.-M., Yang, P. E., Lee, C.-I., Lee, T.-H., & Lee, M.-S. (2025). Asynchrony Between Endometrial miRNA- and mRNA-Based Receptivity Stages Associated with Impaired Receptivity in Recurrent Implantation Failure. International Journal of Molecular Sciences, 26(15), 7349. https://doi.org/10.3390/ijms26157349







