Association of Lipoprotein Lipase (LPL) Variants rs8176337, rs303, and rs304 with Body Mass Index and Total Cholesterol
Abstract
1. Introduction
2. Results
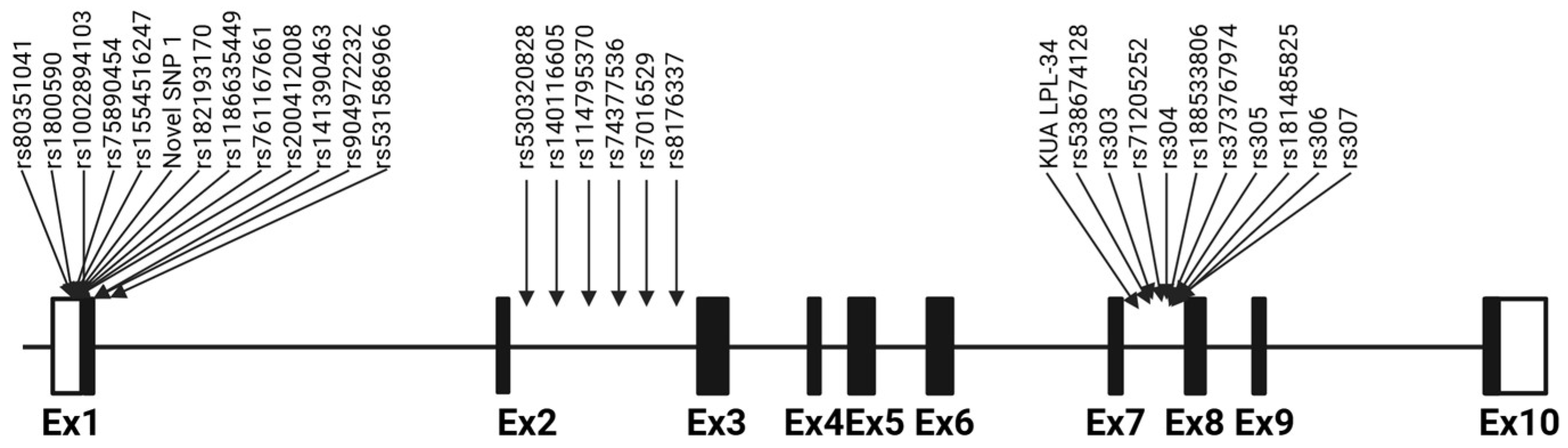
2.1. Sequence Analysis and Variant Identification
2.2. Genotype Distribution and Haplotypes
2.3. Genetic Association
3. Discussion
4. Materials and Methods
4.1. Sample Description
4.2. DNA Extraction
4.3. Amplification and Sequencing of the Target Region
4.4. Sequence Data Analysis and Variant Identification
4.5. Linkage Disequilibrium and Haplotype Construction
4.6. Genotype and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SNP | Single-nucleotide Polymorphism |
| LPL | Lipoprotein Lipase |
| BMI | Body Mass Index |
| MAF | Minor Allele Frequency |
| HWE | Hardy–Weinberg Equilibrium |
| TG | Triglyceride |
| VLDL | Very-low-density Lipoprotein |
| HDL | High-density Lipoprotein |
| UTR | Untranslated Region |
| LD | Linkage Disequilibrium |
References
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef]
- Nickerson, D.A.; Taylor, S.L.; Weiss, K.M.; Clark, A.G.; Hutchinson, R.G.; Stengard, J.; Salomaa, V.; Vartiainen, E.; Boerwinkle, E.; Sing, C.F. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat. Genet. 1998, 19, 233–240. [Google Scholar] [CrossRef]
- Mead, J.R.; Irvine, S.A.; Ramji, D.P. Lipoprotein lipase: Structure, function, regulation, and role in disease. J. Mol. Med. 2002, 80, 753–769. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Strauss, J.G.; Haemmerle, G.; Schoiswohl, G.; Birner-Gruenberger, R.; Riederer, M.; Lass, A.; Neuberger, G.; Eisenhaber, F.; Hermetter, A.; et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004, 306, 1383–1386. [Google Scholar] [CrossRef]
- Merkel, M.; Eckel, R.H.; Goldberg, I.J. Lipoprotein lipase: Genetics, lipid uptake, and regulation. J. Lipid Res. 2002, 43, 1997–2006. [Google Scholar] [CrossRef]
- Deeb, S.S.; Peng, R.L. Structure of the human lipoprotein lipase gene. Biochemistry 1989, 28, 4131–4135. [Google Scholar] [CrossRef]
- Kirchgessner, T.G.; Chuat, J.C.; Heinzmann, C.; Etienne, J.; Guilhot, S.; Svenson, K.; Ameis, D.; Pilon, C.; d’Auriol, L.; Andalibi, A.; et al. Organization of the human lipoprotein lipase gene and evolution of the lipase gene family. Proc. Natl. Acad. Sci. USA 1989, 86, 9647–9651. [Google Scholar] [CrossRef] [PubMed]
- Pirim, D.; Wang, X.; Radwan, Z.H.; Niemsiri, V.; Bunker, C.H.; Barmada, M.M.; Kamboh, M.I.; Demirci, F.Y. Resequencing of LPL in African Blacks and associations with lipoprotein-lipid levels. Eur. J. Hum. Genet. 2015, 23, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- Pirim, D.; Wang, X.; Radwan, Z.H.; Niemsiri, V.; Hokanson, J.E.; Hamman, R.F.; Barmada, M.M.; Demirci, F.Y.; Kamboh, M.I. Lipoprotein lipase gene sequencing and plasma lipid profile. J. Lipid Res. 2014, 55, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Rabacchi, C.; Pisciotta, L.; Cefalu, A.B.; Noto, D.; Fresa, R.; Tarugi, P.; Averna, M.; Bertolini, S.; Calandra, S. Spectrum of mutations of the LPL gene identified in Italy in patients with severe hypertriglyceridemia. Atherosclerosis 2015, 241, 79–86. [Google Scholar] [CrossRef]
- Al-Shammari, R.T.; Al-Serri, A.E.; Barhoush, S.A.; Al-Bustan, S.A. Identification and Characteriza-tion of variants in intron 6 of the LPL Gene Locus among a Sample of the Kuwaiti Population. Genes 2022, 13, 664. [Google Scholar] [CrossRef] [PubMed]
- Malek, S.H.; Al-Serri, A.E.; Al-Bustan, S.A. Genetic association of LPL rs326 with BMI among the Kuwaiti population. Cardiovasc. Endocrinol. Metab. 2021, 10, 215–221. [Google Scholar] [CrossRef]
- Al-Serri, A.; Al-Bustan, S.A.; Al-Sabah, S.K.; Annice, B.G.; Alnaqeeb, M.A.; Mojiminiyi, O.A. As-sociation between the lipoprotein lipase rs1534649 gene polymorphism in intron one with body mass index and high density lipoprotein cholesterol. Saudi. J. Biol. Sci. 2021, 28, 4717–4722. [Google Scholar] [CrossRef] [PubMed]
- Al-Bustan, S.A.; Al-Serri, A.; Annice, B.G.; Alnaqeeb, M.A.; Al-Kandari, W.Y.; Dashti, M. A novel LPL intronic variant: g. 18704C>A identified by re-sequencing Kuwaiti Arab samples is associated with high-density lipoprotein, very low-density lipoprotein and triglyceride lipid levels. PLoS ONE 2018, 13, e0192617. [Google Scholar] [CrossRef] [PubMed]
- He, P.P.; Jiang, T.; OuYang, X.P.; Liang, Y.Q.; Zou, J.Q.; Wang, Y.; Shen, Q.Q.; Liao, L.; Zheng, X.L. Lipoprotein lipase: Biosynthesis, regulatory factors, and its role in atherosclerosis and other diseases. Clin. Chim. Acta 2018, 480, 126–137. [Google Scholar] [CrossRef]
- Heid, I.M.; Boes, E.; Muller, M.; Kollerits, B.; Lamina, C.; Coassin, S.; Gieger, C.; Doring, A.; Klopp, N.; Frikke-Schmidt, R.; et al. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA study sheds new light on intergenic regions. Circ. Cardiovasc. Genet. 2008, 1, 10–20. [Google Scholar] [CrossRef]
- Johansen, C.T.; Wang, J.A.; Lanktree, M.B.; Cao, H.N.; McIntyre, A.D.; Ban, M.R.; Martins, R.A.; Kennedy, B.A.; Hassell, R.G.; Visser, M.E.; et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010, 42, 684–687. [Google Scholar] [CrossRef]
- Al-Bustan, S.A.; Al-Serri, A.; Alnaqeeb, M.A.; Annice, B.G.; Mojiminiyi, O. Genetic association of LPL rs1121923 and rs258 with plasma TG and VLDL levels. Sci. Rep. 2019, 9, 5572. [Google Scholar] [CrossRef]
- Yang, Y.; Mu, Y.; Zhao, Y.; Liu, X.; Zhao, L.; Wang, J.; Xie, Y. Genetic screening of the lipoprotein lipase gene for mutations in Chinese subjects with or without hypertriglyceridemia. J. Genet. Genom. 2007, 34, 381–391. [Google Scholar] [CrossRef]
- Bentley, A.R.; Chen, G.; Shriner, D.; Doumatey, A.P.; Zhou, J.; Huang, H.; Mullikin, J.C.; Blakesley, R.W.; Hansen, N.F.; Bouffard, G.G.; et al. Gene-based sequencing identifies lipid-influencing variants with ethnicity-specific effects in African Americans. PLoS Genet. 2014, 10, e1004190. [Google Scholar] [CrossRef] [PubMed]
- Radha, V.; Vimaleswaran, K.S.; Ayyappa, K.A.; Mohan, V. Association of lipoprotein lipase gene polymorphisms with obesity and type 2 diabetes in an Asian Indian population. Int. J. Obes. 2007, 31, 913–918. [Google Scholar] [CrossRef]
- Gao, R.R.; Wang, M.; Hu, Y.; Xu, C.Y.; Li, Y.C.; Zhang, Z.Y.; Chen, S.Y.; Mao, X.Y. Impact of LPL gene rs283 polymorphism on exercise-induced changes in metabolism of obese adolescents and the regulatory mechanisms behind it. Exp. Physiol. 2015, 100, 698–707. [Google Scholar] [CrossRef]
- Aulchenko, Y.S.; Ripatti, S.; Lindqvist, I.; Boomsma, D.; Heid, I.M.; Pramstaller, P.P.; Penninx, B.W.; Janssens, A.C.; Wilson, J.F.; Spector, T.; et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009, 41, 47–55. [Google Scholar] [CrossRef]
- Akua, T.; Shaul, O. The Arabidopsis thaliana MHX gene includes an intronic element that boosts translation when localized in a 5’ UTR intron. J. Exp. Bot. 2013, 64, 4255–4270. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, M.O.; Wong, H.; Quinones, M.J.; Taylor, K.D.; Guo, X.; Castellani, L.W.; Antoine, H.J.; Yang, H.; Hsueh, W.A.; Rotter, J.I. The 3’ untranslated region of the lipoprotein lipase gene: Haplotype structure and association with post-heparin plasma lipase activity. J. Clin. Endocrinol. Metab. 2005, 90, 4816–4823. [Google Scholar] [CrossRef] [PubMed]
- Schinkel, J.; Spaan, W.J.; Kroes, A.C. Meta-analysis of mutations in the NS5A gene and hepatitis C virus resistance to interferon therapy: Uniting discordant conclusions. Antivir. Ther. 2004, 9, 275–286. [Google Scholar] [CrossRef]
- Desmet, F.O.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Beroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef]
- Chen, Q.; Razzaghi, H.; Demirci, F.Y.; Kamboh, M.I. Functional significance of lipoprotein lipase HindIII polymorphism associated with the risk of coronary artery disease. Atherosclerosis 2008, 200, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Marchadier, D.; Rader, D.J. Lipases and HDL metabolism. Trends Endocrinol. Metab. 2002, 13, 174–178. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; De-Felice, M.; Lochner, A.; Faggart, M.; et al. The structure of haplotype blocks in the human ge-nome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed]
| LPL Novels | SNP ID Number | Position Within Gene | Consequence | Global MAF | Ancestral Allele | Variant Allele | HWE p-Value |
|---|---|---|---|---|---|---|---|
| KUA LPL-16 | rs1554516247 | 5202 Ins/CC | 5′ UTR Variant | NA | 0.997 | 0.003 | 0.940 |
| KUA LPL-26 | rs530320828 | 14866_14876 del/An | Intron Variant | AA = 0.0407 (n = 204) | 0.500 | 0.500 | <0.001 * |
| KUA LPL-36 | rs71205952 | 25708–25710 del/Tn | Intron Variant | delTTT = 0.01779 (n = 206) | 0.500 | 0.500 | <0.001 * |
| SNP | Variable ^ | W/W | n | W/M | n | M/M | n | W/M + M/M | n | p-Value * |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1800590 | BMI | 27.07 ± 6.66 | 421 | 28.18 ± 6.85 | 49 | 24.45 ± 0.78 | 2 | 28.03 ± 6.75 | 51 | 0.330 |
| (T‹G) | TC | 4.73 ± 0.94 | 610 | 4.73 ± 0.87 | 76 | 4.80 ± 1.27 | 2 | 4.72 ± 0.87 | 78 | 0.890 |
| TG | 1.05 ± 0.79 | 610 | 1.16 ± 0.82 | 76 | 1.12 ± 0.79 | 2 | 1.16 ± 0.82 | 78 | 0.349 | |
| VLDL | 0.44 ± 0.33 | 610 | 0.47 ± 0.34 | 76 | 0.45 ± 0.33 | 2 | 0.48 ± 0.34 | 78 | 0.348 | |
| HDL-C | 1.14 ± 0.32 | 610 | 1.07 ± 0.28 | 76 | 1.29 ± 0.13 | 2 | 1.08 ± 0.29 | 78 | 0.207 | |
| LDL-C | 3.12 ± 0.83 | 610 | 3.13 ± 0.70 | 76 | 3.10 ± 1.41 | 2 | 3.13 ± 0.71 | 78 | 0.576 | |
| rs74377536 | BMI | 27.33 ± 7.08 | 342 | 26.73 ± 7.08 | 124 | 27.38 ± 8.09 | 6 | 26.76 ± 5.47 | 130 | 0.415 |
| (C‹A) | TC | 4.75 ± 0.94 | 503 | 4.62 ± 0.89 | 176 | 5.46 ± 1.45 | 9 | 4.67 ± 0.94 | 185 | 0.388 |
| TG | 1.07 ± 0.83 | 503 | 1.04 ± 0.70 | 176 | 1.23 ± 0.65 | 9 | 1.06 ± 0.70 | 185 | 0.495 | |
| VLDL | 0.44 ± 0.35 | 503 | 0.42 ± 0.28 | 176 | 0.51 ± 0.26 | 9 | 0.43 ± 0.28 | 185 | 0.550 | |
| HDL-C | 1.13 ± 0.30 | 503 | 1.11 ± 0.35 | 176 | 1.08 ± 0.39 | 9 | 1.11 ± 0.35 | 185 | 0.191 | |
| LDL-C | 3.13 ± 0.81 | 503 | 3.04 ± 0.78 | 176 | 3.82 ± 1.24 | 9 | 3.09 ± 0.83 | 185 | 0.435 | |
| rs8176337 | BMI | 26.69 ± 6.09 | 302 | 27.92 ± 7.27 | 145 | 28.66 ± 9.09 | 25 | 28.03 ± 7.54 | 170 | 0.036 |
| (C‹G) | TC | 4.71 ± 0.97 | 437 | 4.73 ± 0.90 | 218 | 4.94 ± 0.72 | 33 | 4.75 ± 0.88 | 251 | 0.494 |
| TG | 1.08 ± 0.85 | 437 | 1.03 ± 0.70 | 218 | 1.06 ± 0.80 | 33 | 1.04 ± 0.70 | 251 | 0.430 | |
| VLDL | 0.45 ± 0.37 | 437 | 0.42 ± 0.28 | 218 | 0.45 ± 0.26 | 33 | 0.43 ± 0.28 | 251 | 0.372 | |
| HDL-C | 1.13 ± 0.33 | 437 | 1.12 ± 0.29 | 218 | 1.10 ± 0.34 | 33 | 1.12 ± 0.30 | 251 | 0.956 | |
| LDL-C | 3.09 ± 0.82 | 437 | 3.15 ± 0.83 | 218 | 3.24 ± 0.68 | 33 | 3.17 ± 0.82 | 251 | 0.257 | |
| rs303 | BMI | 27.10 ± 6.56 | 396 | 27.40 ± 7.32 | 71 | 29.46 ± 6.60 | 5 | 27.54 ± 7.25 | 76 | 0.601 |
| (G‹C) | TC | 4.69 ± 0.93 | 562 | 4.88 ± 0.944 | 118 | 4.96 ± 1.07 | 8 | 4.89 ± 0.95 | 126 | 0.023 |
| TG | 1.07 ± 0.82 | 562 | 1.08 ± 0.73 | 118 | 0.73 ± 0.38 | 8 | 1.06 ± 0.72 | 126 | 0.838 | |
| VLDL | 0.44 ± 0.34 | 562 | 0.44 ± 0.29 | 118 | 0.31 ± 0.17 | 8 | 0.43 ± 0.29 | 126 | 0.879 | |
| HDL-C | 1.12 ± 0.31 | 562 | 1.13 ± 0.31 | 118 | 1.20 ± 0.46 | 8 | 1.14 ± 0.33 | 126 | 0.738 | |
| LDL-C | 3.08 ± 0.81 | 562 | 3.25 ± 0.84 | 118 | 3.41 ± 0.85 | 8 | 3.27 ± 0.84 | 126 | 0.039 | |
| rs304 | BMI | 26.98 ± 6.46 | 300 | 27.17 ± 6.12 | 147 | 29.44 ± 10.99 | 25 | 27.50± 7.04 | 172 | 0.418 |
| (T‹G) | TC | 4.66 ± 0.95 | 428 | 4.82 ± 0.91 | 224 | 4.88 ± 0.82 | 36 | 4.83 ± 0.91 | 260 | 0.034 |
| TG | 1.09 ± 0.87 | 428 | 1.02 ± 0.68 | 224 | 1.00 ± 0.53 | 36 | 1.02 ± 0.66 | 260 | 0.868 | |
| VLDL | 0.45 ± 0.36 | 428 | 0.41 ± 0.27 | 224 | 0.41 ± 0.21 | 36 | 0.42 ± 0.27 | 260 | 0.806 | |
| HDL-C | 1.12 ± 0.32 | 428 | 1.14 ± 0.31 | 224 | 1.13 ± 0.28 | 36 | 1.15 ± 0.31 | 260 | 0.195 | |
| LDL-C | 3.06 ± 0.80 | 428 | 3.21 ± 0.83 | 224 | 3.27 ± 0.78 | 36 | 3.22 ± 0.83 | 260 | 0.034 |
| Variable | β-Coefficient | 95% CI | p-Value |
|---|---|---|---|
| rs8176337 | 1.41 | 0.22–2.60 | 0.020 |
| Age (Female) | 0.13 | 0.04–0.14 | <0.001 |
| Sex | −0.48 | −1.7–0.73 | 0.438 |
| TG | 2.32 | 1.40–3.25 | <0.001 |
| A. rs303 | ||||
| Lipid | Variable | β-Coefficient | 95% CI | p-Value |
| TC | rs303 | 0.210 | 0.034–0.386 | 0.020 |
| Age | 0.017 | 0.012–0.022 | <0.001 | |
| Sex (Female) | 0.001 | −0.139–0.140 | 0.992 | |
| LDL-C | rs303 | 0.195 | 0.039–0.351 | 0.014 |
| Age | 0.010 | 0.006–0.014 | <0.001 | |
| Sex | −0.116 | −0.239–0.007 | 0.065 | |
| B. rs304 | ||||
| Lipid | Variable | β-Coefficient | 95% CI | p-Value |
| TC | rs304 | 0.174 | 0.034–0.314 | 0.015 |
| Age | 0.017 | 0.012–0.022 | <0.001 | |
| Sex (Female) | 0.004 | −0.136–0.143 | 0.959 | |
| LDL-C | rs304 | 0.174 | 0.050–0.298 | 0.006 |
| Age | 0.010 | 0.006–0.014 | <0.001 | |
| Sex | −0.113 | −0.237–0.010 | 0.071 | |
| Variables | Participants (n = 688) |
|---|---|
| Age (years) | 32.92 ± 14.26 |
| Sex (Female%) | 414 (60.2%) |
| BMI (kg/m2) | 27.17 ± 6.67 |
| Cholesterol (mmol/L) | 4.73 ± 0.94 |
| Triglyceride (mmol/L) | 1.06 ± 0.80 |
| VLDL (mmol/L) | 0.44 ± 0.33 |
| HDL-C (mmol/L) | 1.13 ± 0.32 |
| LDL-C (mmol/L) | 3.11 ± 0.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Bustan, S.A.; Al-Serri, A.E.; Al-Adsani, A.M.; Miranda, L.; Annice, B.G.; Hamdan, H.; Alnaqeeb, M.A. Association of Lipoprotein Lipase (LPL) Variants rs8176337, rs303, and rs304 with Body Mass Index and Total Cholesterol. Int. J. Mol. Sci. 2025, 26, 7282. https://doi.org/10.3390/ijms26157282
Al-Bustan SA, Al-Serri AE, Al-Adsani AM, Miranda L, Annice BG, Hamdan H, Alnaqeeb MA. Association of Lipoprotein Lipase (LPL) Variants rs8176337, rs303, and rs304 with Body Mass Index and Total Cholesterol. International Journal of Molecular Sciences. 2025; 26(15):7282. https://doi.org/10.3390/ijms26157282
Chicago/Turabian StyleAl-Bustan, Suzanne A., Ahmad E. Al-Serri, Amani M. Al-Adsani, Lavina Miranda, Babitha G. Annice, Hala Hamdan, and Majed A. Alnaqeeb. 2025. "Association of Lipoprotein Lipase (LPL) Variants rs8176337, rs303, and rs304 with Body Mass Index and Total Cholesterol" International Journal of Molecular Sciences 26, no. 15: 7282. https://doi.org/10.3390/ijms26157282
APA StyleAl-Bustan, S. A., Al-Serri, A. E., Al-Adsani, A. M., Miranda, L., Annice, B. G., Hamdan, H., & Alnaqeeb, M. A. (2025). Association of Lipoprotein Lipase (LPL) Variants rs8176337, rs303, and rs304 with Body Mass Index and Total Cholesterol. International Journal of Molecular Sciences, 26(15), 7282. https://doi.org/10.3390/ijms26157282







