Antagonistic Interactions Between Dickeya solani and Bacillus subtilis
Abstract
1. Introduction
2. Results
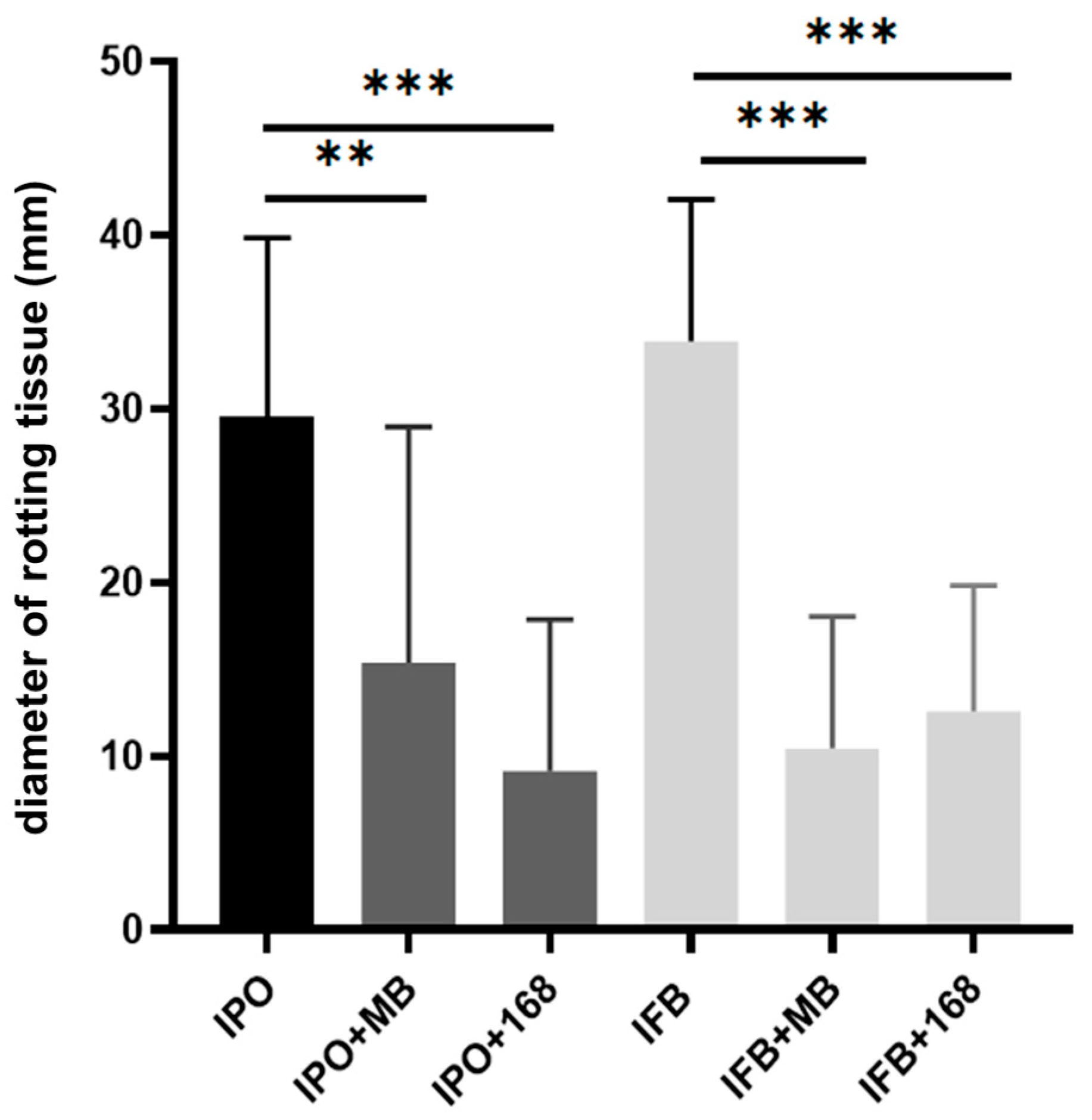
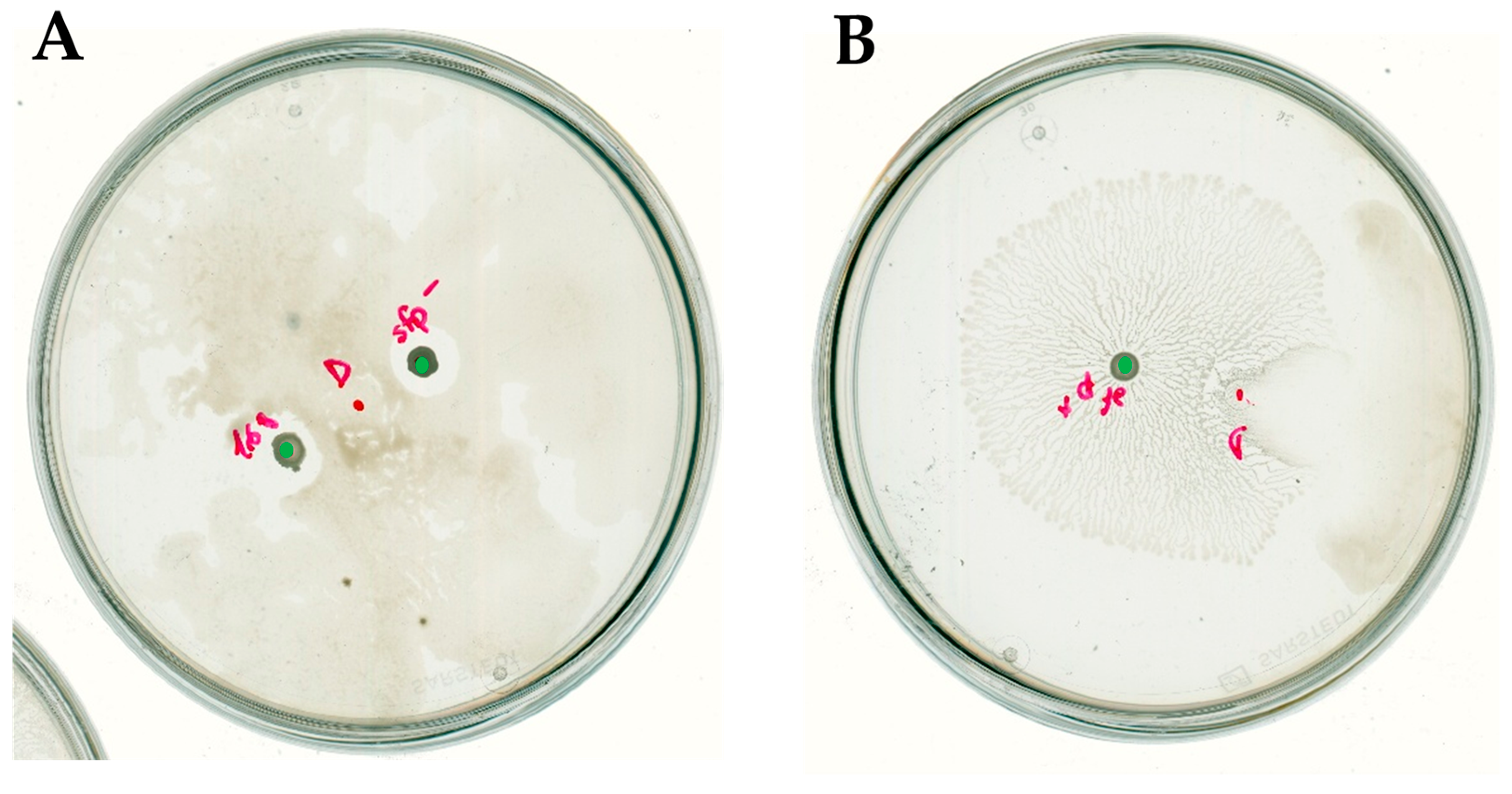
2.1. Bacillus Subtilis Suppresses In Vitro Soft-Rotting of Potatoes Caused by Dickeya solani
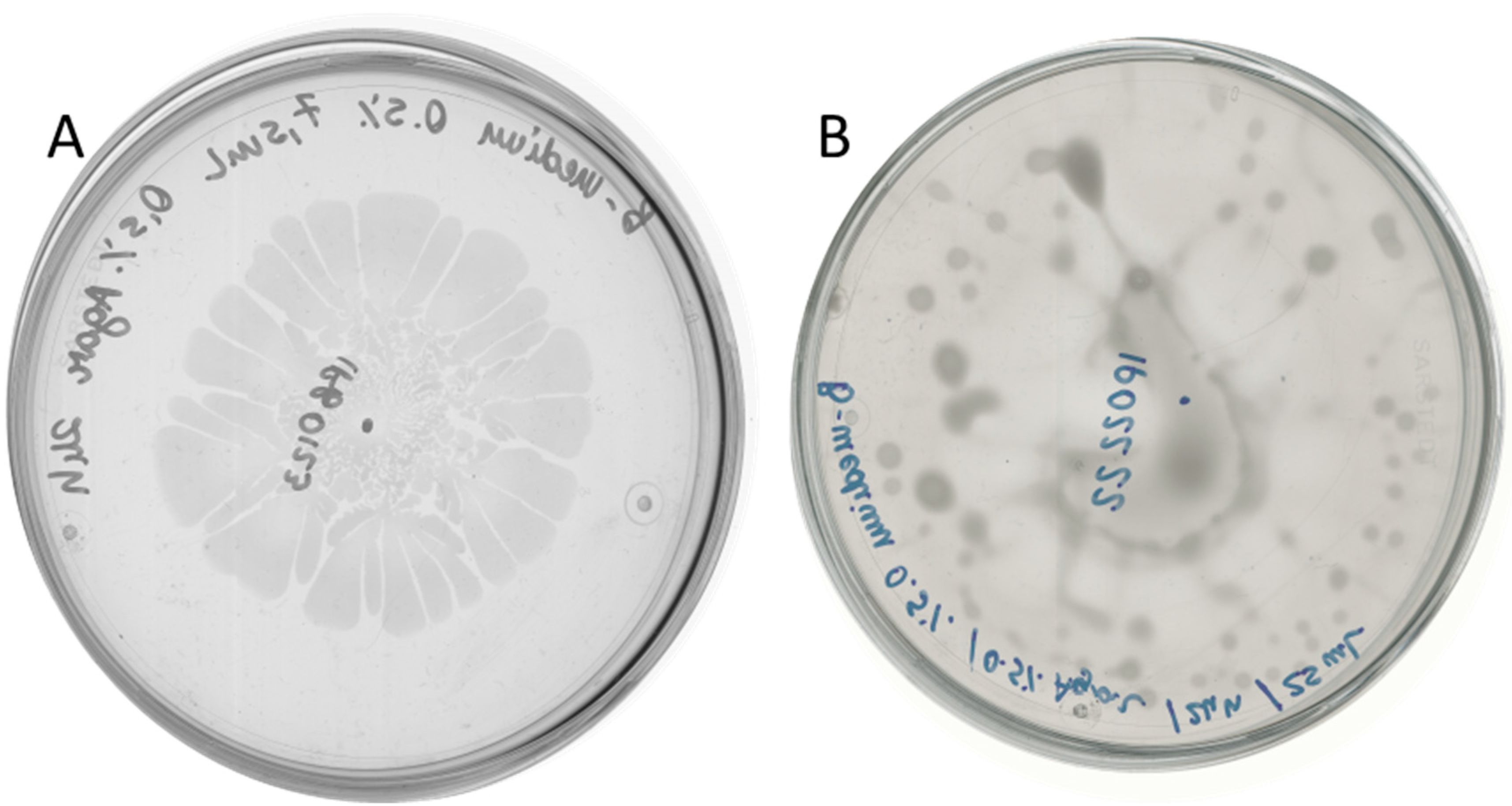
2.2. D. solani Strains IFB0102 and IPO2222 Exhibit Different Swarming Properties on Semi-Solid Medium
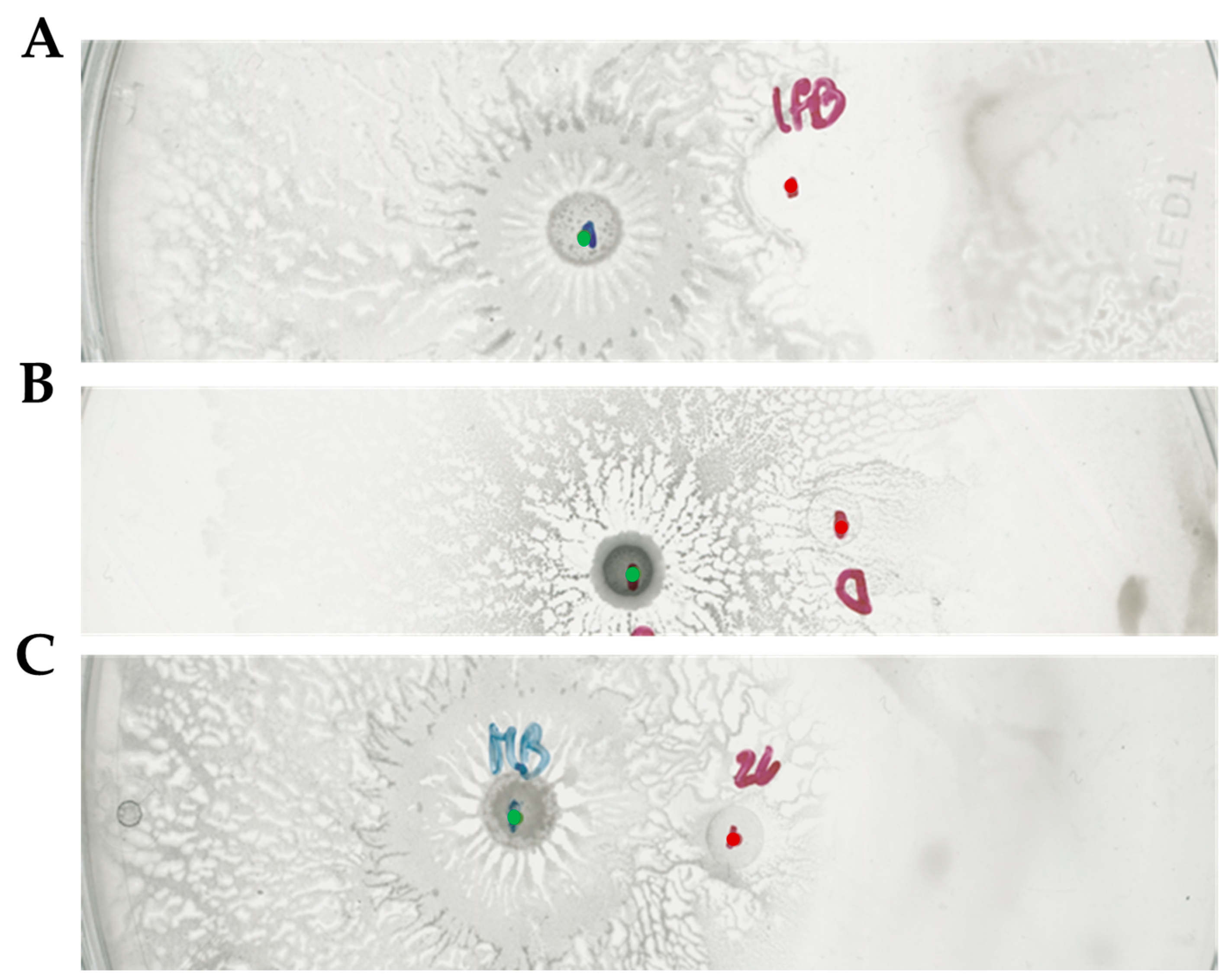
2.3. Bacillus subtilis and Dickeya solani Antagonize on Semi-Solid Medium
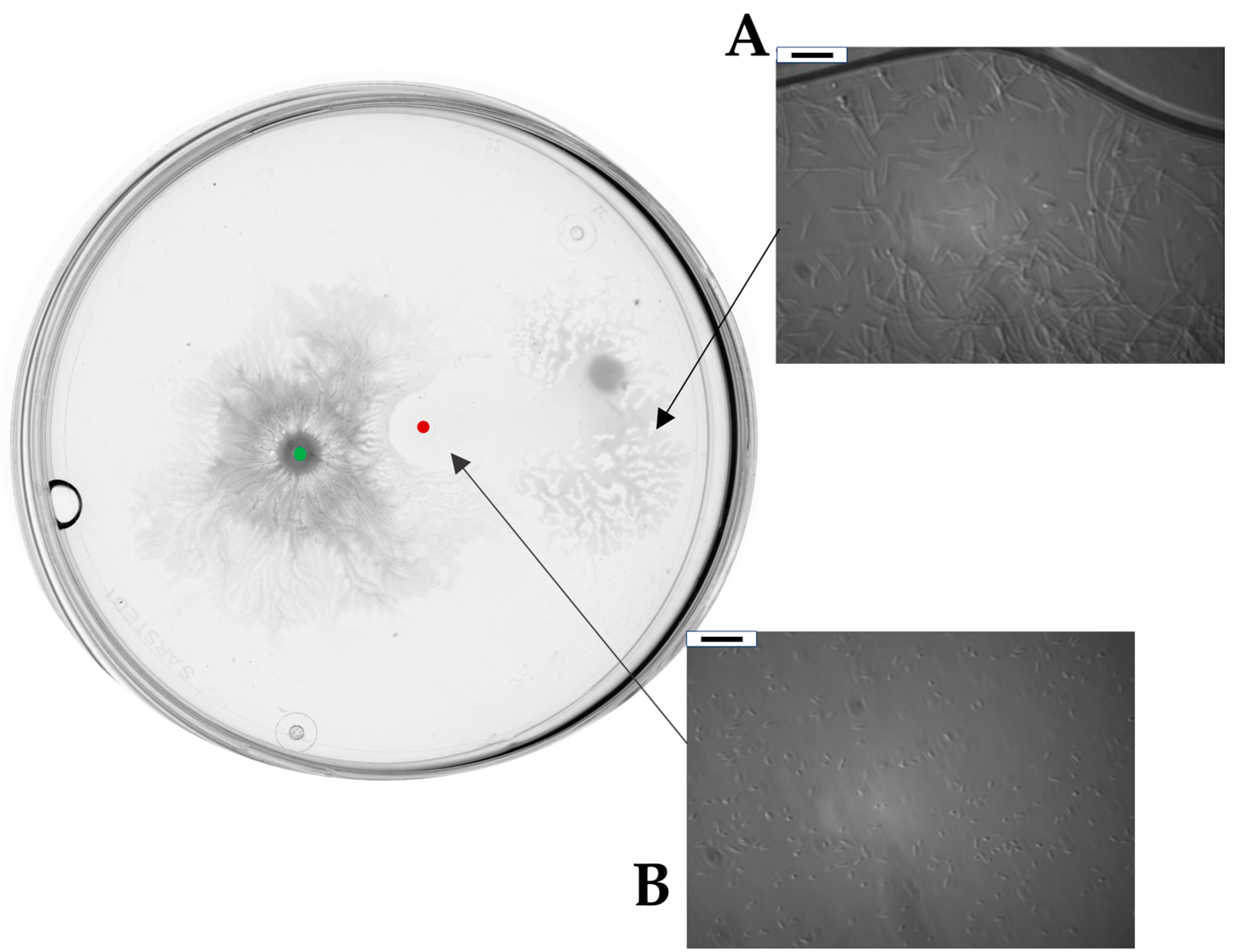
2.4. Surfactin Is Required for Directional Escape of D. solani
2.5. The sRNA ArcZ Is Not Responsible for Differences in Interaction of Investigated D. solani Strains with B. subtilis
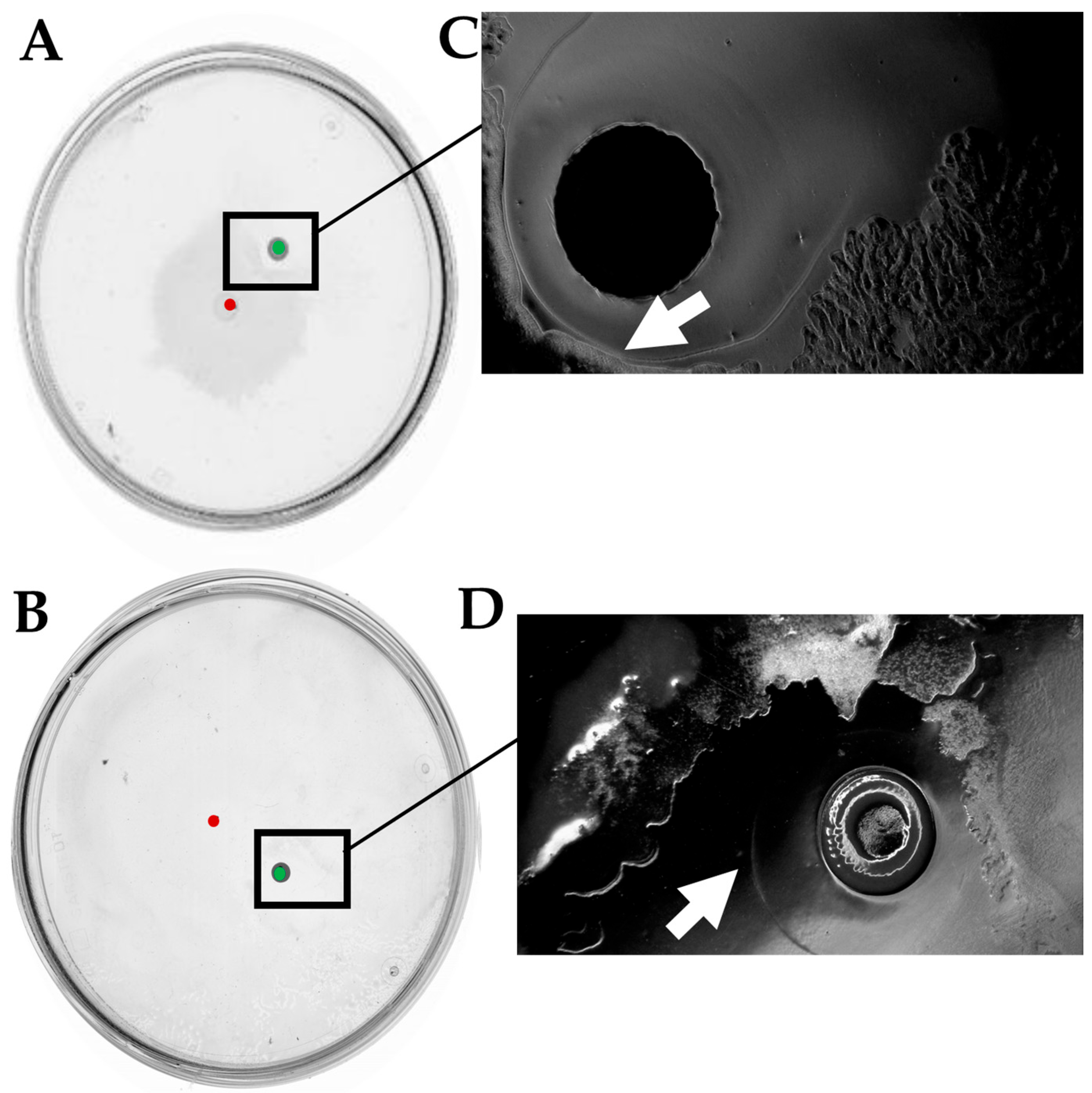
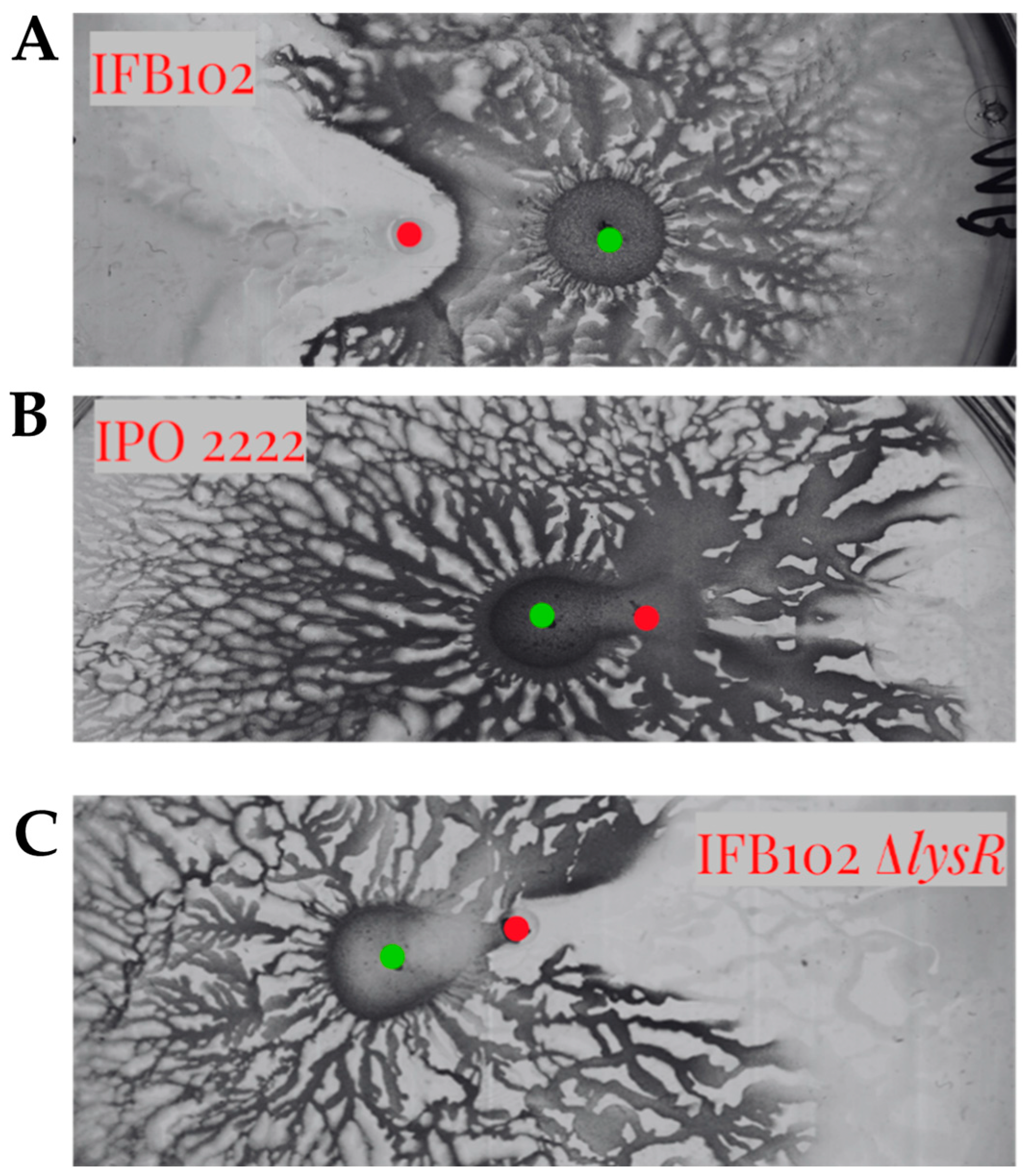
2.6. The LysR Transcriptional Regulator Is Required for Antagonism Between D. solani and B. subtilis
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Media
4.2. Screening for Antagonistic Interaction on Potato Slices
4.3. Swarming Motility
4.4. Swarming Motility Screening of Antagonistic Interaction
4.5. A Drop-Collapsing Test for Biosurfactant Production
4.6. Construction of Plasmids
4.7. B. subtilis Transformation
4.8. Preparation of D. salami Competent Cells
4.9. D. solani Electroporation
4.10. D. solani IFB0102 Genome Sequencing and Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pierce, E.C.; Dutton, R.J. Putting microbial interactions back into community contexts. Curr. Opin. Microbiol. 2022, 65, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2015, 66, 5575–5599. [Google Scholar]
- Perombelon, M.C.M.; Kelman, A. Ecology of the soft rot Erwinias. Annu. Rev. Phytopathol. 1980, 18, 361–387. [Google Scholar] [CrossRef]
- Fikowicz-Krosko, J.; Wszalek-Rozek, K.; Smolarska, A.; Czajkowski, R. First report of isolation of soft rot Pectobacterium carotovorum subsp carotovorum from symptomless bittersweet nightshade occuing in rural area of Poland. J. Plant Pathol. 2017, 99, 1. [Google Scholar]
- Charkowski, A.O. The Changing Face of Bacterial Soft-Rot Diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux-Cotte-Pattat, N.; Pédron, J.; Van Gijsegem, F. Insight into biodiversity of the recently rearranged genus Dickeya. Front. Plant Sci. 2023, 14, 1168480. [Google Scholar] [CrossRef] [PubMed]
- Toth, I.K.; van der Wolf, J.M.; Saddler, G.; Lojkowska, E.; Hélias, V.; Pirhonen, M.; Tsror, L.; Elphinstone, J.G. Dickeya species: An emerging problem for potato production in Europe. Plant Pathol. 2011, 60, 385–399. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- González, J.E.; Keshavan, N.D. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 2006, 70, 859–875. [Google Scholar] [CrossRef] [PubMed]
- des Essarts, R.Y.; Cigna, J.; Quêtu-Laurent, A.; Caron, A.; Munier, E.; Beury-Cirou, A.; Hélias, V.; Faure, D. Biocontrol of the Potato Blackleg and Soft Rot Diseases Caused by Dickeya dianthicola. Appl. Environ. Microbiol. 2015, 82, 268–278. [Google Scholar]
- Husin, N.; Sapak, Z. Bacillus cereus for Controlling Bacterial Heart Rot in Pineapple var. MD2. Trop. Life Sci. Res. 2022, 33, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Luo, J.; Ali, M.A.; Chai, R.; Shahid, M.; Ahmed, T.; Hassan, M.M.; Kadi, R.H.; An, Q.; Li, B.; et al. Synergistic Action of Biosynthesized Silver Nanoparticles and Culture Supernatant of Bacillus amyloliquefacience against the Soft Rot Pathogen Dickeya dadantii. Plants 2023, 12, 1817. [Google Scholar] [CrossRef] [PubMed]
- Krzyżanowska, D.; Obuchowski, M.; Bikowski, M.; Rychłowski, M.; Jafra, S. Colonization of Potato Rhizosphere by GFP-Tagged Bacillus subtilis MB73/2, Pseudomonas sp. P482 and Ochrobactrum sp. A44 Shown on Large Sections of Roots Using Enrichment Sample Preparation and Confocal Laser Scanning Microscopy. Sensors 2012, 12, 17608–17619. [Google Scholar] [CrossRef] [PubMed]
- Krzyżanowska, D.M.; Iwanicki, A.; Ossowicki, A.; Obuchowski, M.; Jafra, S. Genome Sequence of Bacillus subtilis MB73/2, a Soil Isolate Inhibiting the Growth of Plant Pathogens Dickeya spp. and Rhizoctonia solani. Genome Announc. 2013, 1, e00238-13. [Google Scholar] [CrossRef] [PubMed]
- Tsror (Lahkim), L.; Erlich, O.; Lebiush, S.; Hazanovsky, M.; Zig, U.; Slawiak, M.; Grabe, G.; van der Wolf, J.M.; van de Haar, J.J. Assessment of recent outbreaks of Dickeya sp. (syn. Erwinia chrysanthemi) slow wilt in potato crops in Israel. Eur. J. Plant Pathol. 2009, 123, 311–320. [Google Scholar] [CrossRef]
- Gatta, R.; Wiese, A.; Iwanicki, A.; Obuchowski, M. Influence of glucose on swarming and quorum sensing of Dickeya solani. PLoS ONE 2022, 17, e0263124. [Google Scholar] [CrossRef] [PubMed]
- Reuter, K.; Mofid, M.R.; Marahiel, M.A.; Ficner, R. Crystal structure of the surfactin synthetase-activating enzyme sfp: A prototype of the 4′-phosphopantetheinyl transferase superfamily. EMBO J. 1999, 18, 6823–6831. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.K.; Collins-Thompson, D.L.; Lee, H.; Trevors, J.T. A drop-collapsing test for screening surfactant-producing microorganisms. J. Microbiol. Methods 1991, 13, 271–279. [Google Scholar] [CrossRef]
- Julkowska, D.; Obuchowski, M.; Holland, I.B.; Séror, S.J. Comparative analysis of the development of swarming communities of Bacillus subtilis 168 and a natural wild type: Critical effects of surfactin and the composition of the medium. J. Bacteriol. 2005, 187, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Brual, T.; Effantin, G.; Baltenneck, J.; Attaiech, L.; Grosbois, C.; Royer, M.; Cigna, J.; Faure, D.; Hugouvieux-Cotte-Pattat, N.; Gueguen, E. A natural single nucleotide mutation in the small regulatory RNA ArcZ of Dickeya solani switches off the antimicrobial activities against yeast and bacteria. PLoS Genet. 2023, 19, e1010725. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology (Reading) 2008, 154, 3609–3623. [Google Scholar] [CrossRef] [PubMed]
- Demeester, W.; De Paepe, B.; De Mey, M. Fundamentals and Exceptions of the LysR-type Transcriptional Regulators. ACS Synth. Biol. 2024, 13, 3069–3092. [Google Scholar] [CrossRef] [PubMed]
- van der Wolf, J.; Boer, S.; Czajkowski, R.; Cahill, G.; Van Gijsegem, F.; Davey, T.; Dupuis, B.; Ellicott, J.; Jafra, S.; Kooman, M.; et al. Management of Diseases Caused by Pectobacterium and Dickeya Species. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Van Gijsegem, F., van der Wolf, J.M., Toth, I.K., Eds.; Springer: Cham, Switzerland, 2021; pp. 175–214. [Google Scholar]
- Borriss, R. Bacillus, a plant-beneficial bacterium. In Principles of Plant-Microbe Interactions; Lugtenberg, B., Ed.; Springer International Publishing: Leiden, The Netherlands, 2015; pp. 379–391. [Google Scholar]
- Motyka-Pomagruk, A.; Babinska-Wensierska, W.; Sledz, W.; Kaczorowska, A.K.; Lojkowska, E. Phyloproteomic study by MALDI-TOF MS in view of intraspecies variation in a significant homogenous phytopathogen Dickeya solani. Sci. Rep. 2023, 13, 18863. [Google Scholar] [CrossRef] [PubMed]
- Golanowska, M.; Potrykus, M.; Motyka-Pomagruk, A.; Kabza, M.; Bacci, G.; Galardini, M.; Bazzicalupo, M.; Makalowska, I.; Smalla, K.; Mengoni, A.; et al. Comparison of Highly and Weakly Virulent Dickeya solani Strains, With a View on the Pangenome and Panregulon of This Species. Front. Microbiol. 2018, 9, 1940. [Google Scholar] [CrossRef] [PubMed]
- Hérault, E.; Reverchon, S.; Nasser, W. Role of the LysR-type transcriptional regulator PecT and DNA supercoiling in the thermoregulation of pel genes, the major virulence factors in Dickeya dadantii. Environ. Microbiol. 2014, 16, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.K.; Pamma, R.; Hernday, A.D.; Bickham, J.E.; Braaten, B.A.; Low, D.A. Contact-dependent inhibition of growth in Escherichia coli. Science 2005, 309, 1245–1248. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.K.; Webb, J.S.; Braaten, B.A.; Low, D.A. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J. Bacteriol. 2009, 191, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.K.; Diner, E.J.; de Roodenbeke, C.T.; Burgess, B.R.; Poole, S.J.; Braaten, B.A.; Jones, A.M.; Webb, J.S.; Hayes, C.S.; Cotter, P.A.; et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 2010, 468, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Ham, J.H.; Schechter, L.M.; Kim, J.F.; Beer, S.V.; Collmer, A. The Erwinia chrysanthemi EC16 hrp/hrc gene cluster encodes an active Hrp type III secretion system that is flanked by virulence genes functionally unrelated to the Hrp system. Mol. Plant Microbe Interact. 2004, 17, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Potrykus, M.; Golanowska, M.; Hugouvieux-Cotte-Pattat, N.; Lojkowska, E. Regulators involved in Dickeya solani virulence, genetic conservation, and functional variability. Mol. Plant Microbe Interact. 2014, 27, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Hommais, F.; Oger-Desfeux, C.; Van Gijsegem, F.; Castang, S.; Ligori, S.; Expert, D.; Nasser, W.; Reverchon, S. PecS is a global regulator of the symptomatic phase in the phytopathogenic bacterium Erwinia chrysanthemi 3937. J. Bacteriol. 2008, 190, 7508–7522. [Google Scholar] [CrossRef] [PubMed]
- Mhedbi-Hajri, N.; Malfatti, P.; Pédron, J.; Gaubert, S.; Reverchon, S.; Van Gijsegem, F. PecS is an important player in the regulatory network governing the coordinated expression of virulence genes during the interaction between Dickeya dadantii 3937 and plants. Environ. Microbiol. 2011, 13, 2901–2914. [Google Scholar] [CrossRef] [PubMed]
- Budnick, J.A.; Sheehan, L.M.; Ginder, M.J.; Failor, K.C.; Perkowski, J.M.; Pinto, J.F.; Kohl, K.A.; Kang, L.; Michalak, P.; Luo, L.; et al. A central role for the transcriptional regulator VtlR in small RNA-mediated gene regulation in Agrobacterium tumefaciens. Sci. Rep. 2020, 10, 14968. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, J.; Kraus, A.; Ronge, C.; Jagst, M.; Brandenburg, V.B.; Narberhaus, F. A LysR-type transcriptional regulator controls the expression of numerous small RNAs in Agrobacterium tumefaciens. Mol. Microbiol. 2021, 116, 126–139. [Google Scholar] [CrossRef] [PubMed]
- García-Tomsig, N.I.; Robledo, M.; DiCenzo, G.C.; Mengoni, A.; Millán, V.; Peregrina, A.; Uceta, A.; Jiménez-Zurdo, J.I. Pervasive RNA Regulation of Metabolism Enhances the Root Colonization Ability of Nitrogen-Fixing Symbiotic α-Rhizobia. mBio 2022, 13, e03576-21. [Google Scholar] [CrossRef] [PubMed]
- Heroven, A.K.; Dersch, P. RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol. Microbiol. 2006, 62, 1469–1483. [Google Scholar] [CrossRef] [PubMed]
- Rodionova, I.A.; Gao, Y.; Monk, J.; Hefner, Y.; Wong, N.; Szubin, R.; Lim, H.G.; Rodionov, D.A.; Zhang, Z.; Saier, M.H., Jr.; et al. A systems approach discovers the role and characteristics of seven LysR type transcription factors in Escherichia coli. Sci. Rep. 2022, 12, 7274. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zeng, Q.; Khokhani, D.; Tian, F.; Severin, G.B.; Waters, C.M.; Xu, J.; Zhou, X.; Sundin, G.W.; Ibekwe, A.M.; et al. A feed-forward signalling circuit controls bacterial virulence through linking cyclic di-GMP and two mechanistically distinct sRNAs, ArcZ and RsmB. Environ. Microbiol. 2019, 21, 2755–2771. [Google Scholar] [CrossRef] [PubMed]
- Watkins, D.; Arya, D. Models of Hfq interactions with small non-coding RNA in Gram-negative and Gram-positive bacteria. Front. Cell Infect. Microbiol. 2023, 13, 1282258. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-H.; Gavilanes-Ruiz, M.; Okinaka, Y.; Vedel, R.; Berthuy, I.; Boccara, M.; Chen, J.W.; Perna, N.T.; Keen, N.T. hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol. Plant Microbe Interact. 2002, 15, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.; Blanco, C.; Condemine, G.; Expert, D.; Franza, T.; Hayes, C.; Hugouvieux-Cotte-Pattat, N.; López Solanilla, E.; Low, D.; Moleleki, L.; et al. The role of secretion systems and small molecules in soft-rot Enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 2012, 50, 425–449. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Techniques for transformation of E. coli. In DNA Cloning: A Practical Approach; Glover, D.M., Ed.; IRL Press: Washington, DC, USA, 1985; Volume 1, pp. 109–135. [Google Scholar]
- Krzyżanowska, D.M.; Ossowicki, A.; Rajewska, M.; Maciąg, T.; Jabłońska, M.; Obuchowski, M.; Heeb, S.; Jafra, S. When Genome-Based Approach Meets the “Old but Good”: Revealing Genes Involved in the Antibacterial Activity of Pseudomonas sp. P482 against Soft Rot Pathogens. Front. Microbiol. 2016, 7, 782. [Google Scholar] [CrossRef] [PubMed]
- Anagnostopoulos, C.; Crawford, I.P. Transformation studies on the linkage of markers in the tryptophan pathway in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1961, 47, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Sławiak, M.; van Beckhoven, J.R.C.M.; Speksnijder, A.G.C.L.; Czajkowski, R.; Grabe, G.; van der Wolf, J.M. Biochemical and genetical analysis reveal a new clade of biovar 3 Dickeya spp. strains isolated from potato in Europe. Eur. J. Plant Pathol. 2009, 125, 245–261. [Google Scholar] [CrossRef]
- Vagner, V.; Dervyn, E.; Ehrlich, S.D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology (Reading) 1998, 144, 3097–3104. [Google Scholar] [CrossRef] [PubMed]
- Norrander, J.; Kempe, T.; Messing, J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 1983, 26, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Alexeyev, M.F. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram-negative bacteria. Biotechniques 1999, 26, 824–826, 828. [Google Scholar] [CrossRef] [PubMed]
- Antelmann, H.; Engelmann, S.; Schmid, R.; Sorokin, A.; Lapidus, A.; Hecker, M. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor sigmaB in Bacillus subtilis. J. Bacteriol. 1997, 179, 7251–7256. [Google Scholar] [CrossRef] [PubMed]
- Jafra, S.; Przysowa, J.; Czajkowski, R.; Michta, A.; Garbeva, P.; van der Wolf, J.M. Detection and characterization of bacteria from the potato rhizosphere degrading N-acyl-homoserine lactone. Can. J. Microbiol. 2006, 52, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; Volume 2. [Google Scholar]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]


| SNPs | Position | IPO2222 | IFB102 | Product |
|---|---|---|---|---|
| 1 | 2530087 | T | C | ArcZ |
| 2 | 2621920 | C | T | Hypothetical protein kinase |
| 3 | 3554462 | C | A | cytochrome d terminal oxidase subunit 1 |
| 4 | 3674549 | A | G | HNH/endonuclease VII fold putative polymorphic toxin |
| 5 | 4039850 | G | A | intergenic region |
| 6 | 4513052 | A | G | Hypothetical protein |
| 7 | 4635450 | C | T | transcriptional regulator-LysR family |
| Strain | Relevant Characteristics | Reference or Source |
|---|---|---|
| E. coli | ||
| DH5α | F-gyrA96 recA1 relA1 endA1 thi-1 hsdR17 glnV44 deoR D (lacZYA-argF) U169[f80dD(lacZ)M15] | [45] |
| B. subtilis | ||
| MB73/2 | Natural isolate | [46] |
| MB73/2 sfp− | sfp::pMutin4 | This work |
| 168 | trpC2 | [47] |
| 168 sfp+ | trpC2 amyE::sfp | [19] |
| D. solani | ||
| IFB0102 | Natural isolate | [48] |
| IFB0102 lysR | lysR::pMutin4 | This work |
| IPO2222 | Natural isolate | [15] |
| D s0432-1 | Natural isolate | [20] |
| Plasmid | ||
| pMutin4 | Integration vector used for gene inactivation; Ampr Eryr | [49] |
| pMutin-sfp | pMutin4 derivative carrying a PCR product internal to sfp | This work |
| pUC19 | Cloning vector; Ampr | [50] |
| pKNOCK-Gm | Suicide vector for gene knockout; Gmr | [51] |
| pUC-ΔlysR | pUC derivative carrying a gentamycin resistance cassette flanked by PCR products of upstream and downstream regions of lysR gene | This work |
| Amplified Gene | Name | Sequence |
|---|---|---|
| sfp | sfp-F sfp-R | attaGGATCCACGGTTCATGTCTTTCATATC attaGTCGACGATATAGCATGGGGAATGG |
| gentamycin | Gent-F Gent-R | gagaggattcgagAGGACGCGTCAATTCTCG gtattacaaggctTAACAGATGAGGGCAAGC |
| LysR-Left | LysRleft-F LysReft-R | cggccagtgaattcgagctcggtacAAGCTTGTTTCGGTGTTGttgacgcgtcctCTCGAATCCTCTCGTATTATTTTC |
| LysR-Right | LysRright-F LysRright-R | cctcatctgttaAGCCTTGTAATACGGTCC gcatgcctgcaggtcgactctagagCTATTCTAATTCGTTCCGTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gatta, R.; Iwanicki, A.; Czajkowski, R.; Obuchowski, M. Antagonistic Interactions Between Dickeya solani and Bacillus subtilis. Int. J. Mol. Sci. 2025, 26, 7193. https://doi.org/10.3390/ijms26157193
Gatta R, Iwanicki A, Czajkowski R, Obuchowski M. Antagonistic Interactions Between Dickeya solani and Bacillus subtilis. International Journal of Molecular Sciences. 2025; 26(15):7193. https://doi.org/10.3390/ijms26157193
Chicago/Turabian StyleGatta, Roberta, Adam Iwanicki, Robert Czajkowski, and Michał Obuchowski. 2025. "Antagonistic Interactions Between Dickeya solani and Bacillus subtilis" International Journal of Molecular Sciences 26, no. 15: 7193. https://doi.org/10.3390/ijms26157193
APA StyleGatta, R., Iwanicki, A., Czajkowski, R., & Obuchowski, M. (2025). Antagonistic Interactions Between Dickeya solani and Bacillus subtilis. International Journal of Molecular Sciences, 26(15), 7193. https://doi.org/10.3390/ijms26157193







