Extracellular Vesicles-Induced Cell Homing and Odontogenesis via microRNA Signaling for Dentin Regeneration
Abstract
1. Introduction
2. Results
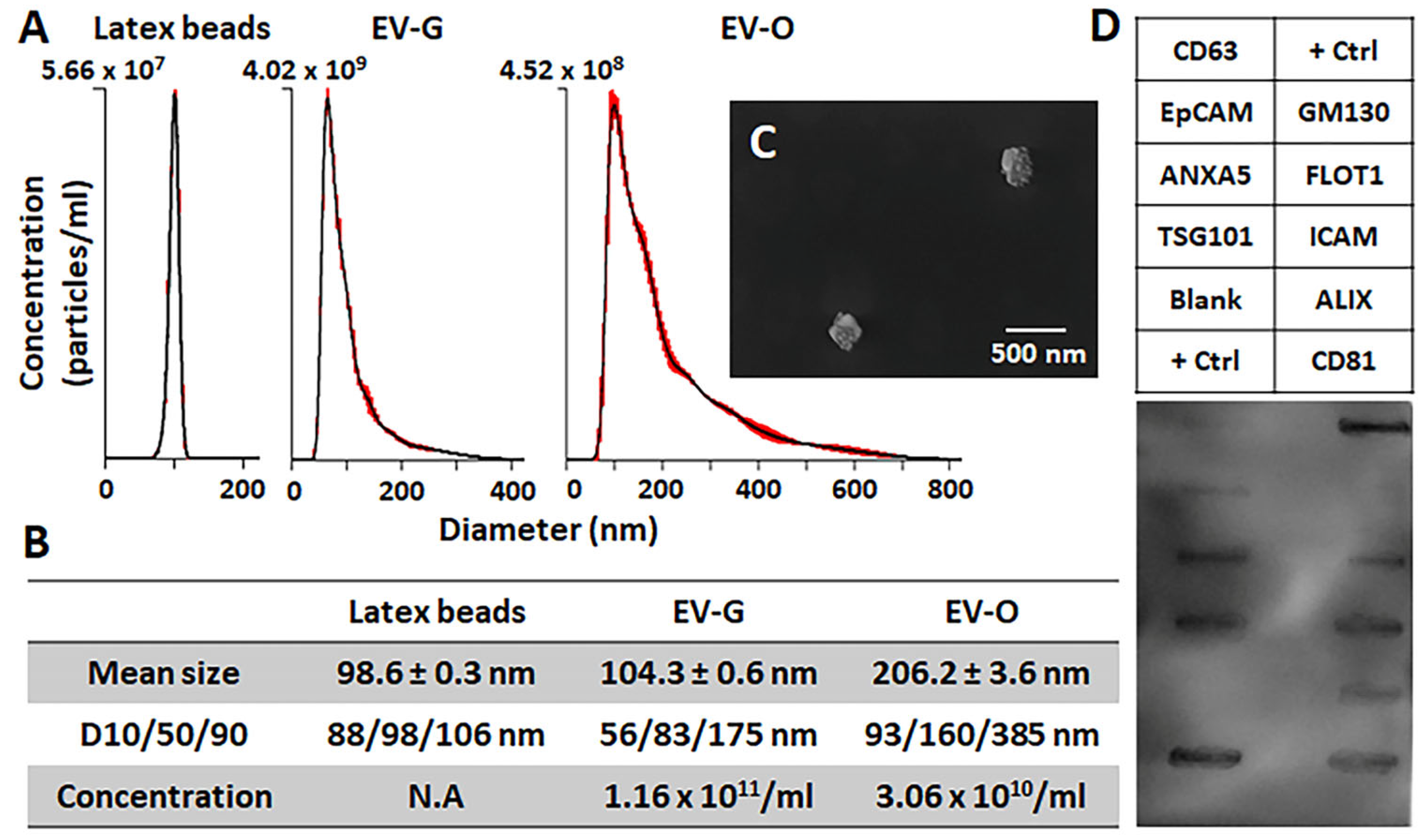
2.1. Characterization of DPSC-EVs
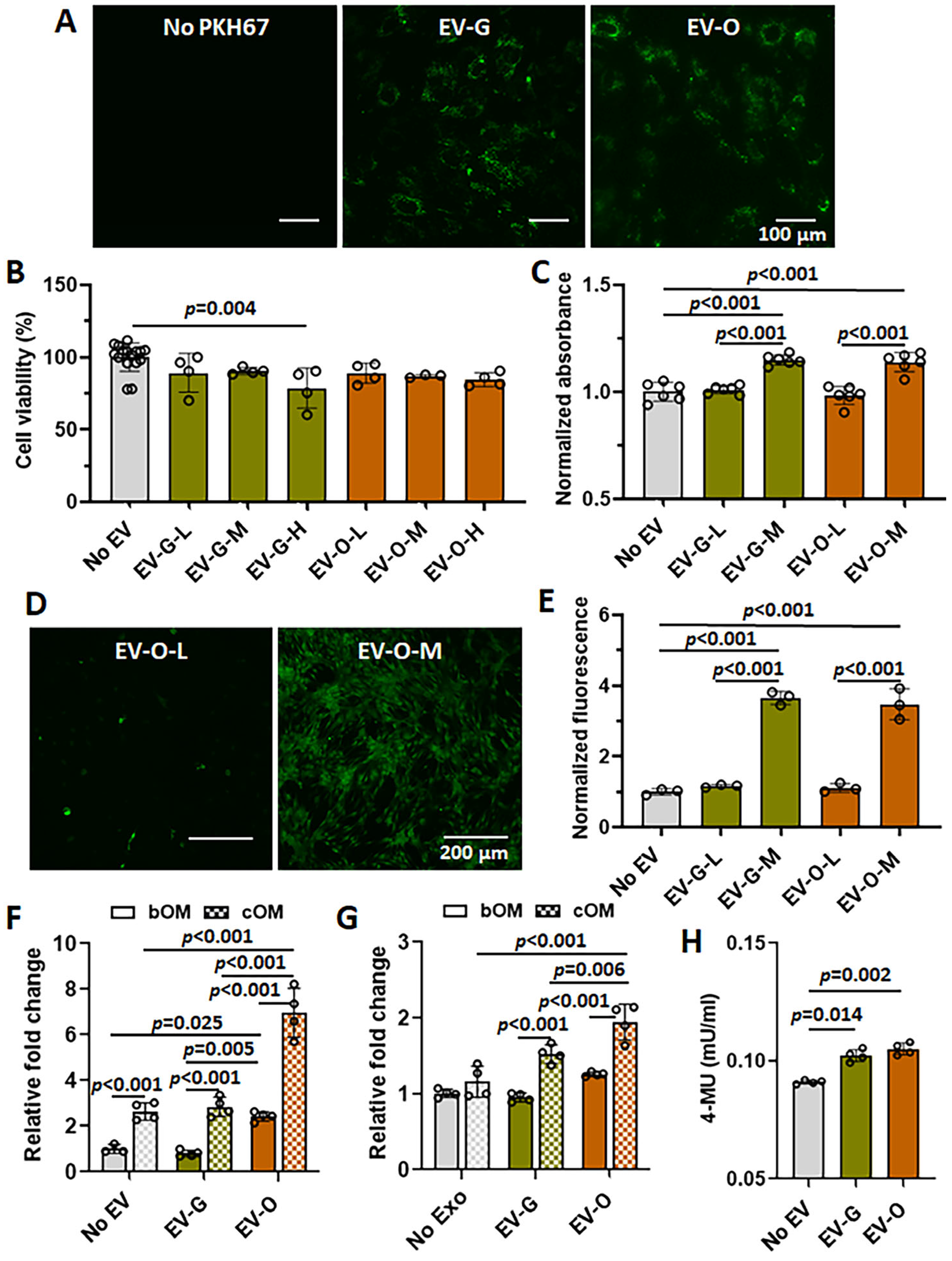
2.2. In Vitro Efficacies of EV-O on Cell Viability, Proliferation, Chemotaxis, and Odontogenesis
2.3. miRNA Profiles
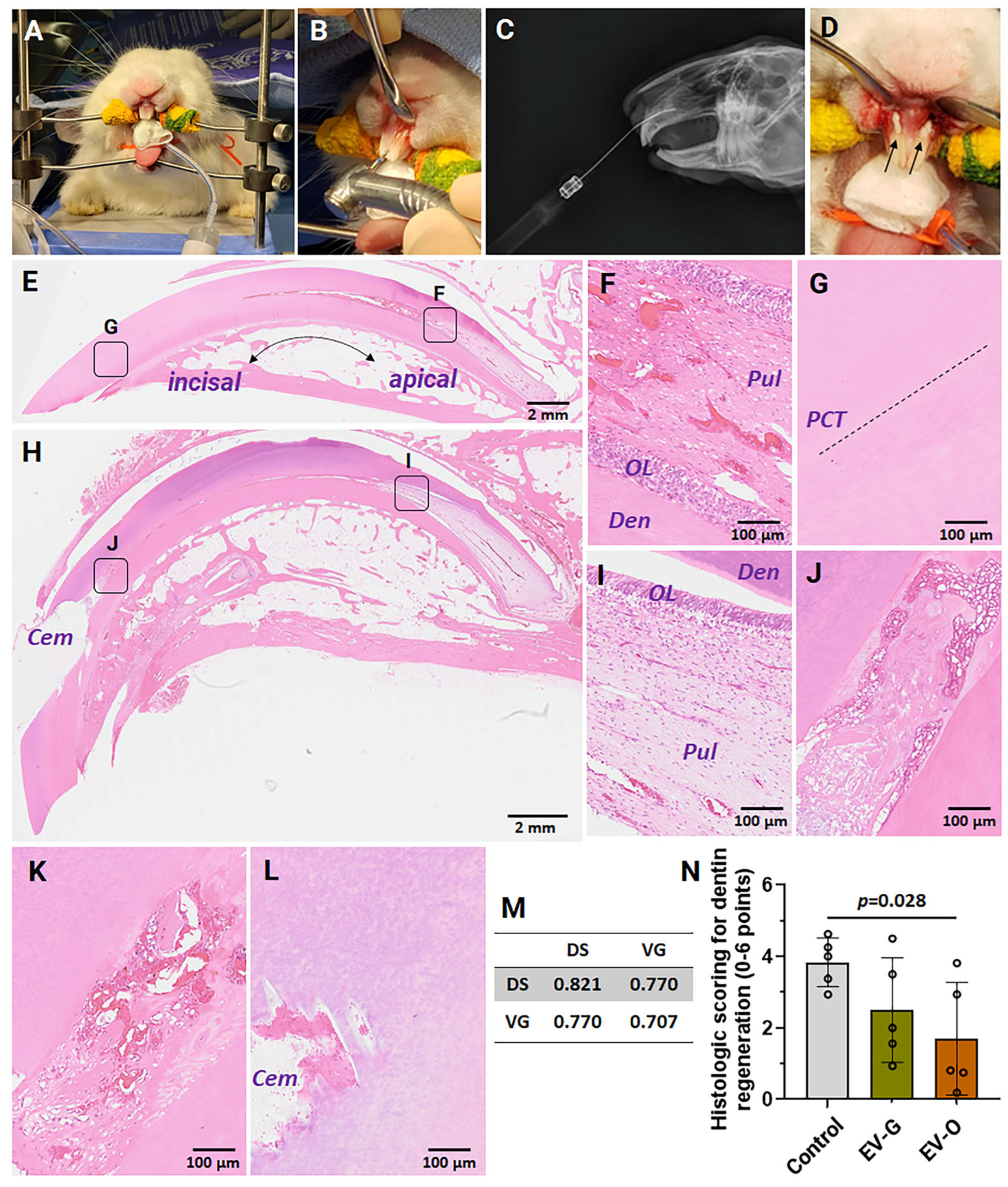
2.4. In Vivo Efficacies
3. Discussion
4. Materials and Methods
4.1. Isolation of DPSCs
4.2. Isolation and Characterization of DPSC-EVs
4.3. Cell Viability, Proliferation, and Chemotaxis
4.4. Odontogenesis
4.5. Next-Generation Sequencing (NGS)
4.6. Rabbit Partial Dentinotomy/Pulpotomy Model
4.7. Histological Evaluation of Dentinogenesis
4.8. Statistics
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linde, A.; Goldberg, M. Dentinogenesis. Crit. Rev. Oral Biol. Med. 1993, 4, 679–728. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Azaryan, E.; Razavi, F.E.; Hanafi-Bojd, M.Y.; Alemzadeh, E.; Naseri, M. Dentin regeneration based on tooth tissue engineering: A review. Biotechnol. Prog. 2023, 39, e3319. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, Y.S.; Shon, W.J.; Park, J.C. Physiologic dentin regeneration: Its past, present, and future perspectives. Front. Physiol. 2023, 14, 1313927. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.C.; Yuan, T.; Zhang, Y.L.; Yin, W.J.; Guo, S.C.; Zhang, C.Q. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics 2017, 7, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Tetta, C.; Consiglio, A.L.; Bruno, S.; Tetta, E.; Gatti, E.; Dobreva, M.; Cremonesi, F.; Camussi, G. The role of microvesicles derived from mesenchymal stem cells in tissue regeneration; a dream for tendon repair? Muscles Ligaments Tendons J. 2012, 2, 212–221. [Google Scholar] [PubMed]
- Zhang, B.; Wang, M.; Gong, A.; Zhang, X.; Wu, X.; Zhu, Y.; Shi, H.; Wu, L.; Zhu, W.; Qian, H.; et al. HucMSC-Exosome Mediated-Wnt4 Signaling Is Required for Cutaneous Wound Healing. Stem Cells 2015, 33, 2158–2168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chu, W.C.; Lai, R.C.; Lim, S.K.; Hui, J.H.; Toh, W.S. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthr. Cartil. 2016, 24, 2135–2140. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhong, Y.; Kong, Y.; Chen, Y.; Feng, J.; Zheng, J. Lineage-specific exosomes promote the odontogenic differentiation of human dental pulp stem cells (DPSCs) through TGFβ1/smads signaling pathway via transfer of microRNAs. Stem Cell Res. Ther. 2019, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, V.; Seol, D.; Gomez-Contreras, P.C.; Keen, H.L.; Shin, K.; Martin, J.A. Exosome-Based Cell Homing and Angiogenic Differentiation for Dental Pulp Regeneration. Int. J. Mol. Sci. 2022, 24, 466. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Heair, H.M.; Kemper, A.G.; Roy, B.; Lopes, H.B.; Rashid, H.; Clarke, J.C.; Afreen, L.K.; Ferraz, E.P.; Kim, E.; Javed, A.; et al. MicroRNA 665 Regulates Dentinogenesis through MicroRNA-Mediated Silencing and Epigenetic Mechanisms. Mol. Cell. Biol. 2015, 35, 3116–3130. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xu, X.; Gao, S.; Huo, S.; Wan, M.; Zhou, X.; Zhou, X.; Zheng, L.; Zhou, Y. MicroRNA-93-5p regulates odontogenic differentiation and dentin formation via KDM6B. J. Transl. Med. 2024, 22, 54. [Google Scholar] [CrossRef] [PubMed]
- Coughlan, C.; Bruce, K.D.; Burgy, O.; Boyd, T.D.; Michel, C.R.; Garcia-Perez, J.E.; Adame, V.; Anton, P.; Bettcher, B.M.; Chial, H.J.; et al. Exosome Isolation by Ultracentrifugation and Precipitation and Techniques for Downstream Analyses. Curr. Protoc. Cell Biol. 2020, 88, e110. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Sędzik, M.; Rakoczy, K.; Sleziak, J.; Kisiel, M.; Kraska, K.; Rubin, J.; Łuniewska, W.; Choromańska, A. Comparative Analysis of Exosomes and Extracellular Microvesicles in Healing Pathways: Insights for Advancing Regenerative Therapies. Molecules 2024, 29, 3681. [Google Scholar] [CrossRef] [PubMed]
- Song, I.; Glass Nas, E.A.; Martin, J.A.; Hlas, A.; Hall, M.M.; Kruse, R.C.; Seol, D.; Duchman, K.R.; Buckwalter, J.A. Exosome Size in Platelet-Rich Plasma is Associated with Effectiveness in Patients Treated for Knee Osteoarthritis. Orthop. J. Sports Med. 2025. [Google Scholar]
- Ishimatsu, H.; Kitamura, C.; Morotomi, T.; Tabata, Y.; Nishihara, T.; Chen, K.K.; Terashita, M. Formation of dentinal bridge on surface of regenerated dental pulp in dentin defects by controlled release of fibroblast growth factor-2 from gelatin hydrogels. J. Endod. 2009, 35, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Lee, C.H.; Chen, M.; Zhao, W.; Fu, S.Y.; Qi, J.J.; Chotkowski, G.; Eisig, S.B.; Wong, A.; Mao, J.J. Induced migration of dental pulp stem cells for in vivo pulp regeneration. J. Dent. Res. 2011, 90, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Mattuella, L.G.; de Figueiredo, J.A.P.; Nor, J.E.; de Araujo, F.B.; Fossati, A.C.M. Vascular endothelial growth factor receptor-2 expression in the pulp of human primary and young permanent teeth. J. Endod. 2007, 33, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, K.; Motani, R.; Sakuta, T.; Yamaguchi, N.; Koga, T.; Matsuo, K.; Nagaoka, S.; Abeyama, K.; Maruyama, I.; Torii, M. The role of vascular endothelial growth factor in human dental pulp cells: Induction of chemotaxis, proliferation, and differentiation and activation of the AP-1-dependent signaling pathway. J. Dent. Res. 2000, 79, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, N.; Hayashi, Y.; Murakami, M.; Alvarez, F.J.; Horibe, H.; Iohara, K.; Nakata, K.; Nakamura, H.; Nakashima, M. Similar in vitro effects and pulp regeneration in ectopic tooth transplantation by basic fibroblast growth factor and granulocyte-colony stimulating factor. Oral Dis. 2015, 21, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, G.M.; Abouauf, E.A.; AbuBakr, N.; Fouad, A.M.; Dörfer, C.E.; El-Sayed, K.M.F. Cell-Based Transplantation versus Cell Homing Approaches for Pulp-Dentin Complex Regeneration. Stem Cells Int. 2021, 2021, 8483668. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.H.; Parent, C.A.; Weaver, A.M. Extracellular vesicles: Critical players during cell migration. Dev. Cell 2021, 56, 1861–1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Peng, Y.; Xue, H.; Liu, G.; Wang, N.; Shao, Z. Correction: MiR-21 regulating PVT1/PTEN/IL-17 axis towards the treatment of infectious diabetic wound healing by modified GO-derived biomaterial in mouse models. J. Nanobiotechnol. 2023, 21, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Luan, S.; Chen, J.; Zhou, Y.; Wang, T.; Li, Z.; Fu, Y.; Zhai, A.; Bi, C. The MSC-Derived Exosomal lncRNA H19 Promotes Wound Healing in Diabetic Foot Ulcers by Upregulating PTEN via MicroRNA-152-3p. Mol. Ther. Nucleic Acids 2020, 19, 814–826. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Deng, J.; Chen, Y.; Wang, Y.; Liu, B.; Liu, J. Engineered Human Adipose Stem-Cell-Derived Exosomes Loaded with miR-21-5p to Promote Diabetic Cutaneous Wound Healing. Mol. Pharm. 2020, 17, 1723–1733. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Xu, J.; Huang, P.; Bai, Y.; Chen, H.; Xu, X.; Hu, Y.; Liu, J.; Zhang, H. miR-9-5p and miR-221-3p Promote Human Mesenchymal Stem Cells to Alleviate Carbon Tetrachloride-Induced Liver Injury by Enhancing Human Mesenchymal Stem Cell Engraftment and Inhibiting Hepatic Stellate Cell Activation. Int. J. Mol. Sci. 2024, 25, 7235. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wang, Q.; Zhang, J.; Sun, L.; Hong, X.; Du, W.; Duan, R.; Jiang, J.; Ji, Y.; Wang, H.; et al. Exosomes derived from mir-214-3p overexpressing mesenchymal stem cells promote myocardial repair. Biomater. Res. 2023, 27, 77. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kim, T.; Choi, G.E.; Park, A.; Yoon, J.; Yu, J.; Suh, N. miR-29a-3p orchestrates key signaling pathways for enhanced migration of human mesenchymal stem cells. Cell Commun. Signal. 2024, 22, 365. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, R.; Huang, C.C.; Ravindran, S. Hijacking the cellular mail: Exosome mediated differentiation of mesenchymal stem cells. Stem Cells Int. 2016, 2016, 3808674. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, N.J.; Lutz, C.S. miR-708 Negatively Regulates TNFα/IL-1β Signaling by Suppressing NF-κB and Arachidonic Acid Pathways. Mediat. Inflamm. 2021, 2021, 5595520. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Liu, S.; Zou, C.; Ai, Y. Correction: miR-708-3p targetedly regulates LSD1 to promote osteoblast differentiation of hPDLSCs in periodontitis. Odontology 2025, 113, 231. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, S.; Bian, C.; Yang, Z.; Zhou, H.; Zeng, Y.; Li, H.; Han, Q.; Zhao, R.C. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012, 21, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Wang, G.; Hu, C.; Shi, Y.; Liao, L.; Shi, S.; Cai, Y.; Cheng, S.; Wang, X.; Liu, Y.; et al. Tumor necrosis factor α suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J. Bone Miner. Res. 2013, 28, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yun, H.J.; Elkin, K.; Guo, Y.; Ding, Y.; Li, G. MicroRNA-29b Suppresses Inflammation and Protects Blood-Brain Barrier Integrity in Ischemic Stroke. Mediat. Inflamm. 2022, 2022, 1755416. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Gao, J.; Gao, F.; Zhao, Y.; Deng, B.; Mu, X.; Xu, L. Extracellular vesicles-encapsulated microRNA-29b-3p from bone marrow-derived mesenchymal stem cells promotes fracture healing via modulation of the PTEN/PI3K/AKT axis. Exp. Cell Res. 2022, 412, 113026. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Wang, Y.; Gao, J.; Yan, Z.; Li, Z.; Zou, X.; Li, Y.; Wang, J.; Guo, Y. miR-29b-3p regulated osteoblast differentiation via regulating IGF-1 secretion of mechanically stimulated osteocytes. Cell. Mol. Biol. Lett. 2019, 24, 11. [Google Scholar] [CrossRef] [PubMed]
- Likitpongpipat, N.; Sangmaneedet, S.; Klanrit, P.; Noisombut, R.; Krisanaprakornkit, S.; Chailertvanitkul, P. Promotion of Dental Pulp Wound Healing in New Zealand White Rabbits’ Teeth by Thai Propolis Product. J. Veter. Dent. 2019, 36, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Wyss, F.; Müller, J.; Clauss, M.; Kircher, P.; Geyer, H.; von Rechenberg, B.; Hatt, J.M. Measuring Rabbit (Oryctolagus cuniculus) Tooth Growth and Eruption by Fluorescence Markers and Bur Marks. J. Veter. Dent. 2016, 33, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, V.; He, R.; Keen, H.L.; Salem, A.K.; Sander, E.A.; Shin, K.; Martin, J.A.; Seol, D. Profiles of Exosomal microRNAs in Joint Cells and Candidate microRNAs for Cartilage Regeneration. Tissue Eng. Part A 2025. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.W.; Kim, J.Y.; Kim, M.J.; Kim, G.H.; Yi, J.K.; Lee, D.W.; Kum, K.Y.; Kim, E.C. Combined effects of mineral trioxide aggregate and human placental extract on rat pulp tissue and growth, differentiation and angiogenesis in human dental pulp cells. Acta Odontol. Scand. 2016, 74, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Chevallier, M.; Guerret, S.; Chossegros, P.; Gerard, F.; Grimaud, J.A. A histological semiquantitative scoring system for evaluation of hepatic fibrosis in needle liver biopsy specimens: Comparison with morphometric studies. Hepatology 1994, 20, 349–355. [Google Scholar] [CrossRef] [PubMed]
| EV-G | EV-O | ||||
|---|---|---|---|---|---|
| miRNA | Reads | % | miRNA | Reads | % |
| ocu-miR-146a-5p | 296,149.58 | 13.32 | ocu-miR-21-5p | 320,322.82 | 17.51 |
| ocu-miR-199a-3p | 217,998.94 | 9.80 | ocu-miR-199a-3p | 197,071.34 | 10.77 |
| ocu-miR-122-5p | 190,513.25 | 8.57 | ocu-miR-221-3p | 188,002.21 | 10.28 |
| ocu-miR-221-3p | 185,003.32 | 8.32 | ocu-miR-24-3p | 90,800.24 | 4.96 |
| ocu-miR-143-3p | 143,493.52 | 6.45 | ocu-miR-214-3p | 86,211.23 | 4.71 |
| ocu-miR-24-3p | 137,071.09 | 6.16 | ocu-miR-23b-3p | 70,641.32 | 3.86 |
| ocu-miR-21-5p | 122,765.06 | 5.52 | ocu-miR-92a-3p | 67,659.80 | 3.70 |
| ocu-miR-92a-3p | 61,986.35 | 2.79 | ocu-miR-29a-3p | 65,430.40 | 3.58 |
| ocu-miR-23b-3p | 49,174.99 | 2.21 | ocu-miR-143-3p | 63,079.22 | 3.45 |
| ocu-miR-27b-3p | 47,342.73 | 2.13 | ocu-miR-22-3p | 58,819.72 | 3.22 |
| ocu-miR-214-3p | 42,716.43 | 1.92 | ocu-miR-27b-3p | 47,713.49 | 2.61 |
| ocu-let-7a-5p | 38,945.78 | 1.75 | ocu-miR-125b-5p | 38,903.85 | 2.13 |
| ocu-miR-16b-5p | 38,384.95 | 1.73 | ocu-miR-34a-5p | 27,086.65 | 1.48 |
| ocu-miR-378-3p | 38,061.67 | 1.71 | ocu-let-7i-5p | 24,071.41 | 1.32 |
| ocu-miR-16a-5p | 35,936.55 | 1.62 | ocu-miR-423-5p | 21,867.29 | 1.20 |
| ocu-miR-34a-5p | 34,561.72 | 1.55 | ocu-miR-16a-5p | 20,585.01 | 1.13 |
| ocu-miR-93-5p | 31,709.98 | 1.43 | ocu-miR-146a-5p | 20,118.16 | 1.10 |
| ocu-miR-152-3p | 28,732.49 | 1.29 | ocu-miR-100-5p | 19,766.88 | 1.08 |
| ocu-miR-100-5p | 27,396.54 | 1.23 | ocu-miR-152-3p | 19,489.91 | 1.07 |
| ocu-let-7b-5p | 25,971.89 | 1.17 | ocu-miR-29c-3p | 18,416.89 | 1.01 |
| ocu-miR-25-3p | 25,817.53 | 1.16 | ocu-let-7b-5p | 18,332.57 | 1.00 |
| ocu-miR-29a-3p | 23,709.67 | 1.07 | ocu-let-7a-5p | 17,266.05 | 0.94 |
| ocu-miR-423-5p | 23,685.38 | 1.07 | ocu-miR-222-3p | 17,009.25 | 0.93 |
| ocu-let-7i-5p | 23,552.89 | 1.06 | ocu-miR-423-3p | 14,179.10 | 0.78 |
| ocu-miR-22-3p | 21,947.65 | 0.99 | ocu-miR-122-5p | 13,182.98 | 0.72 |
| ocu-let-7f-5p | 21,592.76 | 0.97 | ocu-miR-30d-5p | 10,777.27 | 0.59 |
| ocu-miR-6529-5p | 17,416.41 | 0.78 | ocu-miR-30e-5p | 10,171.98 | 0.56 |
| ocu-miR-361-5p | 15,483.81 | 0.70 | ocu-miR-378-3p | 10,125.44 | 0.55 |
| ocu-miR-128a-3p | 14,697.51 | 0.66 | ocu-miR-93-5p | 9997.79 | 0.55 |
| ocu-miR-128b-3p | 14,697.51 | 0.66 | ocu-miR-25-3p | 8869.77 | 0.48 |
| miRNA | log2 (Fold Change) | p-Value | miRNA | log2 (Fold Change) | p-Value |
|---|---|---|---|---|---|
| ocu-miR-708-5p | 4.99 | 1.95 × 10−13 | ocu-miR-146a-5p | −3.88 | 0.00084 |
| ocu-miR-708-3p | 4.51 | 4.72 × 10−14 | ocu-miR-122-5p | −3.85 | 4.10 × 10−6 |
| ocu-miR-885-5p | 3.64 | 2.82 × 10−13 | ocu-miR-503-5p | −2.45 | 1.89 × 10−7 |
| ocu-miR-24-2-5p | 3.40 | 1.27 × 10−10 | ocu-miR-16b-5p | −2.18 | 1.15 × 10−6 |
| ocu-miR-29c-3p | 3.29 | 2.13 × 10−9 | ocu-miR-378-3p | −1.91 | 0.00022 |
| ocu-miR-874-3p | 3.13 | 7.78 × 10−10 | ocu-miR-93-5p | −1.67 | 0.00634 |
| ocu-miR-574-3p | 2.80 | 3.30 × 10−10 | ocu-miR-127-3p | −1.62 | 0.00245 |
| ocu-miR-222-3p | 2.68 | 9.00 × 10−11 | ocu-let-7f-5p | −1.60 | 0.00120 |
| ocu-miR-125b-5p | 2.28 | 1.63 × 10−6 | ocu-miR-6529-5p | −1.55 | 0.00129 |
| ocu-miR-30d-5p | 1.91 | 3.23 × 10−5 | ocu-miR-25-3p | −1.54 | 0.00091 |
| ocu-miR-502a-3p | 1.76 | 0.00029 | ocu-miR-361-5p | −1.38 | 0.00283 |
| ocu-miR-181b-5p | 1.71 | 9.27× 10−5 | ocu-miR-128b-3p | −1.34 | 0.00193 |
| ocu-miR-342-3p | 1.63 | 0.00018 | ocu-miR-128a-3p | −1.34 | 0.00193 |
| ocu-miR-214-5p | 1.62 | 0.00057 | ocu-miR-130b-3p | −1.34 | 0.00689 |
| ocu-miR-30e-5p | 1.57 | 0.00059 | ocu-miR-26b-5p | −1.28 | 0.00516 |
| ocu-miR-136-3p | 1.47 | 0.00920 | ocu-miR-100-3p | −1.26 | 0.00707 |
| ocu-miR-29a-3p | 1.46 | 0.00226 | ocu-miR-143-3p | −1.19 | 0.02610 |
| ocu-miR-31-5p | 1.49 | 0.00329 | ocu-let-7a-5p | −1.17 | 0.01272 |
| ocu-miR-1296-5p | 1.43 | 0.03054 | ocu-miR-151-3p | −1.15 | 0.01897 |
| ocu-miR-22-3p | 1.42 | 0.00380 | |||
| ocu-miR-21-5p | 1.38 | 0.00112 | |||
| ocu-miR-365-3p | 1.34 | 0.00225 | |||
| ocu-miR-328-3p | 1.38 | 0.01729 | |||
| ocu-miR-181a-5p | 1.26 | 0.00350 | |||
| ocu-miR-744-5p | 1.23 | 0.01199 | |||
| ocu-miR-12090-5p | 1.15 | 0.02734 | |||
| ocu-miR-101-3p | 1.12 | 0.00961 | |||
| ocu-miR-542-3p | 1.12 | 0.01808 | |||
| ocu-miR-29b-3p | 1.11 | 0.01377 | |||
| ocu-miR-214-3p | 1.01 | 0.03077 |
| Dentin Formation (0–3) | Inflammatory Cell Response (0–3) |
| 0 = completely covered exposure site | 0 = no inflammation at or beneath the exposure site |
| 1 = >50% covered exposure site | 1 = few scattered inflammatory cells at or beneath the exposure site |
| 2 = 10–50% covered exposure site | 2 = general or localized moderate inflammatory cell infiltration at or beneath the exposure site |
| 3 = no covered exposure site | 3 = severe inflammation and/or abscess formation at or beneath the exposure site |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganesh, V.; Fredericks, D.C.; Petersen, E.B.; Keen, H.L.; He, R.; Turner, J.D.; Martin, J.A.; Salem, A.K.; Shin, K.; Parolia, A.; et al. Extracellular Vesicles-Induced Cell Homing and Odontogenesis via microRNA Signaling for Dentin Regeneration. Int. J. Mol. Sci. 2025, 26, 7182. https://doi.org/10.3390/ijms26157182
Ganesh V, Fredericks DC, Petersen EB, Keen HL, He R, Turner JD, Martin JA, Salem AK, Shin K, Parolia A, et al. Extracellular Vesicles-Induced Cell Homing and Odontogenesis via microRNA Signaling for Dentin Regeneration. International Journal of Molecular Sciences. 2025; 26(15):7182. https://doi.org/10.3390/ijms26157182
Chicago/Turabian StyleGanesh, Venkateswaran, Douglas C. Fredericks, Emily B. Petersen, Henry L. Keen, Rui He, Jordon D. Turner, James A. Martin, Aliasger K. Salem, Kyungsup Shin, Abhishek Parolia, and et al. 2025. "Extracellular Vesicles-Induced Cell Homing and Odontogenesis via microRNA Signaling for Dentin Regeneration" International Journal of Molecular Sciences 26, no. 15: 7182. https://doi.org/10.3390/ijms26157182
APA StyleGanesh, V., Fredericks, D. C., Petersen, E. B., Keen, H. L., He, R., Turner, J. D., Martin, J. A., Salem, A. K., Shin, K., Parolia, A., & Seol, D. (2025). Extracellular Vesicles-Induced Cell Homing and Odontogenesis via microRNA Signaling for Dentin Regeneration. International Journal of Molecular Sciences, 26(15), 7182. https://doi.org/10.3390/ijms26157182







