Unveiling Adulteration in Herbal Markets: MassARRAY iPLEX Assay for Accurate Identification of Plumbago indica L.
Abstract
1. Introduction
2. Results
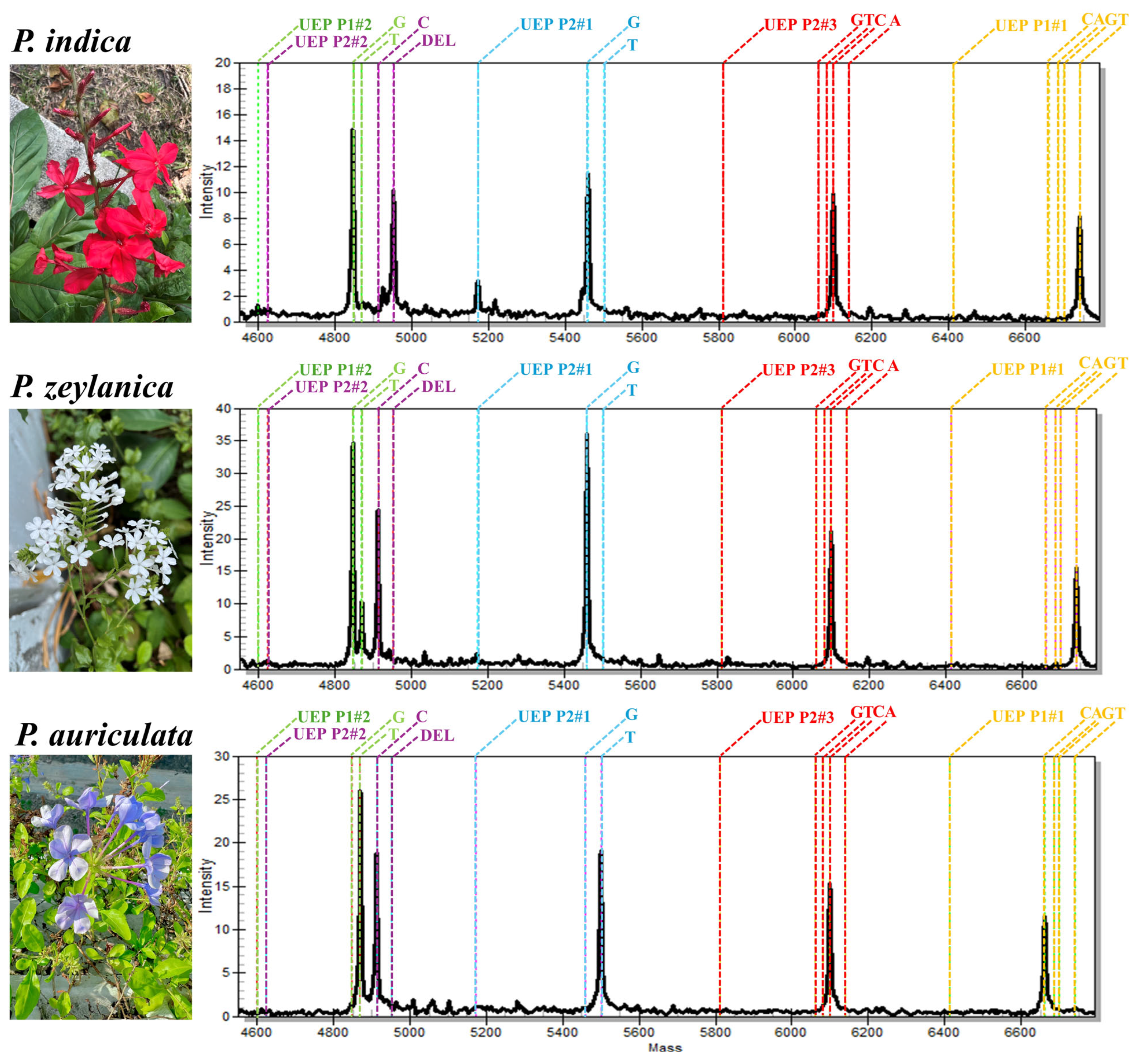
2.1. MALDI-TOF MS Analysis Differentiated P. indica from P. zeylanica and P. auriculata
2.2. Sensitivity of MassArray Technique for the Identification of P. indica, P. zeylanica, and P. auriculata
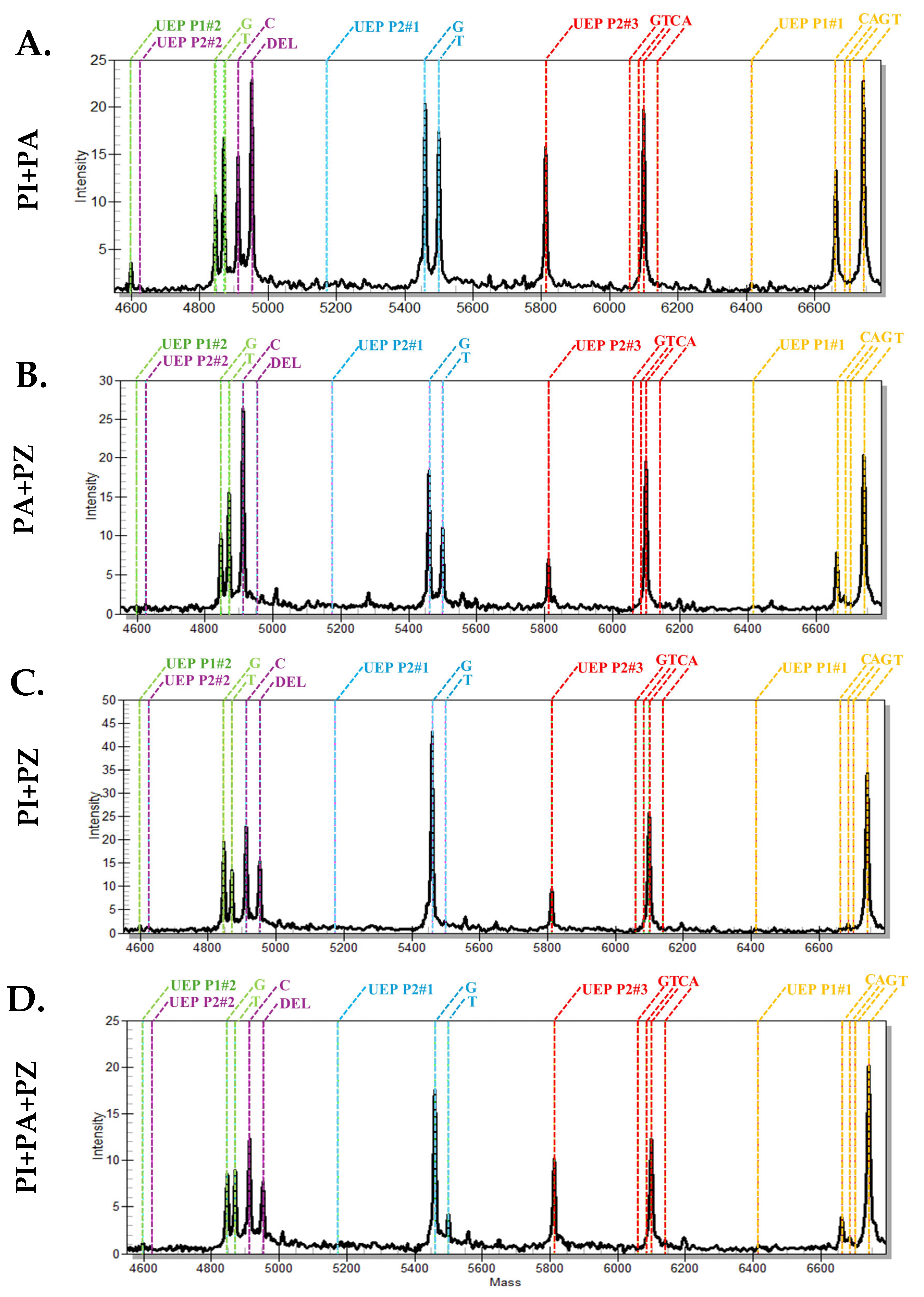
2.3. Identification of P. indica in Plumbago-Mixed Samples the MassARRAY Analysis Successfully Identified Plumbago indica in the Mixed Plumbago Sample
2.4. Identification of Jettamoon Pleung Daeng Crude Drugs and the Crude Drug Composed in the Traditional Thai Medicinal Formulations
3. Discussion
4. Materials and Methods
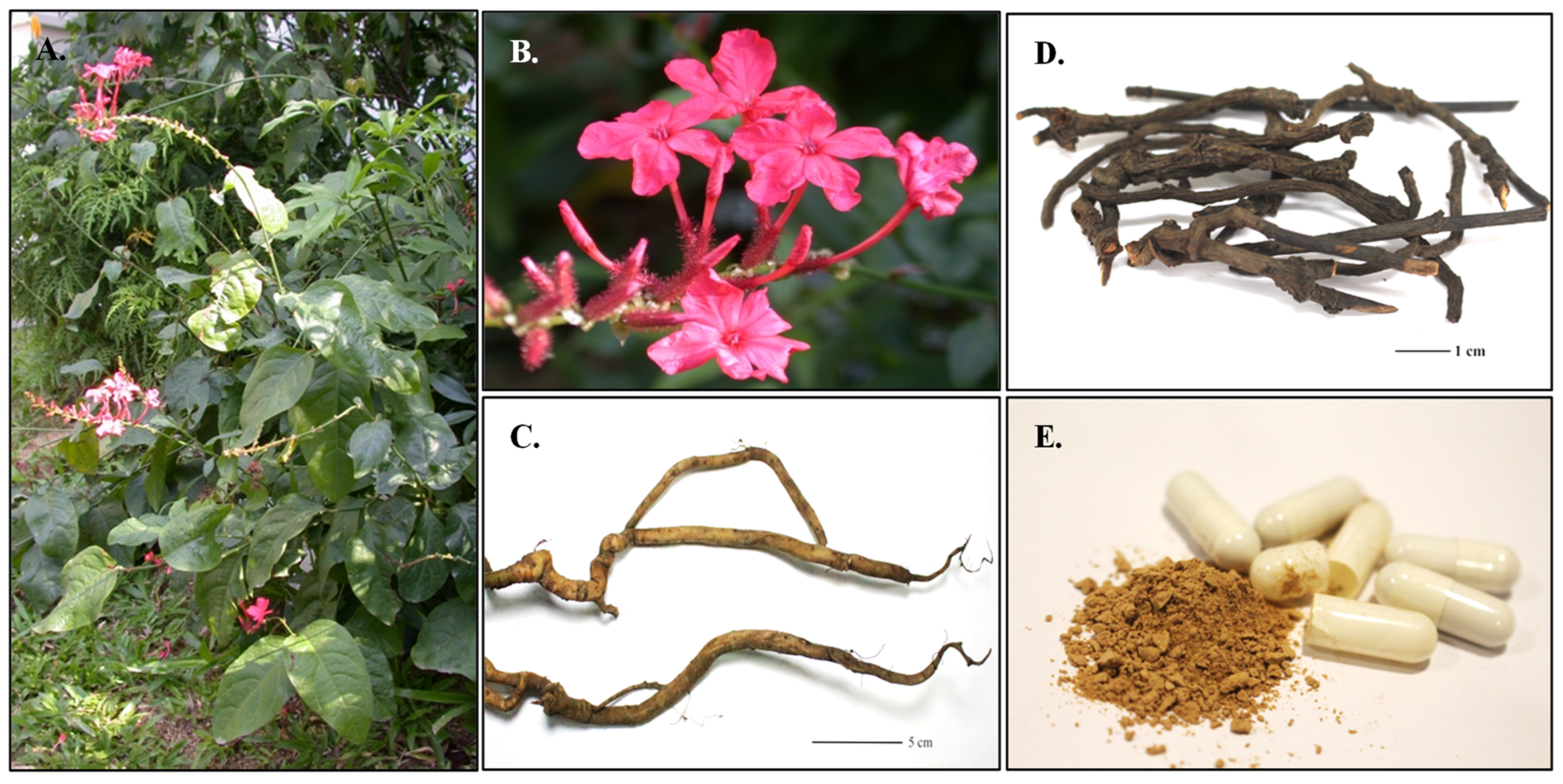
4.1. Plant Materials, Commercial Crude Drugs, and Traditional Formulations
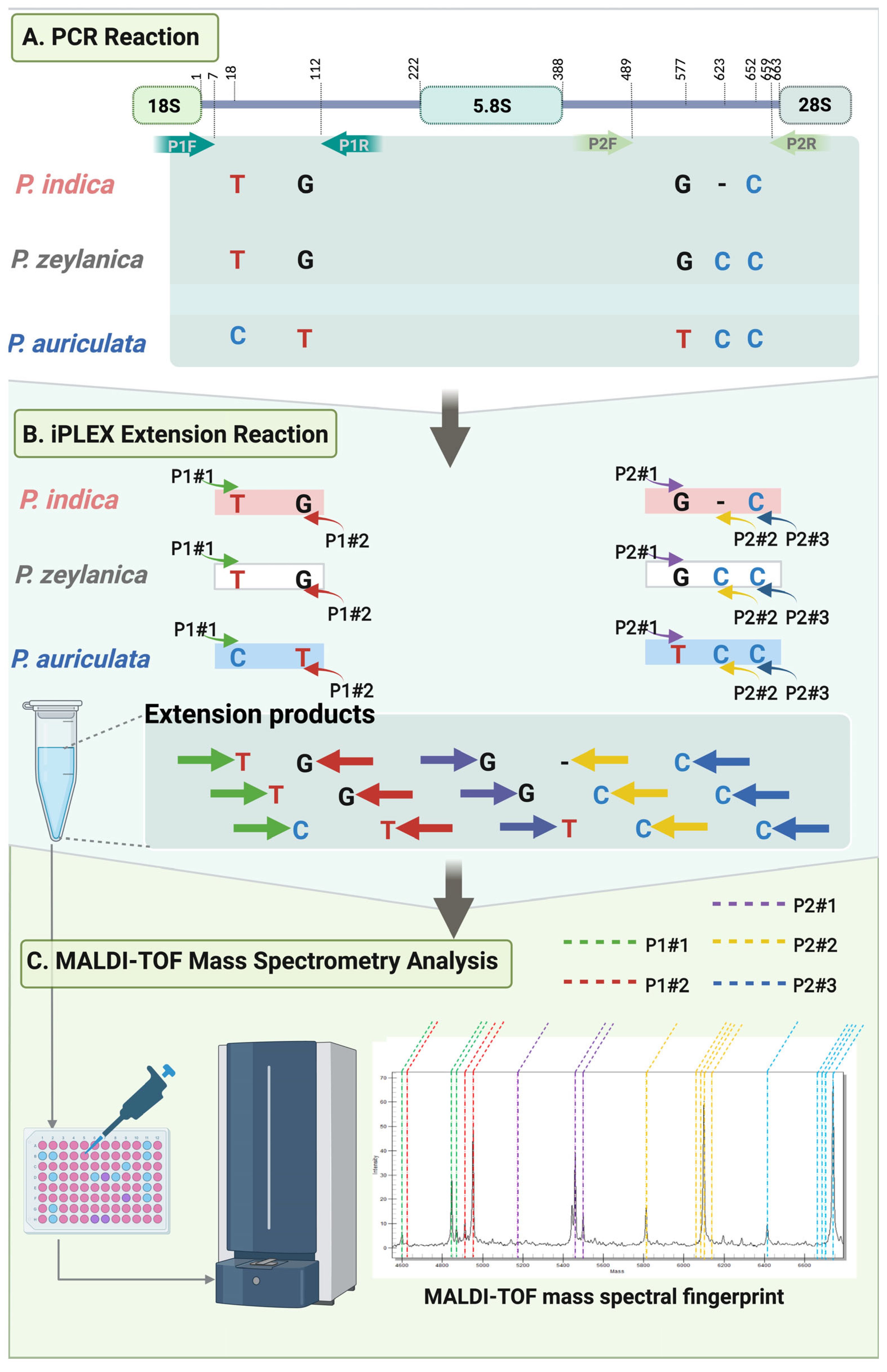
4.2. Primer Sets and Design
4.3. DNA Extraction, Amplification, and Nucleotide Sequencing
4.4. The iPLEX Assay on the MassARRAY System for the P. indica Identification
4.5. Sensitivity of the iPLEX Extension Assay for the Differentiation of P. indica from Its Related Species
4.6. Identification of P. indica in Plumbago-Mixed Samples
4.7. Authentication of the Commercial P. indica Crude Drugs and the Polyherbal Products
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BLAST | Basic Local Alignment Search Tool |
| Da | Dalton |
| ddNTPs | Dideoxynucleotides |
| DEL | Deletion |
| FDA | Food and Drug Administration |
| GC-MS | Gas Chromatography–Mass Spectrometry |
| HPLC | High-Performance Liquid Chromatography |
| HPTLC | High-Performance Thin-Layer Chromatography |
| INDEL | Insertion/Deletion |
| ITS | Internal Transcribed Spacer |
| LC-MS/MS | Liquid Chromatography–Tandem Mass Spectrometry |
| LFA | Lateral-Flow Immunochromatographic Assay |
| LOD | Limit of Detection |
| LOH | Loss of Heterozygosity |
| MALDI-TOF MS | Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry |
| NGS | Next-Generation Sequencing |
| PCR | Polymerase Chain Reaction |
| SAP | Shrimp Alkaline Phosphatase |
| SBE | Single-Base Extension |
| SNP | Single Nucleotide Polymorphism |
| UEP | Unextended Primer |
| WHO | World Health Organization |
References
- Abdu, A.; Prakash, A.; Kondal, R.; Sharma, S.; Bhagat, M.; Pal, R.; Singh, H.; Singh, B.; Kaur, S. Comprehensive review on Plumbago indica: Traditional, pharmacological insights and conservation strategies. J. Appl. Pharm. Sci. 2024, 14, 1–16. [Google Scholar] [CrossRef]
- Thitiorul, S.; Ratanavalachai, T.; Tanuchit, S.; Itharat, A.; Sakpakdeejaroen, I. Genotoxicity and interference with cell cycle activities by an ethanolic extract from Thai Plumbago indica roots in human lymphocytes in vitro. Asian Pac. J. Cancer Prev. 2013, 14, 2487–2490. [Google Scholar] [CrossRef]
- Sharma, B.; Dhiman, C.; Hasan, G.M.; Shamsi, A.; Hassan, M.I. Pharmacological Features and Therapeutic Implications of Plumbagin in Cancer and Metabolic Disorders: A Narrative Review. Nutrients 2024, 16, 3033. [Google Scholar] [CrossRef] [PubMed]
- Makchuchit, S.; Rattarom, R.; Itharat, A. The anti-allergic and anti-inflammatory effects of Benjakul extract (a Thai traditional medicine), its constituent plants and its some pure constituents using in vitro experiments. Biomed. Pharmacother. 2017, 89, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Duenngai, K.; Promraksa, B.; Janthamala, S.; Thanee, M.; Sirithawat, P.; Wisungre, S.; Deechan, S.; Meechai, N.; Paratang, P.; Techasen, A. Antioxidant and Anticancer Potentials and Metabolic Profiling of Benjakul, A Thai Herbal Preparation. Trop. J. Nat. Prod. Res. 2024, 8, 6877–6883. [Google Scholar] [CrossRef]
- Threrapanithan, C.; Jaiaree, N.; Itharat, A.; Makchuchit, S.; Thongdeeying, P.; Panthong, S. Anti-inflammatory and antioxidant activities of the Thai traditional remedy called “Leard-ngam” and its plant ingredients. Thammasat Med. J. 2015, 15, 376–383. [Google Scholar]
- Shukla, B.; Saxena, S.; Usmani, S.; Kushwaha, P. Phytochemistry and pharmacological studies of Plumbago zeylanica L.: A medicinal plant review. Clin. Phytosci. 2021, 7, 34. [Google Scholar] [CrossRef]
- Melk, M.M.; Melek, F.R.; El-Sayed, A.F. Enzymes inhibitory capabilities of phenolics from Plumbago indica L. and Plumbago auriculata Lam.: In-vitro studies and molecular docking. Process Biochem. 2024, 136, 1–13. [Google Scholar] [CrossRef]
- Sherif, A.E.; Amen, Y.; Shimizu, K. Validation of the potential anti-inflammatory activity of Plumbago auriculata Lam. S. Afr. J. Bot. 2022, 147, 467–471. [Google Scholar] [CrossRef]
- Thongkhao, K.; Pongkittiphan, V.; Phadungcharoen, T.; Tungphatthong, C.; Urumarudappa, S.K.J.; Pengsuparp, T.; Sutanthavibul, N.; Wiwatcharakornkul, W.; Kengtong, S.; Sukrong, S. Differentiation of Cyanthillium cinereum, a smoking cessation herb, from its adulterant Emilia sonchifolia using macroscopic and microscopic examination, HPTLC profiles and DNA barcodes. Sci. Rep. 2020, 10, 14753. [Google Scholar] [CrossRef]
- Khwanchum, L.; Intharuksa, A.; Yanaso, S.; Thongkhao, K. Differentiation of Centella asiatica (L.) Urb. from Hydrocotyle umbellata L. using the trnH-psbA region, species-specific bands and anisocytic stomata as markers for quality control of raw materials and their products. J. Appl. Res. Med. Aromat. Plants 2023, 37, 100504. [Google Scholar] [CrossRef]
- Urumarudappa, S.K.J.; Rosario, S.; G, R.; Sukrong, S. A comprehensive review on Saraca asoca (Fabaceae)—Historical perspective, traditional uses, biological activities, and conservation. J. Ethnopharmacol. 2023, 317, 116861. [Google Scholar] [CrossRef] [PubMed]
- Khamnuan, S.; Phrutivorapongkul, A.; Pitchakarn, P.; Buacheen, P.; Karinchai, J.; Chittasupho, C.; Na Takuathung, M.; Theansungnoen, T.; Thongkhao, K.; Intharuksa, A. The Identification and Cytotoxic Evaluation of Nutmeg (Myristica fragrans Houtt.) and Its Substituents. Foods 2023, 12, 4211. [Google Scholar] [CrossRef] [PubMed]
- Lenora, R.D.K.; Dharmadasa, R.M.; Abeysinghe, D.C.; Arawwawala, L.D.A.M.; Dharmadasa, R.M.; Abeysinghe, D.C.; Arawwawala, L.D.A.M. Investigation of Plumbagin Content in Plumbago indica Linn. Grown under Different Growing Systems. Pharmacologia 2012, 3, 57–60. [Google Scholar] [CrossRef]
- Srivastava, S.; Misra, A. Quality Control of Herbal Drugs: Advancements and Challenges. In New Age Herbals: Resource, Quality and Pharmacognosy; Singh, B., Peter, K.V., Eds.; Springer: Singapore, 2018; pp. 189–209. [Google Scholar]
- World Health Organization. Quality Control Methods for Herbal Materials: Updated edition of Quality Control Methods for Medicinal Plant Materials; World Health Organization: Geneva, Switzerland, 1998. [Google Scholar]
- Serrano, R.; Da Silva, G.; Silva, O. Application of light and scanning electron microscopy in the identification of herbal medicines. Microsc. Sci. Technol. Appl. Educ. 2010, 3, 182–190. [Google Scholar]
- Song, J.H.; Yang, S.; Choi, G. Taxonomic Implications of Leaf Micromorphology Using Microscopic Analysis: A Tool for Identification and Authentication of Korean Piperales. Plants 2020, 9, 566. [Google Scholar] [CrossRef]
- Tarassoli, Z.; Faraji, H.; Tajabadi, F.; Shabani, M.; Shahbazi, H. Hollow Fiber-Based Liquid Phase Microextraction Combined with Chemometrics Approach: Determination of Rosmarinic Acid in Lavandula angustifolia Products. J. Anal. Chem. 2024, 79, 1717–1723. [Google Scholar] [CrossRef]
- Lai, K.-M.; Cheng, Y.-Y.; Tsai, T.-H. Integrated LC-MS/MS Analytical Systems and Physical Inspection for the Analysis of a Botanical Herbal Preparation. Molecules 2015, 20, 10641–10656. [Google Scholar] [CrossRef]
- Dubnicka, M.; Cromwell, B.; Levine, M. Investigation of the Adulteration of Essential Oils by GC-MS. Curr. Anal. Chem. 2020, 16, 965–969. [Google Scholar] [CrossRef]
- Tarasoli, Z.; Faraji, H.; Tajabadi, F.; Shabani, M.; Shahbazi, H. Evaluation of adulteration in Lavandula angustifolia Mill. products using GC/MS combined with chemometric methods. J. Med. Plants 2021, 20, 34–46. [Google Scholar] [CrossRef]
- Cao, K.; Chen, J.; Huang, R.; Lu, R.; Zhou, X.; Bu, Y.; Li, L.; Yao, C. Metabolomics analysis reveals the differences between Abrus cantoniensis Hance and Abrus mollis Hance. BMC Plant Biol. 2023, 23, 375. [Google Scholar] [CrossRef]
- Ha, W.-Y.; Wong, K.-L.; Ma, W.-Y.; Lau, Y.-Y.; Chan, W.-H. Enhancing Testing Laboratory Engagement in Plant DNA Barcoding through a Routine Workflow—A Case Study on Chinese Materia Medica (CMM). Plants 2022, 11, 1317. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yin, X.; Han, J.; Sun, W.; Yao, H.; Song, J.; Li, X. DNA barcoding in herbal medicine: Retrospective and prospective. J. Pharm. Anal. 2023, 13, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Thongkhao, K.; Tungphatthong, C.; Sukrong, S. A PCR-lateral flow immunochromatographic assay (PCR-LFA) for detecting Aristolochia species, the plants responsible for aristolochic acid nephropathy. Sci. Rep. 2022, 12, 12188. [Google Scholar] [CrossRef] [PubMed]
- Tungphatthong, C.; Phadungcharoen, T.; Sooksawate, T.; Sukrong, S. PCR combined with lateral flow immunochromatographic assay to differentiate the narcotic Mitragyna speciosa from related species and detect it in forensic evidence. Forensic Sci. Int. 2022, 331, 111149. [Google Scholar] [CrossRef]
- Dechbumroong, P.; Aumnouypol, S.; Denduangboripant, J.; Sukrong, S. DNA barcoding of Aristolochia plants and development of species-specific multiplex PCR to aid HPTLC in ascertainment of Aristolochia herbal materials. PLoS ONE 2018, 13, e0202625. [Google Scholar] [CrossRef]
- Buggs, R.J.; Chamala, S.; Wu, W.; Gao, L.; May, G.D.; Schnable, P.S.; Soltis, D.E.; Soltis, P.S.; Barbazuk, W.B. Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol. Ecol. 2010, 19 (Suppl. 1), 132–146. [Google Scholar] [CrossRef]
- Galeano, C.H.; Cortés, A.J.; Fernández, A.C.; Soler, Á.; Franco-Herrera, N.; Makunde, G.; Vanderleyden, J.; Blair, M.W. Gene-Based Single Nucleotide Polymorphism Markers for Genetic and Association Mapping in Common Bean. BMC Genet. 2012, 13, 48. [Google Scholar] [CrossRef]
- De Silva, A.L.; Kamper, W.; Ogbourne, S.M.; Nichols, J.; Royle, J.W.L.; Peters, T.; Hawkes, D.; Hosseini Bai, S.; Wallace, H.M.; Trueman, S.J. MassARRAY and SABER Analyses of SNPs in Embryo DNA Reveal the Abscission of Self-Fertilised Progeny during Fruit Development of Macadamia (Macadamia integrifolia Maiden & Betche). Int. J. Mol. Sci. 2024, 25, 6419. [Google Scholar] [CrossRef]
- Liang, R.; Li, J.; Zhao, Y.; Qi, H.; Bao, S.; Wang, F.; Duan, H.; Huang, H. A comparative study of MassARRAY and GeneXpert assay in detecting rifampicin resistance in tuberculosis patients’ clinical specimens. Front. Microbiol. 2024, 15, 1287806. [Google Scholar] [CrossRef]
- Intharuksa, A.; Phrutivorapongkul, A.; Thongkhao, K. Integrating DNA barcoding, microscopic, and chemical analyses for precise identification of Plumbago indica L., A prominent medicinal plant. Microchem. J. 2024, 199, 110038. [Google Scholar] [CrossRef]
- Jurinke, C.; Boom, D.; Cantor, C.R.; Köster, H. The Use of MassARRAY Technology for High Throughput Genotyping. Adv. Biochem. Eng. Biotechnol. Adv. 2002, 77, 57–74. [Google Scholar] [CrossRef]
- Ichim, M.C. The DNA-Based Authentication of Commercial Herbal Products Reveals Their Globally Widespread Adulteration. Front. Pharmacol. 2019, 10, 1227. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishnan, K.P.; Raja, S.S.; Balachandran, I. A Reverse Phase HPLC-UV and HPTLC Methods for Determination of Plumbagin in Plumbago indica and Plumbago zeylanica. Indian J. Pharm. Sci. 2008, 70, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Anthoons, B.; Lagiotis, G.; Drouzas, A.D.; de Boer, H.; Madesis, P. Barcoding High Resolution Melting (Bar-HRM) enables the discrimination between toxic plants and edible vegetables prior to consumption and after digestion. J. Food Sci. 2022, 87, 4221–4232. [Google Scholar] [CrossRef]
- Hao, L.; Shi, X.; Wen, S.; Yang, C.; Chen, Y.; Yue, S.; Chen, J.; Luo, K.; Liu, B.; Sun, Y.; et al. Single nucleotide polymorphism-based visual identification of Rhodiola crenulata using the loop-mediated isothermal amplification technique. Front. Plant Sci. 2024, 15, 1492083. [Google Scholar] [CrossRef]
- Svidnicki, M.C.; Silva-Costa, S.M.; Ramos, P.Z.; dos Santos, N.Z.; Martins, F.T.; Castilho, A.M.; Sartorato, E.L. Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iPLEX(R) technology. BMC Med. Genet. 2015, 16, 85. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hewitt, G.M. Nuclear DNA analyses in genetic studies of populations: Practice, problems and prospects. Mol. Ecol. 2003, 12, 563–584. [Google Scholar] [CrossRef]
- Dutta, A.; Schacherer, J. The dynamics of loss of heterozygosity events in genomes. EMBO Rep. 2025, 26, 602–612. [Google Scholar] [CrossRef]
- Intharuksa, A.; Sasaki, Y.; Ando, H.; Charoensup, W.; Suksathan, R.; Kertsawang, K.; Sirisa-Ard, P.; Mikage, M. The combination of ITS2 and psb A-trn H region is powerful DNA barcode markers for authentication of medicinal Terminalia plants from Thailand. J. Nat. Med. 2020, 74, 282–293. [Google Scholar] [CrossRef]
- Wang, M.; Lin, H.; Lin, H.; Du, P.; Zhang, S. From species to varieties: How modern sequencing technologies are shaping Medicinal Plant Identification. Genes 2024, 16, 16. [Google Scholar] [CrossRef]
- Intharuksa, A.; Denduangboripant, J.; Chansakaow, S.; Thongkhao, K.; Sukrong, S. HPLC and DNA barcoding profiles for identification of the selected twelve Mucuna species and its application for detecting prohibited aphrodisiac Mucuna products. Heliyon 2023, 9, e14130. [Google Scholar] [CrossRef]
- Yanaso, S.; Phrutivorapongkul, A.; Hongwiset, D.; Piyamongkol, S.; Intharuksa, A. Verification of Thai ethnobotanical medicine “Kamlang Suea Khrong” driven by multiplex PCR and powerful TLC techniques. PLoS ONE 2021, 16, e0257243. [Google Scholar] [CrossRef]
- Han, K.; Wang, M.; Zhang, L.; Wang, C. Application of molecular methods in the identification of ingredients in Chinese herbal medicines. Molecules 2018, 23, 2728. [Google Scholar] [CrossRef]
- Pinya, T.; Intharuksa, A.; Yanaso, S.; Kamnuan, S.; Phrutivorapongkul, A. Conventional and molecular pharmacognostic characters integrated with chemical profiles of five Piper plants in the Thai herbal pharmacopoeia and their admixture/adulteration/substitution situations in Thailand. J. Nat. Med. 2022, 76, 605–620. [Google Scholar] [CrossRef]
- Stanford, A.M.; Harden, R.; Parks, C.R. Phylogeny and biogeography of Juglans (Juglandaceae) based on matK and ITS sequence data. Am. J. Bot. 2000, 87, 872–882. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
| Primers | Target SNP Position | Primer Sequences (UEP) | Mass (Da) | Expected Masses of the Single-Base Extension Product (Da) | ||||
|---|---|---|---|---|---|---|---|---|
| C | A | G | T | DEL | ||||
| P1#1 | 18 | AAGGATCATTGTCGAAACCTC | 6414.2 | 6661.4 | 6685.4 | 6701.4 | 6741.3 | n.d. |
| P1#2 | 112 | TTGTTCAAGCCTGGG | 4599.0 | n.d. | n.d. | 4846.2 | 4870.2 | n.d. |
| P2#1 | 577 | CCGCGAAGCGTCGTGCC | 5172.4 | n.d. | n.d. | 5459.6 | 5499.5 | n.d. |
| P2#2 | 623 | CCTGGGGTCGCATGG | 4625.0 | 4912.2 | n.d. | n.d. | n.d. | 4952.1 |
| P2#3 | 652 | ATATGCTTAAACTCAGCGG | 5811.8 | 6059.0 | 6083.0 | 6741.3 | 6138.9 | n.d. |
| Species | Target SNP Position | Expected Mass of the Single-Base Extension Products | Mass Profile of the Single-Base Extension Products from Authentic Plants | ||||||
|---|---|---|---|---|---|---|---|---|---|
| C | G | T | INDEL | C | G | T | INDEL | ||
| P. indica | P1#1 | - | - | 6741.3 | - | - | - | 6741.3 | - |
| P1#2 | - | 4846.2 | - | - | - | 4846.2 | - | - | |
| P2#1 | - | 5459.6 | - | - | - | 5459.6 | - | - | |
| P2#2 | - | - | - | 4952.1 | - | - | - | 4952.1 | |
| P2#3 | 6059.0 | - | - | - | 6059.0 | - | - | - | |
| P. zeylanica | P1#1 | - | - | 6741.3 | - | - | - | 6741.3 | - |
| P1#2 | - | 4846.2 | - | - | - | 4846.2 | 4870.2 * | - | |
| P2#1 | - | 5459.6 | - | - | - | 5459.6 | - | - | |
| P2#2 | 4912.2 | - | - | - | 4912.2 | - | - | - | |
| P2#3 | 6059.0 | - | - | - | 6059.0 | - | - | - | |
| P. auriculata | P1#1 | 6661.4 | - | - | - | 6661.4 | - | - | - |
| P1#2 | - | - | 4870.2 | - | - | - | 4870.2 | - | |
| P2#1 | - | - | 5499.5 | - | - | - | 5499.5 | - | |
| P2#2 | 4912.2 | - | - | - | 4912.2 | - | - | - | |
| P2#3 | 6059.0 | - | - | - | 6059.0 | - | - | - | |
| Samples | DNA Concentration (ng/μL) | Extension Primers | ||||
|---|---|---|---|---|---|---|
| P1#1 | P1#2 | P2#1 | P2#2 | P2#3 | ||
| P. indica | 10 | T | G | G | DEL | C |
| 1 | T | G | G | DEL | C | |
| 0.1 | T | G | G | DEL | C | |
| 0.01 | T | G | G | DEL | C | |
| 0.001 | T | G | G | C/DEL | C | |
| 0.0001 | T | G/T | G | C | C | |
| 0.00001 | T | G | G | C | C | |
| 0.000001 | T | G/T | G | C | C | |
| 0 | - | - | - | - | - | |
| P. zeylanica | 10 | T | G | G | C | C |
| 1 | T | G | G | C | C | |
| 0.1 | T | G | G | C | C | |
| 0.01 | T | G | G | C | C | |
| 0.001 | T | T | G | C | C | |
| 0.0001 | T | G/T | G | C | C | |
| 0.00001 | T | G | G | DEL | C | |
| 0.000001 | T | G/T | G | C | C | |
| 0 | - | - | - | - | - | |
| P. auriculata | 10 | C | T | T | C | C |
| 1 | C | T | T | C | C | |
| 0.1 | C | T | T | C | C | |
| 0.01 | C | T | T | C | C | |
| 0.001 | C/T | G/T | G/T | C | C | |
| 0.0001 | T | G | G | C | C | |
| 0.00001 | T | G/T | G | C | C | |
| 0.000001 | T | G/T | G | C | C | |
| 0 | - | - | - | - | - | |
| Samples | Extension Primers | Result | ||||
|---|---|---|---|---|---|---|
| P1#1 | P1#2 | P2#1 | P2#2 | P2#3 | ||
| PI | T | G | G | DEL | C | P. indica |
| PZ | T | T | G | C | C | P. zeylanica |
| PA | C | T | T | C | C | P. auriculata |
| C-1 | T | G | G | DEL | C | P. indica |
| C-2 | T | G | G | DEL | C | P. indica |
| C-3 | T | G | G | DEL | C | P. indica |
| C-4 | T | G | G | DEL | C | P. indica |
| C-5 | T | G | G | DEL | C | P. indica |
| C-6 | T | G | G | DEL | C | P. indica |
| C-7 | T | G | G | DEL | C | P. indica |
| C-8 | T | G | G | DEL | C | P. indica |
| C-9 | T | G | G | DEL | C | P. indica |
| R-1 | T | G | G | DEL | C | P. indica |
| R-2 | T | T | G | C | C | P. zeylanica |
| R-3 | T | G | G | DEL | C | P. indica |
| R-4 | T | G | G | DEL | C | P. indica |
| R-5 | T | G | G | DEL | C | P. indica |
| R-6 | T | T | G | C | C | P. zeylanica |
| Codes | Sample Details | Locality |
|---|---|---|
| Authentic Plumbago species | ||
| PI | P. indica L. | Maerim, Chiang Mai |
| PZ | P. zeylanica L. | Mueang, Chiang Mai |
| PA | P. auriculata Lam. | Mueang, Chiang Mai |
| Crude drugs | ||
| C-1 | Jettamoon Pleung Daeng | Mueang, Phatthalung |
| C-2 | Jettamoon Pleung Daeng | Samphanthawong, Bangkok |
| C-3 | Jettamoon Pleung Daeng | Samphanthawong, Bangkok |
| C-4 | Jettamoon Pleung Daeng | Samphanthawong, Bangkok |
| C-5 | Jettamoon Pleung Daeng | Mueang, Nakhon Pathom |
| C-6 | Jettamoon Pleung Daeng | Hat Yai, Songkhla |
| C-7 | Jettamoon Pleung Daeng | Mueang, Nakhon Pathom |
| C-8 | Jettamoon Pleung Daeng | Samphanthawong, Bangkok |
| C-9 | Jettamoon Pleung Daeng | Mueang, Chiang Mai |
| Thai traditional formulations | ||
| R-1 | Ya Benchakun | Faculty of Pharmacy, Chiang Mai University (In-house preparation) |
| R-2 | Ya Benchakun | Company 1 |
| R-3 | Ya Fai Pralaikan | Faculty of Pharmacy, Chiang Mai University (In-house preparation) |
| R-4 | Ya Fai Pralaikan | Company 2 |
| R-5 | Ya Fai Ha Kong | Faculty of Pharmacy, Chiang Mai University (In-house preparation) |
| R-6 | Ya Fai Ha Kong | Company 3 |
| Primer Set | Primer Name | Target(Region/SNP) | Direction | Primer Sequences (5′→ 3′) | Original Design |
|---|---|---|---|---|---|
| DNA barcode | ITS5A | ITS region | Forward | CCT TAT CAT TTA GAG GAA GGA G | [48] |
| ITS4 | ITS region | Reverse | TCC TCC GCT TAT TGA TAT GC | [49] | |
| Specific primer | P1F | ITS1 region | Forward | ACG TTG GAT GAA CCT GCG GAA GGA TCA TTG | This study |
| P1R | ITS1 region | Reverse | ACG TTG GAT GGC GCC GTG TTT TTG TTC AAG | This study | |
| P2F | ITS2 region | Forward | ACG TTG GAT GCG GTT GGC TTA AAT TCG GG | This study | |
| P2R | ITS2 region | Reverse | ACG TTG GAT GCT TAT TGA TAT GCT TAA ACT | This study | |
| iPLEX extension primer | Ext_P1#1 | SNP18 | Forward | AAG GAT CAT TGT CGA AAC CTC | This study |
| Ext_P1#2 | SNP112 | Reverse | TTG TTC AAG CCT GGG | This study | |
| Ext_P2#1 | SNP577 | Forward | CCG CGA AGC GTC GTG CC | This study | |
| Ext_P2#2 | SNP623 | Reverse | CCT GGG GTC GCA TGG | This study | |
| Ext_P2#3 | SNP652 | Reverse | ATA TGC TTA AAC TCA GCG G | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thongkhao, K.; Intharuksa, A.; Phrutivorapongkul, A. Unveiling Adulteration in Herbal Markets: MassARRAY iPLEX Assay for Accurate Identification of Plumbago indica L. Int. J. Mol. Sci. 2025, 26, 7168. https://doi.org/10.3390/ijms26157168
Thongkhao K, Intharuksa A, Phrutivorapongkul A. Unveiling Adulteration in Herbal Markets: MassARRAY iPLEX Assay for Accurate Identification of Plumbago indica L. International Journal of Molecular Sciences. 2025; 26(15):7168. https://doi.org/10.3390/ijms26157168
Chicago/Turabian StyleThongkhao, Kannika, Aekkhaluck Intharuksa, and Ampai Phrutivorapongkul. 2025. "Unveiling Adulteration in Herbal Markets: MassARRAY iPLEX Assay for Accurate Identification of Plumbago indica L." International Journal of Molecular Sciences 26, no. 15: 7168. https://doi.org/10.3390/ijms26157168
APA StyleThongkhao, K., Intharuksa, A., & Phrutivorapongkul, A. (2025). Unveiling Adulteration in Herbal Markets: MassARRAY iPLEX Assay for Accurate Identification of Plumbago indica L. International Journal of Molecular Sciences, 26(15), 7168. https://doi.org/10.3390/ijms26157168







