Genome-Wide Identification of the SiNHX Gene Family in Foxtail Millet (Setaria Italica) and Functional Characterization of SiNHX7 in Arabidopsis
Abstract
1. Introduction
2. Results
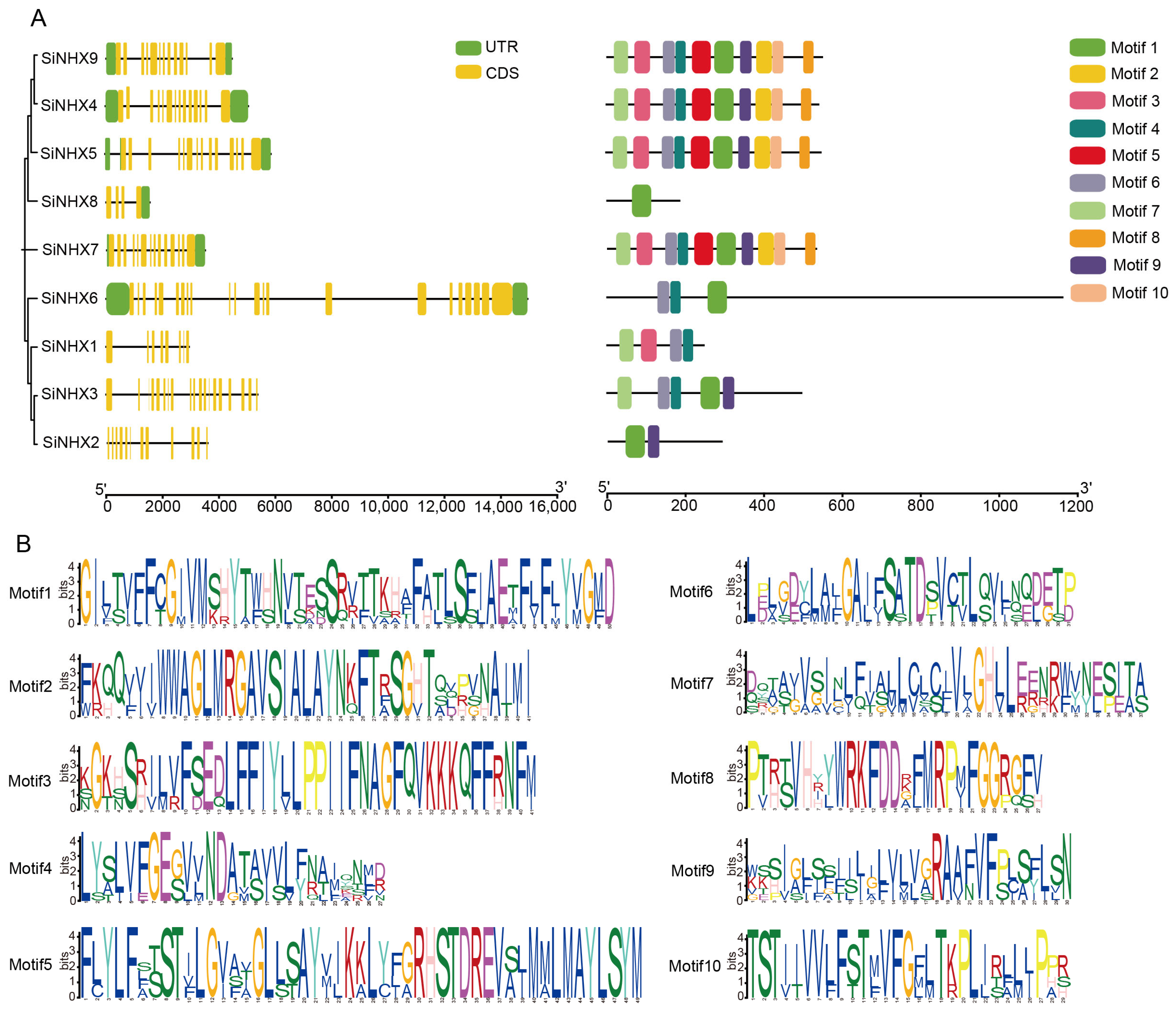
2.1. Phylogenetic and Structural Characterization of NHX Gene Family in Foxtail Millet
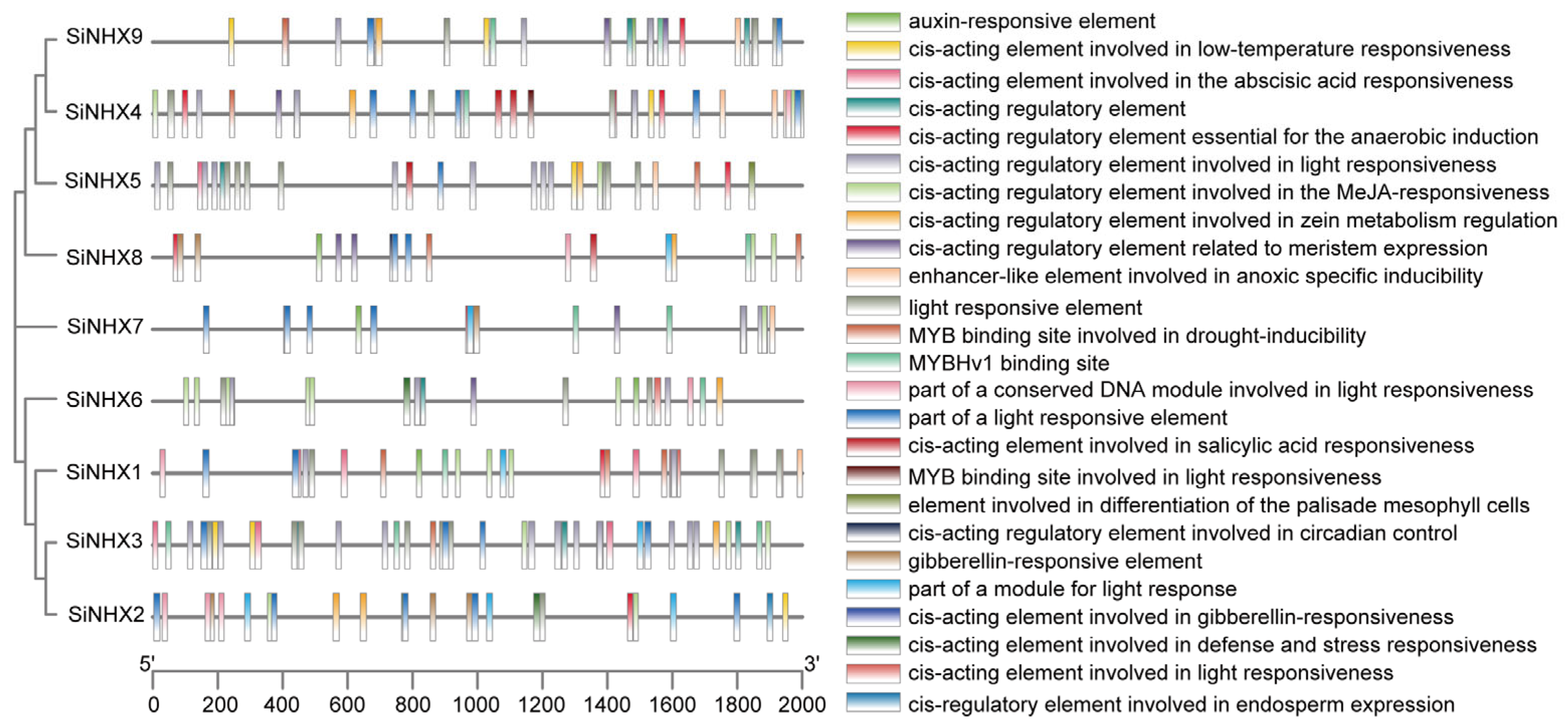
2.2. Analysis of Putative Cis-Acting Regulatory Elements (CAREs or Cis-Elements) in SiNHX Genes
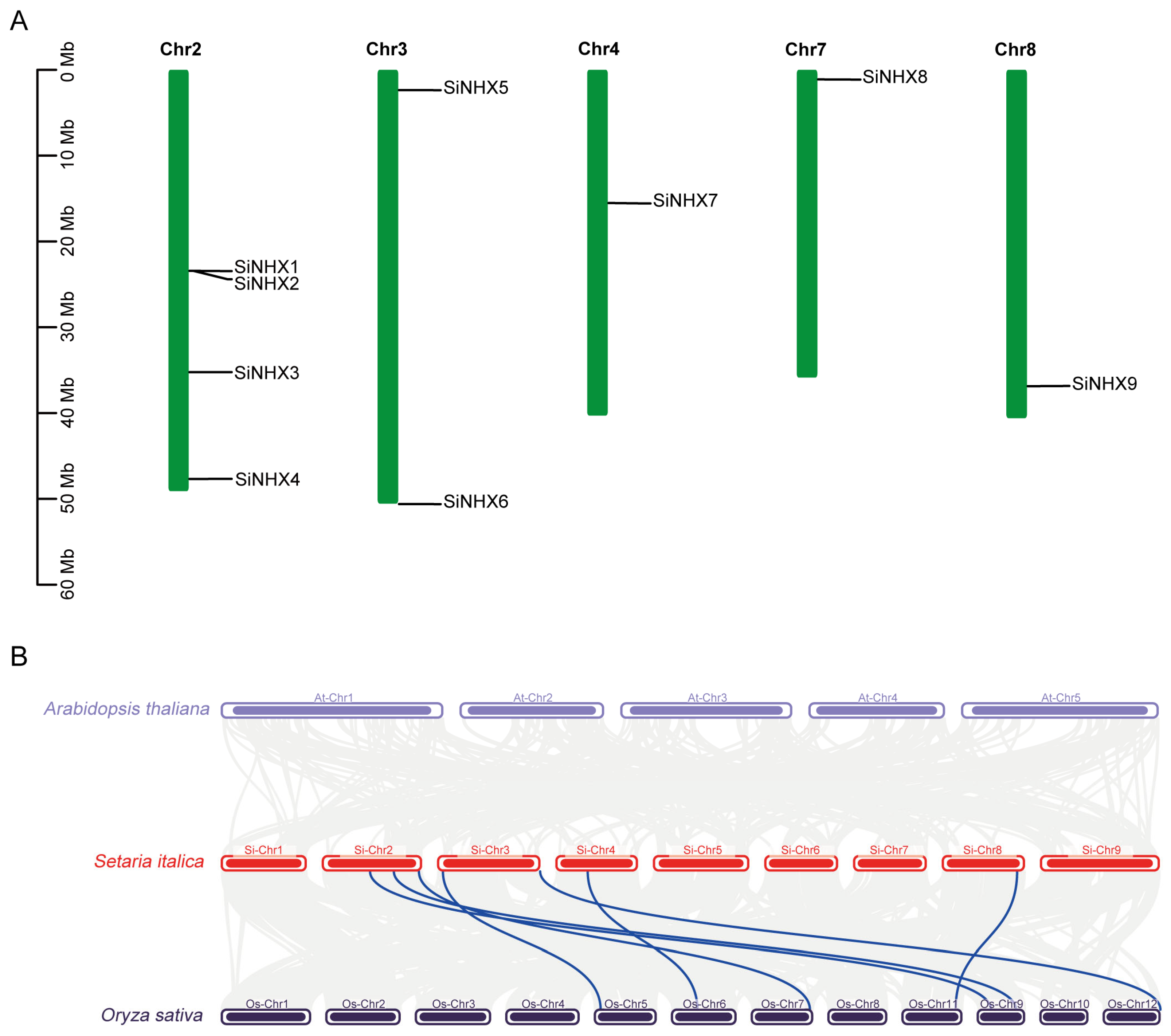
2.3. Chromosome Distribution and Collinearity Analysis of SiNHX Gene Family Members
2.4. Physical and Chemical Properties of NHX Proteins in Foxtail Millet
2.5. Prediction Secondary Structure and Subcellular Localization of SiNHX Proteins
2.6. Tissue Expression Analysis of the SiNHX Gene Family
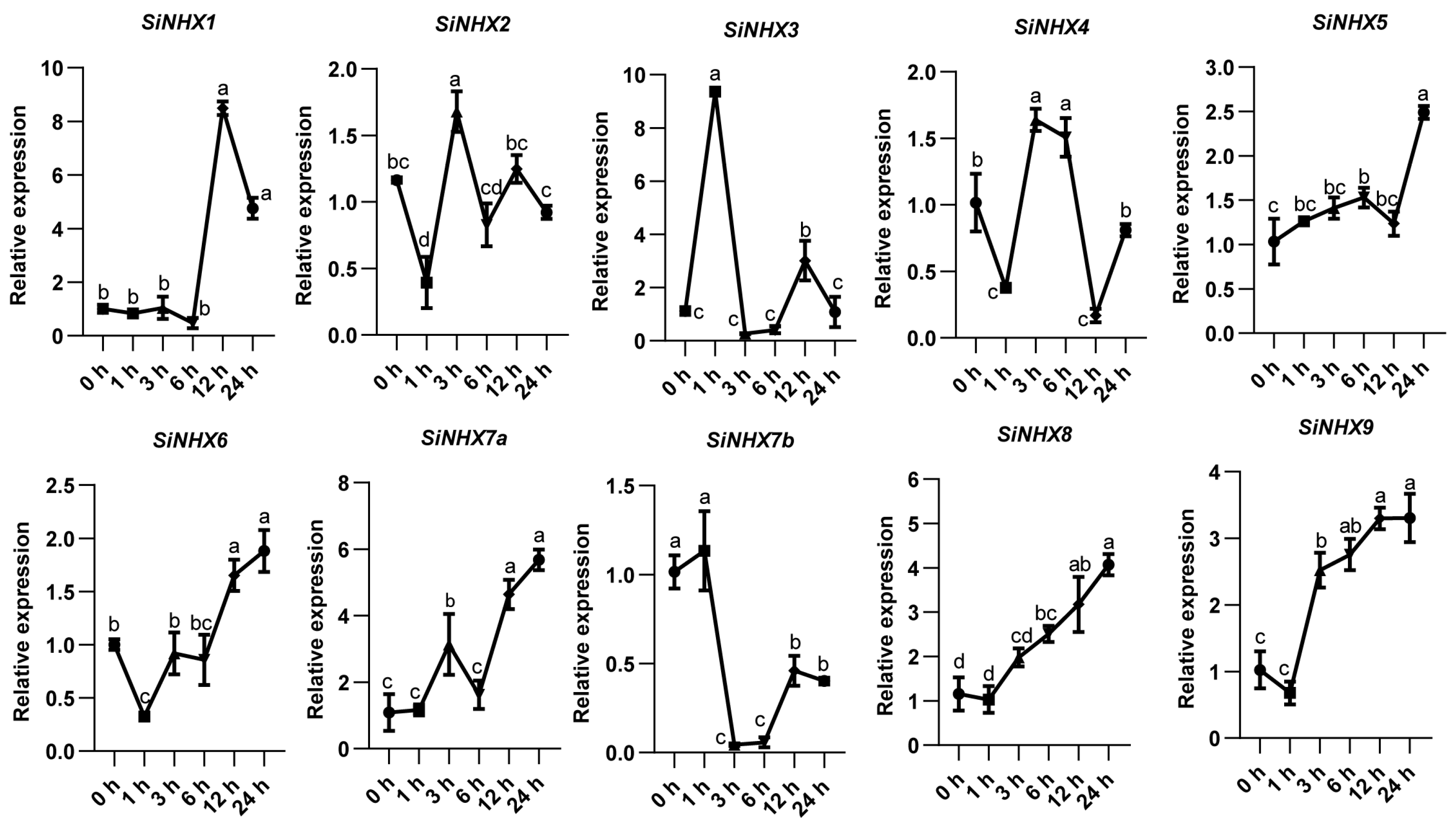
2.7. Analysis of Root Expression of SiNHX Gene Family Treated with Salt Stress
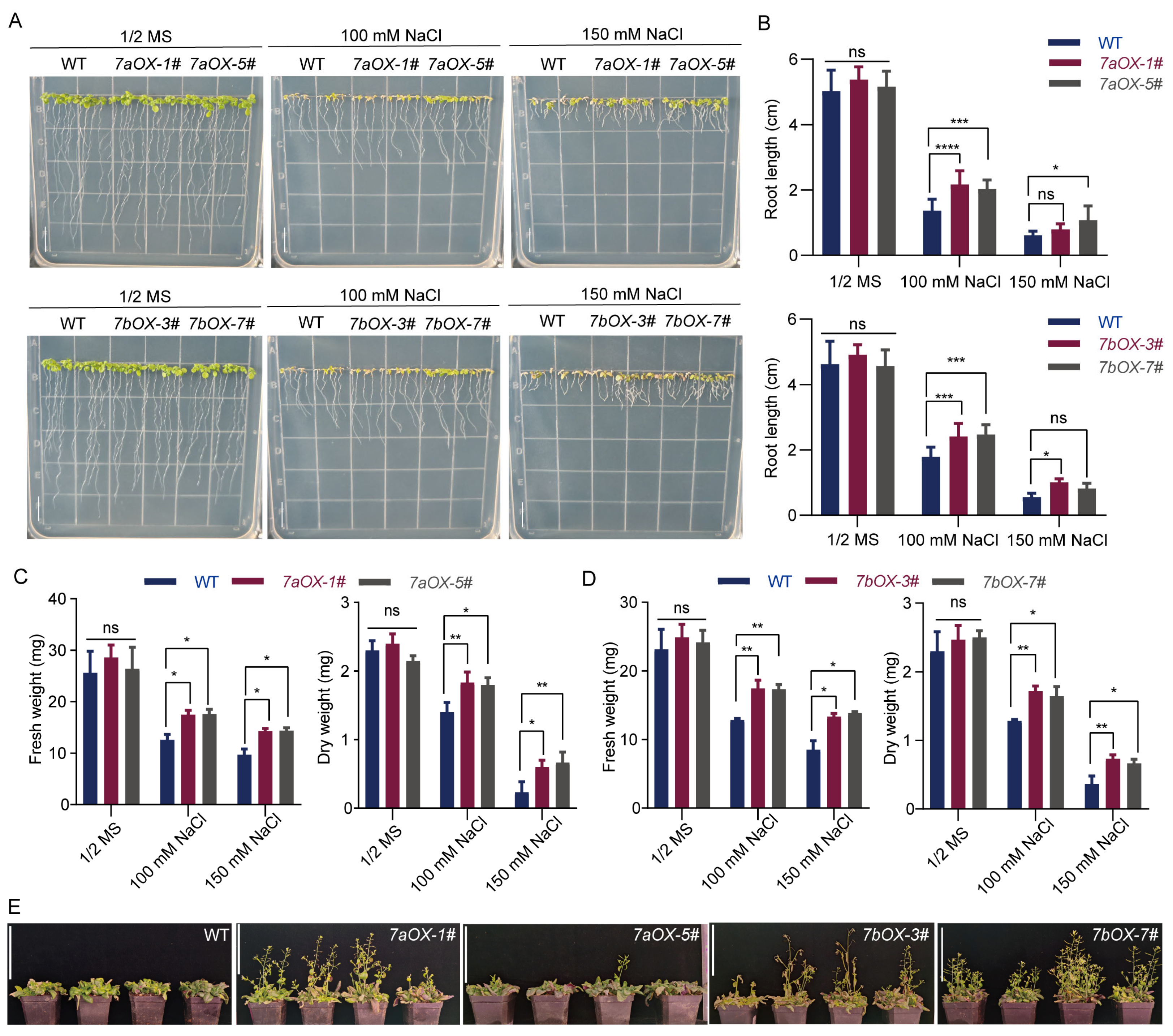
2.8. Overexpression of SiNHX7 Enhances Salt Tolerance of Transgenic Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Nomenclature of NHX Gene Family Members in Foxtail Millet
4.3. Construct the Phylogenetic Tree of the NHX Gene Family
4.4. Examination of Physicochemical Characteristics of NHX Gene Family Proteins in Foxtail Millet
4.5. Chromosome Mapping, Gene Structure, and Conserved Domain Analysis of the SiNHX Gene
4.6. Prediction of Cis-Acting Elements in the SiNHX Gene Promoter
4.7. Collinearity Analysis of the NHX Gene Family in Foxtail Millet
4.8. Construction and Genetic Transformation of Gene Overexpression Vector
4.9. RNA Extraction, cDNA Synthesis, and RT-qPCR
4.10. Analysis of Seed Germination Rate
4.11. Salt Treatment of Transgenic Arabidopsis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Litalien, A.; Zeeb, B. Curing the earth: A review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci. Total Environ. 2020, 698, 134235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Zhou, H. Plant salt response: Perception, signaling, and tolerance. Front. Plant Sci. 2023, 13, 1053699. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Li, J.; Yang, Y.; Jiang, C.; Guo, Y. Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol. 2024, 66, 303–329. [Google Scholar] [CrossRef] [PubMed]
- Haque, T.; Bhaskara, G.B.; Yin, J.; Bonnette, J.; Juenger, T.E. Natural variation in growth and leaf ion homeostasis in response to salinity stress in Panicum hallii. Front. Plant Sci. 2022, 13, 1019169. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R.; Mulet, J.M.; Rios, G.; Marquez, J.A.; De Larrinoa, I.F.; Leube, M.P.; Mendizabal, I.; Pascual-Ahuir, A.; Proft, M.; Ros, R. A glimpse of the mechanisms of ion homeostasis during salt stress. J. Exp. Bot. 1999, 50, 1023–1036. [Google Scholar] [CrossRef]
- Fu, H.; Yang, Y. How plants tolerate salt stress. Curr. Issues Mol. Biol. 2023, 45, 5914–5934. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995, 109, 735. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.J.; Miklavcic, S.J. A comprehensive biophysical model of ion and water transport in plant roots. III. Quantifying the energy costs of ion transport in salt-stressed roots of Arabidopsis. Front. Plant Sci. 2020, 11, 865. [Google Scholar] [CrossRef] [PubMed]
- Dragwidge, J.M.; Ford, B.A.; Ashnest, J.R.; Das, P.; Gendall, A.R. Two endosomal NHX-type Na+/H+ antiporters are involved in auxin-mediated development in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1660–1669. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Song, H.; Zhang, L. New insight into plant saline-alkali tolerance mechanisms and application to breeding. Int. J. Mol. Sci. 2022, 23, 16048. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Pandey, G.K.; Tuteja, N. Calcium-and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 2008, 471, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Petrov, V.; Yun, D.-J.; Gechev, T. Revisiting plant salt tolerance: Novel components of the SOS pathway. Trends Plant Sci. 2023, 28, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Waditee, R.; Hibino, T.; Tanaka, Y.; Nakamura, T.; Incharoensakdi, A.; Takabe, T. Halotolerant Cyanobacterium Aphanothece halophytica Contains an Na+/H+ Antiporter, Homologous to Eukaryotic Ones, with Novel Ion Specificity Affected by C-terminal Tail. J. Biol. Chem. 2001, 276, 36931–36938. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Ohto, M.-a.; Esumi, T.; Tajima, H.; Zhu, Z.; Cagnac, O.; Belmonte, M.; Peleg, Z.; Yamaguchi, T.; Blumwald, E. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 2011, 23, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Tajima, H.; Liang, Y.-C.; Ohto, M.-a.; Ushijima, K.; Nakano, R.; Esumi, T.; Coku, A.; Belmonte, M.; Blumwald, E. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 2011, 23, 3482–3497. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Zhang, S.; Gong, H.; Tajima, H.; Blumwald, E. Cation specificity of vacuolar NHX-type cation/H+ antiporters. Plant physiol. 2019, 179, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Hamaji, K.; Nagira, M.; Yoshida, K.; Ohnishi, M.; Oda, Y.; Uemura, T.; Goh, T.; Sato, M.H.; Morita, M.T.; Tasaka, M. Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2023–2033. [Google Scholar] [CrossRef] [PubMed]
- Dragwidge, J.M.; Scholl, S.; Schumacher, K.; Gendall, A.R. NHX-type Na+ (K+)/H+ antiporters are required for TGN/EE trafficking and endosomal ion homeostasis in Arabidopsis thaliana. J. Cell Sci. 2019, 132, jcs226472. [Google Scholar] [CrossRef] [PubMed]
- Al-Harrasi, I.; Jana, G.A.; Patankar, H.V.; Al-Yahyai, R.; Rajappa, S.; Kumar, P.P.; Yaish, M.W. A novel tonoplast Na+/H+ antiporter gene from date palm (PdNHX6) confers enhanced salt tolerance response in Arabidopsis. Plant Cell Rep. 2020, 39, 1079–1093. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Blumwald, E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 2014, 22, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ratner, A.; Jacoby, B. Effect of K+, its counter anion, and pH on sodium efflux from barley root tips. J. Exp. Bot. 1976, 27, 843–852. [Google Scholar] [CrossRef]
- Fukuda, A.; Nakamura, A.; Tanaka, Y. Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1999, 1446, 149–155. [Google Scholar] [CrossRef]
- Gaxiola, R.A.; Rao, R.; Sherman, A.; Grisafi, P.; Alper, S.L.; Fink, G.R. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 1999, 96, 1480–1485. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-H.; Zhang, B.; Xu, Z.-Q. Salt tolerance conferred by overexpression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1 in common buckwheat (Fagopyrum esculentum). Transgenic Res. 2008, 17, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef] [PubMed]
- Reguera, M.; Bassil, E.; Blumwald, E. Intracellular NHX-type cation/H+ antiporters in plants. Mol. Plant 2014, 7, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Colin, L.; Ruhnow, F.; Zhu, J.-K.; Zhao, C.; Zhao, Y.; Persson, S. The cell biology of primary cell walls during salt stress. Plant Cell 2023, 35, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Reguera, M.; Bassil, E.; Tajima, H.; Wimmer, M.; Chanoca, A.; Otegui, M.S.; Paris, N.; Blumwald, E. pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 2015, 27, 1200–1217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-X.; Blumwald, E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 2001, 19, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Leidi, E.O.; Barragán, V.; Rubio, L.; El-Hamdaoui, A.; Ruiz, M.T.; Cubero, B.; Fernández, J.A.; Bressan, R.A.; Hasegawa, P.M.; Quintero, F.J. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010, 61, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.A.; Zafar, Y.; Iqbal, J.; Iqbal, M.M.; Rashid, U.; Ali, G.M.; Arif, A.; Nazir, F. Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea L.) improves salt and drought tolerence. Mol. Biotechnol. 2011, 49, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Li, Q.; Wang, H.; Zhang, J.; Du, J.; Feng, H.; Blumwald, E.; Yu, L.; Xu, G. Two NHX-type transporters from Helianthus tuberosus improve the tolerance of rice to salinity and nutrient deficiency stress. Plant Biotechnol. J. 2018, 16, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; An, J.; Xu, H.; Chen, J.; Pan, L.; Zhao, R.; Wang, N.; Gai, J.; Li, Y. A soybean sodium/hydrogen exchanger GmNHX6 confers plant alkaline salt tolerance by regulating Na+/K+ homeostasis. Front. Plant Sci. 2022, 13, 938635. [Google Scholar] [CrossRef] [PubMed]
- Barragán, V.; Leidi, E.O.; Andrés, Z.; Rubio, L.; De Luca, A.; Fernandez, J.A.; Cubero, B.; Pardo, J.M. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef] [PubMed]
- Chanroj, S.; Wang, G.; Venema, K.; Zhang, M.W.; Delwiche, C.F.; Sze, H. Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front. Plant Sci. 2012, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Ford, B.A.; Ernest, J.R.; Gendall, A.R. Identification and characterization of orthologs of AtNHX5 and AtNHX6 in Brassica napus. Front. Plant Sci. 2012, 3, 208. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Jia, G.; Zhi, H.; Niu, Z.; Chai, Y.; Li, W.; Wang, Y.; Li, H.; Lu, P.; Zhao, B. Genetic diversity and population structure of Chinese foxtail millet [Setaria italica (L.) Beauv.] landraces. G3 Genes Genomes Genet. 2012, 2, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Brutnell, T.P.; Wang, L.; Swartwood, K.; Goldschmidt, A.; Jackson, D.; Zhu, X.-G.; Kellogg, E.; Van Eck, J. Setaria viridis: A model for C4 photosynthesis. Plant Cell 2010, 22, 2537–2544. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Tang, S.; Zhi, H.; Chen, J.; Zhang, J.; Liang, H.; Alam, O.; Li, H.; Zhang, H.; Xing, L. A graph-based genome and pan-genome variation of the model plant Setaria. Nat. Genet. 2023, 55, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Diao, X.; Schnable, J.; Bennetzen, J.L.; Li, J. Initiation of Setaria as a model plant. Front. Agric. Sci. Eng. 2014, 1, 16–20. [Google Scholar] [CrossRef]
- Sharma, N.; Niranjan, K. Foxtail millet: Properties, processing, health benefits, and uses. Food Rev. Int. 2018, 34, 329–363. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Zhu, J.-K. Thriving under stress: How plants balance growth and the stress response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Muthamilarasan, M.; Prasad, M. Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell Tissue Organ Cult. 2013, 115, 13–22. [Google Scholar] [CrossRef]
- Lu, K.-K.; Song, R.-F.; Guo, J.-X.; Zhang, Y.; Zuo, J.-X.; Chen, H.-H.; Liao, C.-Y.; Hu, X.-Y.; Ren, F.; Lu, Y.-T. CycC1; 1–WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis. Plant Cell 2023, 35, 2570–2591. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|
| SiNHX1 | Seita.2G160100.1 | 26,780.57 | 5.26 | 49.12 | 115.37 | 0.821 |
| SiNHX2 | Seita.2G160200.1 | 32,416.94 | 5.31 | 45.86 | 84.56 | 0.062 |
| SiNHX3 | Seita.2G249400.1 | 53,875.11 | 5.81 | 40.57 | 102.49 | 0.367 |
| SiNHX4 | Seita.2G422800.1 | 59,209.59 | 8.63 | 36.27 | 110.65 | 0.588 |
| SiNHX5 | Seita.3G038800.1 | 59,818.53 | 7.68 | 31.02 | 115.68 | 0.645 |
| SiNHX6 | Seita.3G409000.1 | 128,696.73 | 6.65 | 43.34 | 102.63 | 0.052 |
| SiNHX7 | Seita.4G138500.1 | 58,178.09 | 8.37 | 31.47 | 107.43 | 0.605 |
| SiNHX8 | Seita.7G006000.1 | 21,103.35 | 9.87 | 46.25 | 64.54 | −0.514 |
| SiNHX9 | Seita.8G215400.1 | 59,596.85 | 8.99 | 38.46 | 111.41 | 0.548 |
| Gene Name | Gene ID | Alpha Helix (%) | Extended Strand (%) | Beta Turn (%) | Random Coil (%) | Subcellular Localization |
|---|---|---|---|---|---|---|
| SiNHX1 | Seita.2G160100.1 | 47.97 | 17.89 | 0 | 34.15 | Vacuole |
| SiNHX2 | Seita.2G160200.1 | 54.01 | 12.54 | 0 | 33.45 | Vacuole |
| SiNHX3 | Seita.2G249400.1 | 51.21 | 12.15 | 0 | 36.64 | Vacuole |
| SiNHX4 | Seita.2G422800.1 | 48.98 | 12.80 | 0 | 38.22 | Vacuole |
| SiNHX5 | Seita.3G038800.1 | 53.66 | 12.82 | 0 | 33.52 | Vacuole |
| SiNHX6 | Seita.3G409000.1 | 55.17 | 9.40 | 0 | 35.43 | Cell membrane |
| SiNHX7 | Seita.4G138500.1 | 49.43 | 13.02 | 0 | 37.55 | Vacuole |
| SiNHX8 | Seita.7G006000.1 | 40.98 | 15.85 | 0 | 43.17 | Vacuole |
| SiNHX9 | Seita.8G215400.1 | 45.70 | 12.98 | 0 | 41.32 | Vacuole |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, X.; Chen, D.-Y.; Sun, M.; Zhang, J.; Zhang, M.; Wu, H.; Wang, H.; Dong, S.; Yuan, X.; Li, X.; et al. Genome-Wide Identification of the SiNHX Gene Family in Foxtail Millet (Setaria Italica) and Functional Characterization of SiNHX7 in Arabidopsis. Int. J. Mol. Sci. 2025, 26, 7139. https://doi.org/10.3390/ijms26157139
Chu X, Chen D-Y, Sun M, Zhang J, Zhang M, Wu H, Wang H, Dong S, Yuan X, Li X, et al. Genome-Wide Identification of the SiNHX Gene Family in Foxtail Millet (Setaria Italica) and Functional Characterization of SiNHX7 in Arabidopsis. International Journal of Molecular Sciences. 2025; 26(15):7139. https://doi.org/10.3390/ijms26157139
Chicago/Turabian StyleChu, Xiaoqian, Dan-Ying Chen, Mengmeng Sun, Jiajing Zhang, Minghua Zhang, Hejing Wu, Hongzhi Wang, Shuqi Dong, Xiangyang Yuan, Xiaorui Li, and et al. 2025. "Genome-Wide Identification of the SiNHX Gene Family in Foxtail Millet (Setaria Italica) and Functional Characterization of SiNHX7 in Arabidopsis" International Journal of Molecular Sciences 26, no. 15: 7139. https://doi.org/10.3390/ijms26157139
APA StyleChu, X., Chen, D.-Y., Sun, M., Zhang, J., Zhang, M., Wu, H., Wang, H., Dong, S., Yuan, X., Li, X., Gao, L., Yang, G., & Wang, J.-G. (2025). Genome-Wide Identification of the SiNHX Gene Family in Foxtail Millet (Setaria Italica) and Functional Characterization of SiNHX7 in Arabidopsis. International Journal of Molecular Sciences, 26(15), 7139. https://doi.org/10.3390/ijms26157139






