Optimization of Chromosome Preparation and Karyotype Analysis of Winter Turnip Rape (Brassica rape L.)
Abstract
1. Introduction
2. Results
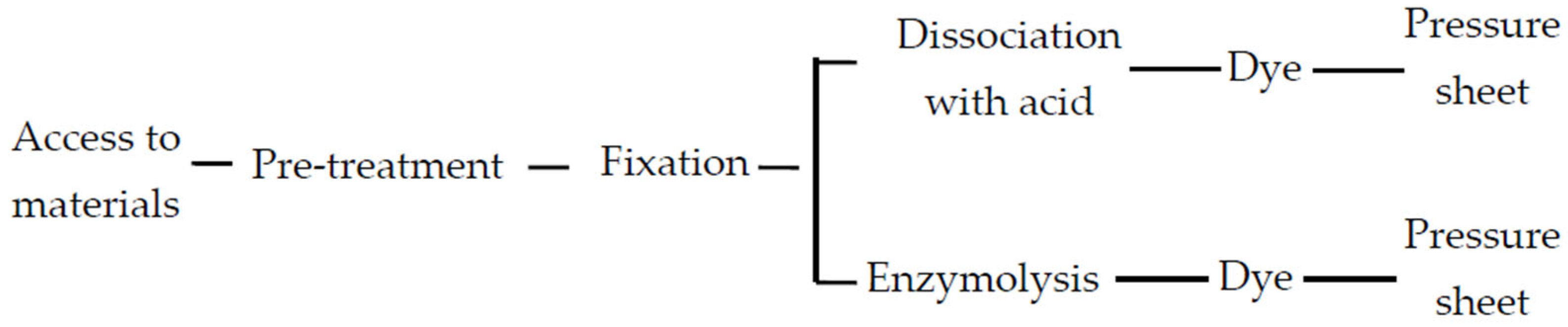
2.1. Optimization of Chromosome Preparation Techniques
2.1.1. Determine the Best Time to Take Materials
2.1.2. Effect of Different Pretreatment Times on Chromosomes
2.1.3. Effect of Fixation Time on Chromosome Preparation Efficiency
2.1.4. Effect of Different Dissociation Methods and Times on Chromosome Preparation Efficiency
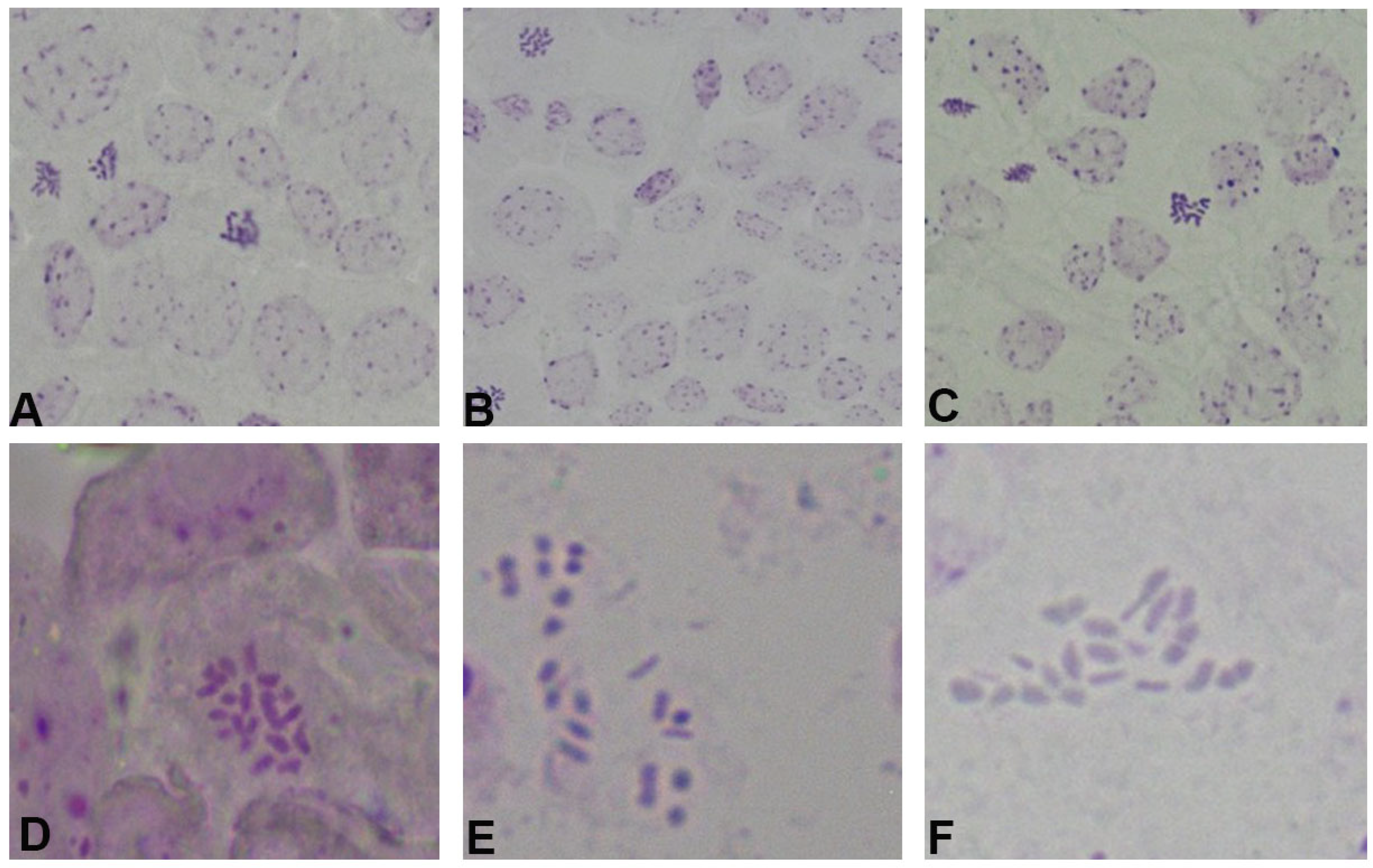
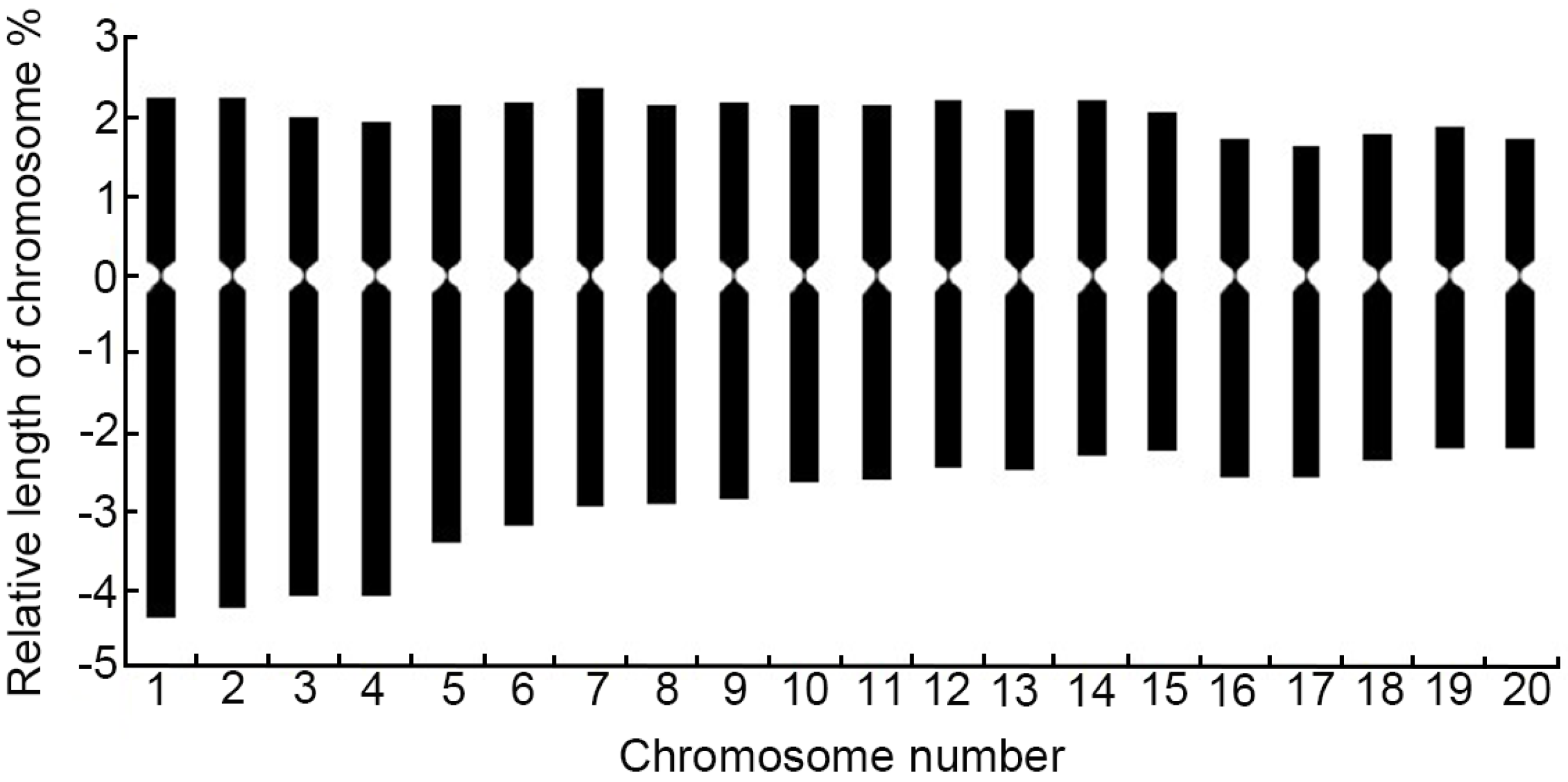
2.2. Karyotype Analysis of Winter Turnip Rape Chromosomes
2.2.1. Number of Chromosomes in Rapeseed
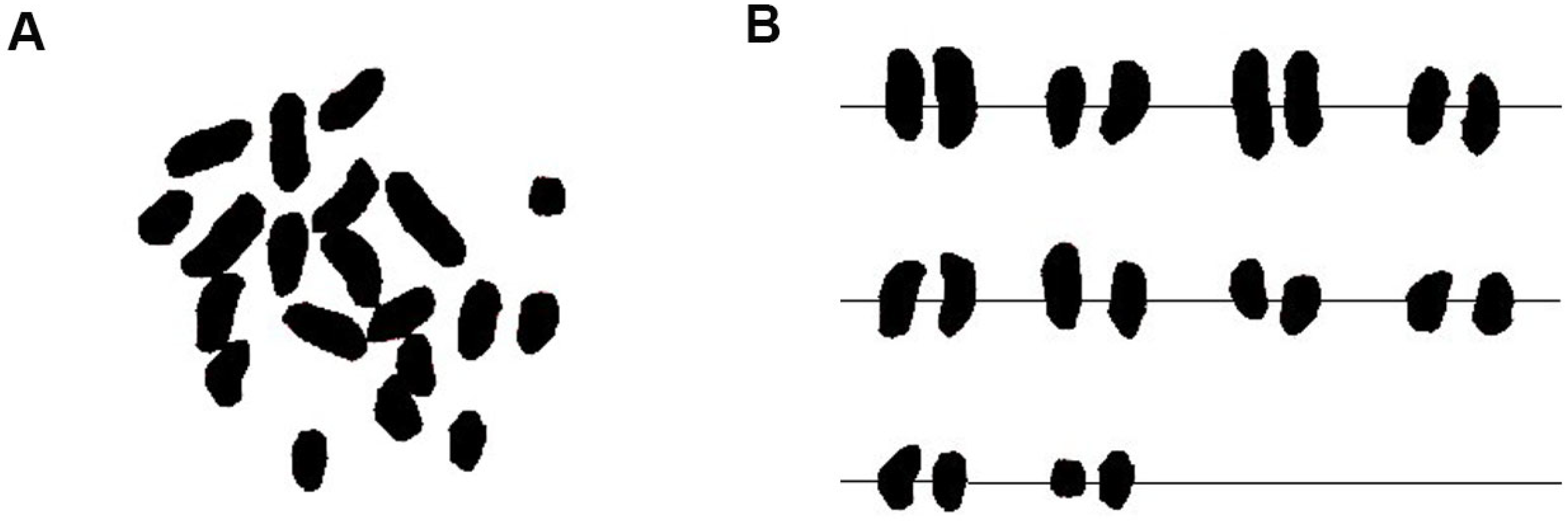
2.2.2. Karyotype Analysis
3. Discussion
3.1. Staining and Slicing Technology
3.2. Chromosome Karyotype Analysis
4. Materials and Methods
4.1. Optimization of Material Collection Time
4.2. Optimization of Preprocessing Time
4.3. Optimization of Fixed Time
4.4. Optimization of the Dissociation Stage
4.5. Staining and Microscopic Examination
4.6. Karyotype Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tao, X.; Zhao, Y.; Ma, L.; Wu, J.; Zeng, R.; Jiao, J.; Li, R.; Ma, W.; Lian, Y.; Wang, W.; et al. Cloning and functional analysis of the BrCUC2 gene in Brassica rapa L. Front. Plant Sci. 2023, 14, 1274567. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Zhang, R.; Sun, W.; Zhang, J.; Wang, H. Study on climatic suitability for winter rapeseed planting in arid and cold regions in North China. Sci. Agri. Sini 2014, 47, 2541–2551. (In Chinese) [Google Scholar]
- Ma, L.; Wang, X.; Pu, Y.; Wu, J.; Coulter, J.A.; Li, X.; Wang, L.; Liu, L.; Fang, Y.; Niu, Z.; et al. Ecological and economic benefits of planting winter rapeseed (Brassica rapa L.) in the wind erosion area of northern China. Sci. Rep. 2019, 9, 20272. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xu, X.D.; Liu, L.; Ma, L.; Pu, Y.; Wang, W.; Hua, X.Y.; Song, J.M.; Liu, K.; Lu, G.; et al. A Chromosome Level Genome Assembly of a Winter Turnip Rape (Brassica rapa L.) to Explore the Genetic Basis of Cold Tolerance. Front. Plant Sci. 2022, 13, 936958. [Google Scholar] [CrossRef]
- Niu, Z.; Liu, L.; Yue, J.; Wu, J.; Wang, W.; Pu, Y.; Ma, L.; Fang, Y.; Sun, W. Genome-Wide Identification of GSTs Gene Family and Functional Analysis of BraGSTF2 of Winter Rapeseed (Brassica rapa L.) under Cold Stress. Genes 2023, 14, 1689. [Google Scholar] [CrossRef]
- Niu, Z.; Liu, L.; Pu, Y.; Ma, L.; Wu, J.; Hu, F.; Fang, Y.; Li, X.; Sun, W.; Wang, W.; et al. iTRAQ-based quantitative proteome analysis insights into cold stress of Winter Rapeseed (Brassica rapa L.) grown in the field. Sci. Rep. 2021, 11, 23434. [Google Scholar] [CrossRef]
- Dong, X.; Liu, Z.; Wei, J.; Zheng, G.; Li, H.; Wang, Y.; Tian, H.; Cui, J.; Wu, Z.; Cao, X.; et al. The BrAFP1 promoter drives gene-specific expression in leaves and stems of winter rapeseed (Brassica rapa L.) under cold induction. Plant Sci. 2023, 331, 111669. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, Y.; Jiang, J.; Zhang, F.; Ma, L.; Wu, D.; Wang, Y.; Sun, W. Identification of cold stress responsive microRNAs in two winter turnip rape (Brassica rapa L.) by high throughput sequencing. BMC Plant Biol. 2018, 18, 52. [Google Scholar] [CrossRef]
- Sattler, M.C.; de Oliveira, S.C.; Mendonça, M.A.C.; Clarindo, W.R. Coffea cytogenetics: From the first karyotypes to the meeting with genomics. Planta 2022, 255, 112. [Google Scholar] [CrossRef]
- Kadluczka, D.; Grzebelus, E. Using carrot centromeric repeats to study karyotype relationships in the genus Daucus (Apiaceae). BMC Genom. 2021, 22, 508. [Google Scholar] [CrossRef]
- Mandáková, T.; Schranz, M.E.; Sharbel, T.F.; de Jong, H.; Lysak, M.A. Karyotype evolution in apomictic Boechera and the origin of the aberrant chromosomes. Plant J. 2015, 82, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Wang, D.; Wu, Y.; Guo, X.; Zhang, Y.; Lu, X. Karyotype analysis of Gazania rigens varieties. Hortc. Plant J. 2016, 2, 279–283. [Google Scholar] [CrossRef]
- Chen, N.; Wang, Y.; He, M.; An, F.; Wang, J.; Song, C. Identification of chromosome ploidy and karyotype analysis of cherries (Prunus pseudocerasus Lindl.) in Guizhou. PeerJ 2024, 12, e18668. [Google Scholar] [CrossRef] [PubMed]
- Kulaz, H.; Najafi, S.; Tuncturk, R.; Tuncturk, M.; Albalawi, M.A.; Alalawy, A.I.; Oyouni, A.A.A.; Alasmari, A.; Poczai, P.; Sayyed, R.Z. Analysis of Nuclear DNA Content and Karyotype of Phaseolus vulgaris L. Genes 2022, 14, 47. [Google Scholar] [CrossRef]
- Jha, T.B. Karyotype diversity in cultivated and wild Indian rice through EMA-based chromosome analysis. J. Genet. 2021, 100, 81. [Google Scholar] [CrossRef]
- Morawetz, W. Systematics and karyoevolution in Magnoliidae: Tetrameranthus as compared with other Annonaceae genera of the same chromosome number. Plant Syst. Evol. 1986, 154, 147–173. [Google Scholar] [CrossRef]
- Yang, G.; Han, Y.; Yin, H.; Li, X.; Wang, H.; Bao, Y. Cytogenetic Identification and Molecular Marker Analysis of Two Wheat-Thinopyrum ponticum Translocations with Stripe Rust Resistance. Plants 2024, 14, 27. [Google Scholar] [CrossRef]
- Yang, Z.J.; Li, G.R.; Feng, J.; Jiang, H.R.; Ren, Z.L. Molecular cytogenetic characterization and disease resistance observation of wheat-Dasypyrum breviaristatum partial amphiploid and its derivatives. Hereditas 2005, 142, 80–85. [Google Scholar] [CrossRef]
- McManus, B.A.; Coleman, D.C. Molecular epidemiology, phylogeny and evolution of Candida albicans. Infect. Genet. Evol. 2014, 21, 166–178. [Google Scholar] [CrossRef]
- Cao, R.B.; Chen, R.; Liao, K.X.; Li, H.; Xu, G.B.; Jiang, X.L. Karyotype and LTR-RTs analysis provide insights into oak genomic evolution. BMC Genom. 2024, 25, 328. [Google Scholar] [CrossRef]
- Weiss-Schneeweiss, H.; Tremetsberger, K.; Schneeweiss, G.M.; Parker, J.S.; Stuessy, T.F. Karyotype diversification and evolution in diploid and polyploid South American Hypochaeris (Asteraceae) inferred from rDNA localization and genetic fingerprint data. Ann. Bot. 2008, 101, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Kitano, J. Tempo and mode in karyotype evolution revealed by a probabilistic model incorporating both chromosome number and morphology. PLoS Genet. 2021, 17, e1009502. [Google Scholar] [CrossRef] [PubMed]
- Moraes, A.P.; Olmos Simões, A.; Ojeda Alayon, D.I.; de Barros, F.; Forni-Martins, E.R. Detecting Mechanisms of Karyotype Evolution in Heterotaxis (Orchidaceae). PLoS ONE 2016, 11, e0165960. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Luo, L.; Yu, Z.; Lei, J.; Zhang, M.; Deng, Z. Repetitive Sequence Barcode Probe for Karyotype Analysis in Tripidium arundinaceum. Int. J. Mol. Sci. 2022, 6, 23. [Google Scholar] [CrossRef]
- Hasterok, R.; Wang, K.; Jenkins, G. Progressive refinement of the karyotyping of Brachypodium genomes. New Phytol. 2020, 227, 1668–1675. [Google Scholar] [CrossRef]
- Wang, J.; Yuan, M.; Feng, Y.; Zhang, Y.; Bao, S.; Hao, Y.; Ding, Y.; Gao, X.; Yu, Z.; Xu, Q.; et al. A common whole-genome paleotetraploidization in Cucurbitales. Plant Physiol. 2022, 190, 2430–2448. [Google Scholar] [CrossRef]
- Liu, D.; Xia, X.; Wu, Y.M.; Gu, J.H.; Tian, Y.X.; Sun, B. Analysis of factors influencing chromosome preparation effectiveness in plants. Zhejiang Agric. Sci. 2015, 56, 1654–1657. (In Chinese) [Google Scholar] [CrossRef]
- Xiao, S.X.; Huang, Y.R.; Gao, L.; Jia, G.X.; Gao, X. Optimization of chromosome staining techniques and karyotype analysis in Althaea rosea root tips. Mol. Plant Breed. 2021, 19, 5800–5807. (In Chinese) [Google Scholar] [CrossRef]
- Vicente, A.R.; Saladié, M.; Rose, J.K.; Labavitch, J.M. The linkage between cell wall metabolism and fruit softening: Looking to the future. J. Sci. Food Agric. 2007, 87, 1435–1448. [Google Scholar] [CrossRef]
- Zhang, T.; Tang, H.; Vavylonis, D.; Cosgrove, D.J. Disentangling loosening from softening: Insights into primary cell wall structure. Plant J. 2019, 100, 1101–1117. [Google Scholar] [CrossRef]
- Fukui, K.; Nakayama, S. Plant Chromosomes: Laboratory Methods; CRC Press: Boca Raton, FL, USA, 1996. [Google Scholar]
- Kirov, I.; Divashuk, M.; Van Laere, K.; Soloviev, A.; Khrustaleva, L. An easy “SteamDrop” method for high quality plant chromosome preparation. Mol. Cytogenet. 2014, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Jiang, C.; Peng, H.; Shi, Q.; Guo, X.; Yuan, Y.; Huang, L. Analysis of the age of Panax ginseng based on telomere length and telomerase activity. Sci. Rep. 2015, 5, 7985. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, S.; Wang, Y.; Jiang, D.; Zhang, Y.; Hu, L.; Zhu, Y.; Jia, Q.; Yin, J.; Liu, Y.; et al. Field performance of disease-free plants of ginger produced by tissue culture and agronomic, cytological, and molecular characterization of the morphological variants. Agronomy 2022, 13, 74. [Google Scholar] [CrossRef]
- Lima-de-Faria, A. Classification of genes, rearrangements and chromosomes according to the chromosome field. Hereditas 1980, 93, 1–46. [Google Scholar] [CrossRef]
- Lei, T.; Zhao, C.Y.; Li, F.; Gao, S.P. Optimization of chromosome staining in Plumbago auriculata and karyotype analysis. In China Ornamental Horticulture Research Progress; China Forestry Publishing House: Beijing, China, 2016. (In Chinese) [Google Scholar]
- Hu, F.R.; Liu, H.H.; Wang, F.; Bao, R.L.; Liu, G.X. Root tip chromosome karyotype analysis of hyacinth cultivars. Genet. Mol. Res. 2015, 14, 10863–10876. [Google Scholar] [CrossRef]
- Vimala, Y.; Lavania, S.; Lavania, U.C. Chromosome change and karyotype differentiation–implications in speciation and plant systematics. Nucleus 2021, 64, 33–54. [Google Scholar] [CrossRef]
- Wang, Y.; Hua, L.Y.; Qian, G.Z. Optimization of chromosome preparation technology and karyotype analysis in root tips of Pingyitiancha. Jiangsu Agric. Sci. 2019, 47, 91–96. (In Chinese) [Google Scholar]
- Xia, X.; Gu, J.H.; Liu, D.; Wu, Y.M.; Tian, Y.X.; Chen, Q.; Sun, B. Optimization of chromosome preparation and karyotype analysis in Brassica alboglabra. J. Zhejiang Univ. 2016, 42, 527–534. (In Chinese) [Google Scholar]
- Sun, Y.; Li, Z.P.; Fang, Y.F.; Jiang, N.N.; Wang, Y. Chromosome preparation and karyotype analysis in Ligustrum lucidum. Shandong For. Sci. Technol. 2019, 49, 1–5. (In Chinese) [Google Scholar]
- Tu, H.Y.; Zhang, A.L.; Xiao, W.; Chen, D.; Zeng, H.M. Optimization of chromosome preparation methods in Zingiberaceae plants. J. Trop. Crops 2016, 37, 1017–1021. (In Chinese) [Google Scholar]
- Eltoum, I.; Fredenburgh, J.; Myers, R.B.; Grizzle, W.E. Introduction to the theory and practice of fixation of tissues. J. Histotechnol. 2001, 24, 173–190. [Google Scholar] [CrossRef]
- Yang, H.B.; Rao, L.B.; Guo, H.Y.; Duan, H.P.; Chen, Y.T. Karyotype analysis of five Alnus species. J. Plant Genet. Resour. 2013, 14, 1203–1207. (In Chinese) [Google Scholar]
- Shahriari, Z.; Heidari, B.; Dadkhodaie, A.; Richards, C.M. Analysis of karyotype, chromosome characteristics, variation in mucilage content and grain yield traits in Plantago ovata and P. psyllium species. Ind. Crop Prod. 2018, 123, 676–686. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef]
- Jiang, X.; Hu, Q.; Mei, D.; Li, X.; Xiang, L.; Al-Shehbaz, I.A.; Song, X.; Liu, J.; Lysak, M.A.; Sun, P. Chromosome fusions shaped karyotype evolution and evolutionary relationships in the model family Brassicaceae. Nat. Commun. 2025, 16, 4631. [Google Scholar] [CrossRef]
- Koenig, D.; Weigel, D. Beyond the thale: Comparative genomics and genetics of Arabidopsis relatives. Nat. Rev. Genet. 2015, 16, 285–298. [Google Scholar] [CrossRef]
- Senavongse, R.; Saensouk, S.; Saensouk, P. Comparative karyotype analysis in five strains of Colocasia esculenta (L.) Schott (Araceae) in Thailand. Cytologia 2018, 83, 169–173. [Google Scholar] [CrossRef]
- Prajitha, V.; Thoppil, J.E. Cytogenetic characterization of Amaranthus caudatus L. and Amaranthus hybridus subsp. cruentus (L.). Thell. Cytotechnology 2018, 70, 95–101. [Google Scholar] [CrossRef]
- Meng, Z.; Wang, F.; Xie, Q.; Li, R.; Shen, H.; Li, H. Reconstruction of karyotypic evolution in Saccharum spontaneum species by comparative oligo-FISH mapping. BMC Plant Biol. 2022, 22, 599. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold: London, UK, 1971. [Google Scholar]
- Wang, L.J.; Gao, M.D.; Sheng, M.Y.; Yin, J. Cluster analysis of karyotype similarity coefficients in Epimedium (Berberidaceae): Insights in the systematics and evolution. PhytoKeys 2020, 161, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.G.; Sun, H.; Li, Z.M. Chromosome data mining and its application in plant diversity research. Plant Sci. J. 2019, 37, 260–269. [Google Scholar]
- Peruzzi, L.; Eroğlu, H.E. Karyotype asymmetry: Again, how to measure and what to measure? Comp. Cytogenet. 2013, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Zhao, H.; Lu, S.; Yu, L.; Zhang, G.; Zhang, Y.; Yang, Q.Y.; Zhou, Y.; Wang, X.; Ma, W.; et al. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol. Plant 2021, 14, 470–487. [Google Scholar] [CrossRef]
- Kakui, Y.; Barrington, C.; Kusano, Y.; Thadani, R.; Fallesen, T.; Hirota, T.; Uhlmann, F. Chromosome arm length, and a species-specific determinant, define chromosome arm width. Cell Rep. 2022, 41, 111753. [Google Scholar] [CrossRef]
- Lin, X.X.; Chen, L.J.; Rong, C.Y.; Chen, K.Q.; Gao, G.J. Optimization of chromosome preparation and karyotype analysis in Panicum repens. Pratacult. Sci. 2021, 38, 89–98. (In Chinese) [Google Scholar]
- Zeng, J.; Geng, X.; Zhao, Z.; Zhou, W. Tipping the balance: The dynamics of stem cell maintenance and stress responses in plant meristems. Curr. Opin. Plant Biol. 2024, 78, 102510. [Google Scholar] [CrossRef]
- Han, C.; Xu, J.M.; Huang, F.F.; Su, Y.T. Optimization of chromosome pressing technology of eucalyptus. Jiangsu Agric. Sci. 2016, 44, 233–235. [Google Scholar]
- Li, M.X.; Zhang, C.F. Techniques for Chromosome Research in Plants; Northeast Forestry University: Harbin, China, 1991. [Google Scholar]
- Wu, M.; Chang, Y.; Hu, H.; Mu, R.; Zhang, Y.; Qin, X.; Duan, X.; Li, W.; Tu, H.; Zhang, W.; et al. LUBAC controls chromosome alignment by targeting CENP-E to attached kinetochores. Nat. Commun. 2019, 10, 273. [Google Scholar] [CrossRef]
- Liu, J.G.; Li, J.H.; Guo, H.L.; Geng, Y.R.; Wang, R.Y.; Hou, L.L. Optimization of chromosome tableting technology and karyotype analysis of Chionanthus retusus root tips. Mol. Plant Breed. 2020, 18, 8223–8232. [Google Scholar]
- Li, M.X.; Chen, R.Y. Standardization issues in plant karyotype analysis. J. Wuhan Bot. Res. 1985, 3, 297–302. (In Chinese) [Google Scholar]
- Arano, H. Cytological Studies in Subfamily Carduoideae of Japan. Bot. Mag. 1963, 76, 32–39. [Google Scholar] [CrossRef]
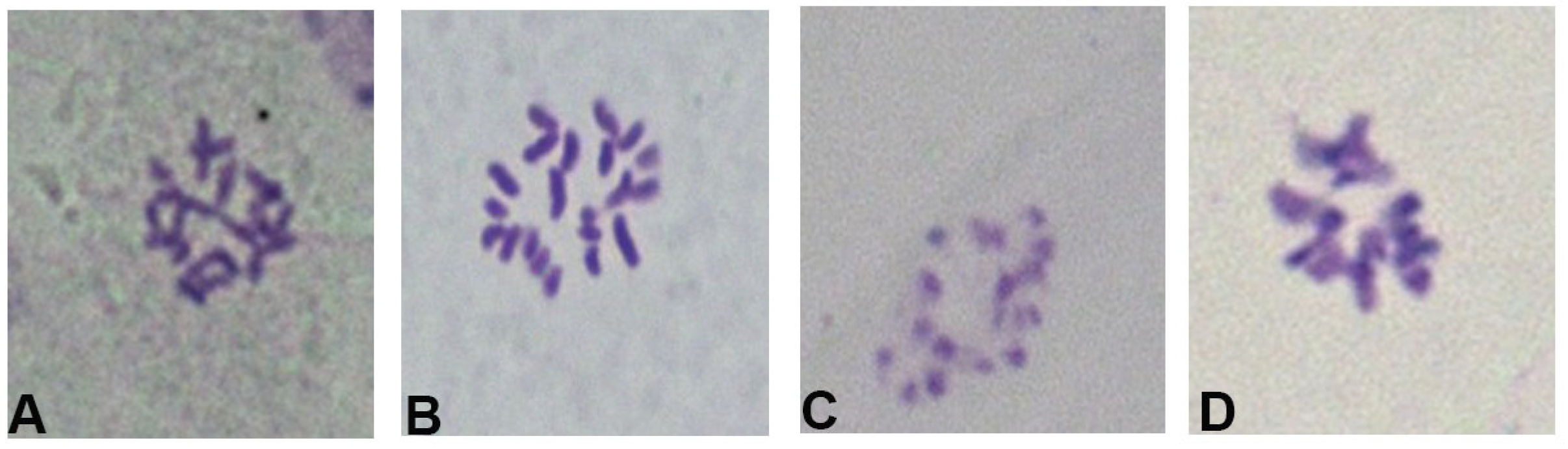
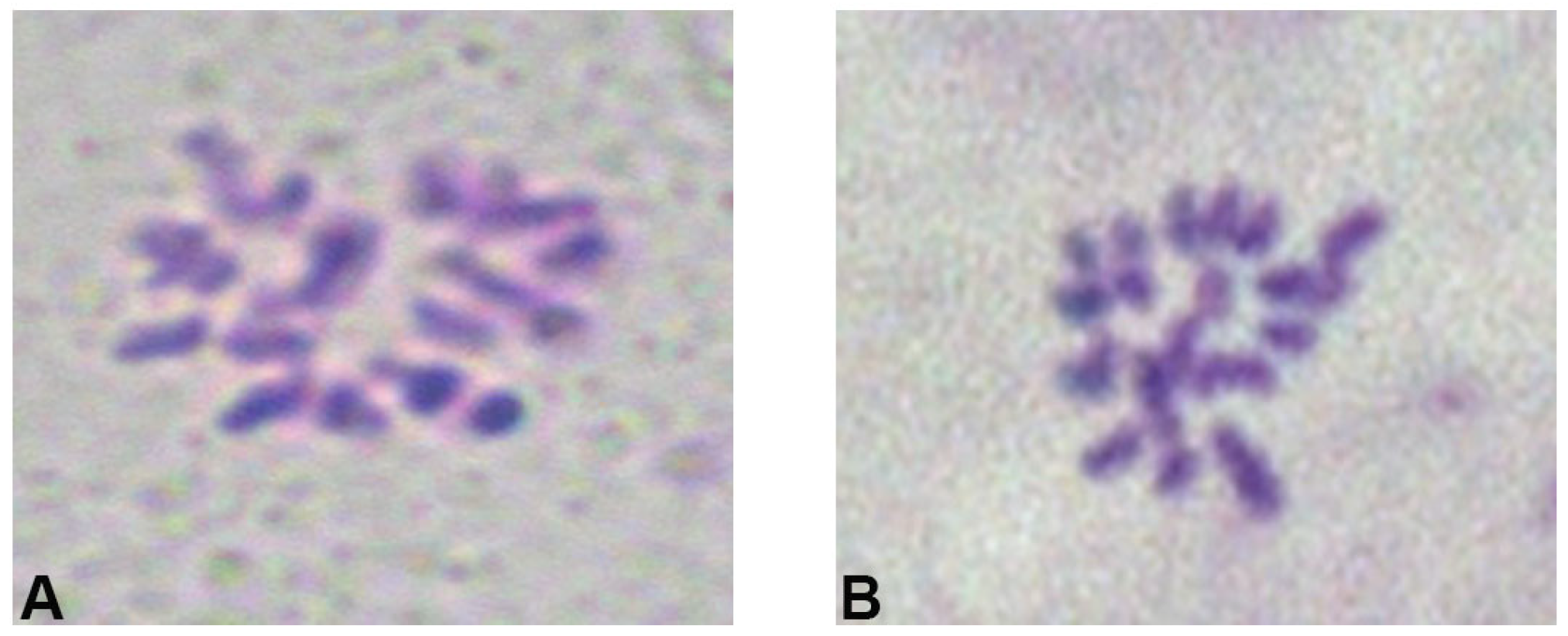
| Collection Time | Proportion of Metaphase Cell/% |
|---|---|
| 8:00–10:00 | 8.57 ± 0.72 |
| 10:00–12:00 | 8.33 ± 2.75 |
| 12:00–14:00 | 8.46 ± 2.33 |
| 14:00–16:00 | 8.45 ± 2.96 |
| 16:00–18:00 | 8.26 ± 0.61 |
| Number of Chromosomes | Relative Length/% | Index of Relative Length | Type of Length | Arm Ratio (Long Arm/Short Arm) | Type of Chromosome | ||
|---|---|---|---|---|---|---|---|
| Long Arm | Short Arm | Total Length | |||||
| 1 | 4.36 | 2.25 | 6.61 | 1.32 | L | 1.94 | sm |
| 2 | 4.26 | 2.25 | 6.51 | 1.30 | L | 1.89 | sm |
| 3 | 4.08 | 2.01 | 6.09 | 1.22 | M2 | 2.04 | sm |
| 4 | 4.08 | 1.94 | 6.02 | 1.20 | M2 | 2.11 | sm |
| 5 | 3.41 | 2.15 | 5.56 | 1.11 | M2 | 1.59 | m |
| 6 | 3.20 | 2.18 | 5.38 | 1.08 | M2 | 1.47 | m |
| 7 | 2.96 | 2.36 | 5.32 | 1.06 | M2 | 1.25 | m |
| 8 | 2.92 | 2.15 | 5.07 | 1.01 | M2 | 1.36 | m |
| 9 | 2.85 | 2.18 | 5.03 | 1.01 | M1 | 1.31 | m |
| 10 | 2.64 | 2.15 | 4.79 | 0.96 | M1 | 1.23 | m |
| 11 | 2.60 | 2.15 | 4.75 | 0.95 | M1 | 1.21 | m |
| 12 | 2.46 | 2.22 | 4.68 | 0.94 | M1 | 1.11 | m |
| 13 | 2.50 | 2.11 | 4.61 | 0.92 | M1 | 1.18 | m |
| 14 | 2.32 | 2.22 | 4.54 | 0.91 | M1 | 1.05 | m |
| 15 | 2.25 | 2.08 | 4.33 | 0.87 | M1 | 1.08 | m |
| 16 | 2.57 | 1.72 | 4.29 | 0.86 | M1 | 1.49 | m |
| 17 | 2.57 | 1.65 | 4.22 | 0.85 | M1 | 1.55 | m |
| 18 | 2.36 | 1.80 | 4.16 | 0.83 | M1 | 1.31 | m |
| 19 | 2.22 | 1.87 | 4.09 | 0.82 | M1 | 1.19 | m |
| 20 | 2.22 | 1.72 | 3.94 | 0.79 | M1 | 1.29 | m |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, T.; Zeng, X.; Xu, Y.; Zhang, F.; Ma, L.; Pu, Y.; Liu, L.; Wang, W.; Wu, J.; Sun, W.; et al. Optimization of Chromosome Preparation and Karyotype Analysis of Winter Turnip Rape (Brassica rape L.). Int. J. Mol. Sci. 2025, 26, 7127. https://doi.org/10.3390/ijms26157127
Fan T, Zeng X, Xu Y, Zhang F, Ma L, Pu Y, Liu L, Wang W, Wu J, Sun W, et al. Optimization of Chromosome Preparation and Karyotype Analysis of Winter Turnip Rape (Brassica rape L.). International Journal of Molecular Sciences. 2025; 26(15):7127. https://doi.org/10.3390/ijms26157127
Chicago/Turabian StyleFan, Tingting, Xiucun Zeng, Yaozhao Xu, Fei Zhang, Li Ma, Yuanyuan Pu, Lijun Liu, Wangtian Wang, Junyan Wu, Wancang Sun, and et al. 2025. "Optimization of Chromosome Preparation and Karyotype Analysis of Winter Turnip Rape (Brassica rape L.)" International Journal of Molecular Sciences 26, no. 15: 7127. https://doi.org/10.3390/ijms26157127
APA StyleFan, T., Zeng, X., Xu, Y., Zhang, F., Ma, L., Pu, Y., Liu, L., Wang, W., Wu, J., Sun, W., & Yang, G. (2025). Optimization of Chromosome Preparation and Karyotype Analysis of Winter Turnip Rape (Brassica rape L.). International Journal of Molecular Sciences, 26(15), 7127. https://doi.org/10.3390/ijms26157127






