Participants in Transcription–Replication Conflict and Their Role in Formation and Resolution of R-Loops
Abstract
1. Introduction
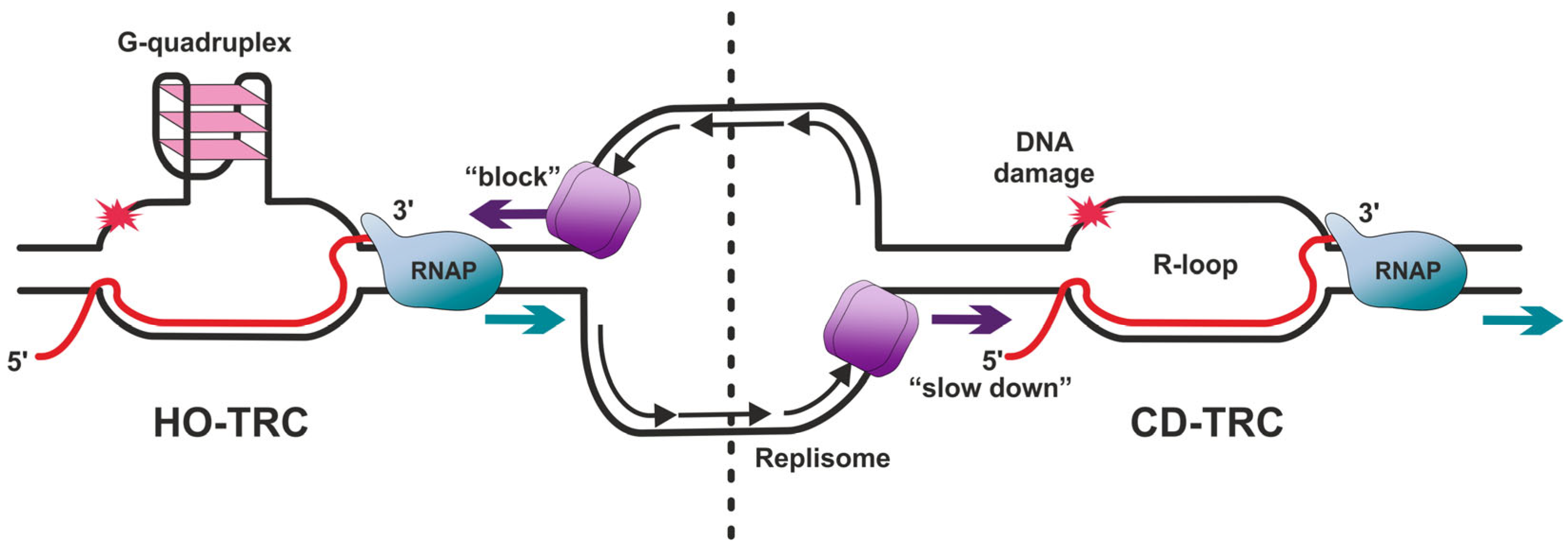
2. Dealing with TRCs
3. TRC-Induced Non-B DNA Structures
3.1. G-Quadruplexes
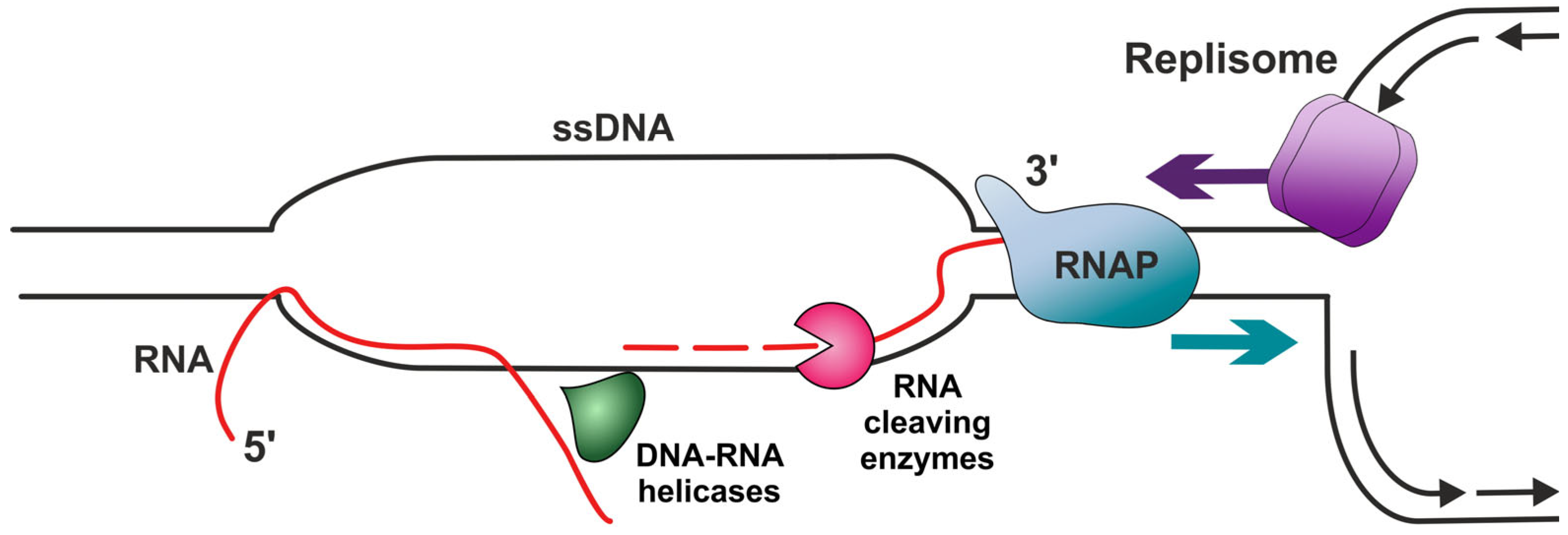
3.2. R-Loops
3.2.1. Factors Promoting R-Loop Formation
| Protein | Protein Function (Known Role in The Organism) | Possible Role in R-Loops Formation | Ref. |
|---|---|---|---|
| RAD51 (RecA *) | Plays an important role in homologous strand exchange, a key step in DNA repair, through homologous recombination (HR). Coats DNA-forming nucleoprotein filaments. Interacts with many partners. Participates in the Fanconi anemia (FA) pathway. | Possibly involved in the formation of trans R-loops. | [87,104] |
| RAD52 | Involved in DSB repair. Plays a key role in genetic recombination and DNA repair, promoting the annealing of complementary single-stranded DNA and stimulating RAD51 recombinase. | ||
| RNAP II (RNAP) | DNA-dependent RNA polymerase which synthesizes mRNA precursors and several functional non-coding RNAs. | Tends to accumulate together with R-loops at HO-TRC sites and acts as a main obstacle to replication fork progression. Extended pausing can stabilize R-loops. RNAP was demonstrated to partially protect short R-loop segments from RNase H1-facilitated cleavage. | [114,116,122] |
| DHX9 (also known as RHA) | RNA helicase A. Participates in the assembly of splicing factors onto nascent RNA. | Promotes the formation of R-loops in cells where splicing factors are absent. | [123] |
| PRC2 | Polycomb repressive complex. Exhibits histone methyltransferase activity and primarily methylates histone H3 on lysine 27. | Opens DNA bubbles and induces the formation of RNA–DNA hybrids. | [124] |
| RPA | Binds and stabilizes ssDNA intermediates that form during DNA replication or upon DNA stress. | Supposably promotes R-loop formation by binding to RNA. | [125] |
| TET1 | DNA dioxygenase. Catalyzes oxidation of epigenetic 5-methylcytosine to 5-hydroxymethylcytosine. | Catalytic activity in the region of transcribed genes leads to preferential formation of R-loops therein. | [86,126] |
3.2.2. Factors Facilitating R-Loop Suppression
Factors Preventing R-Loop Formation
- RNA-coating proteins
- Chromatin structure
- Topoisomerases
- PrimPol
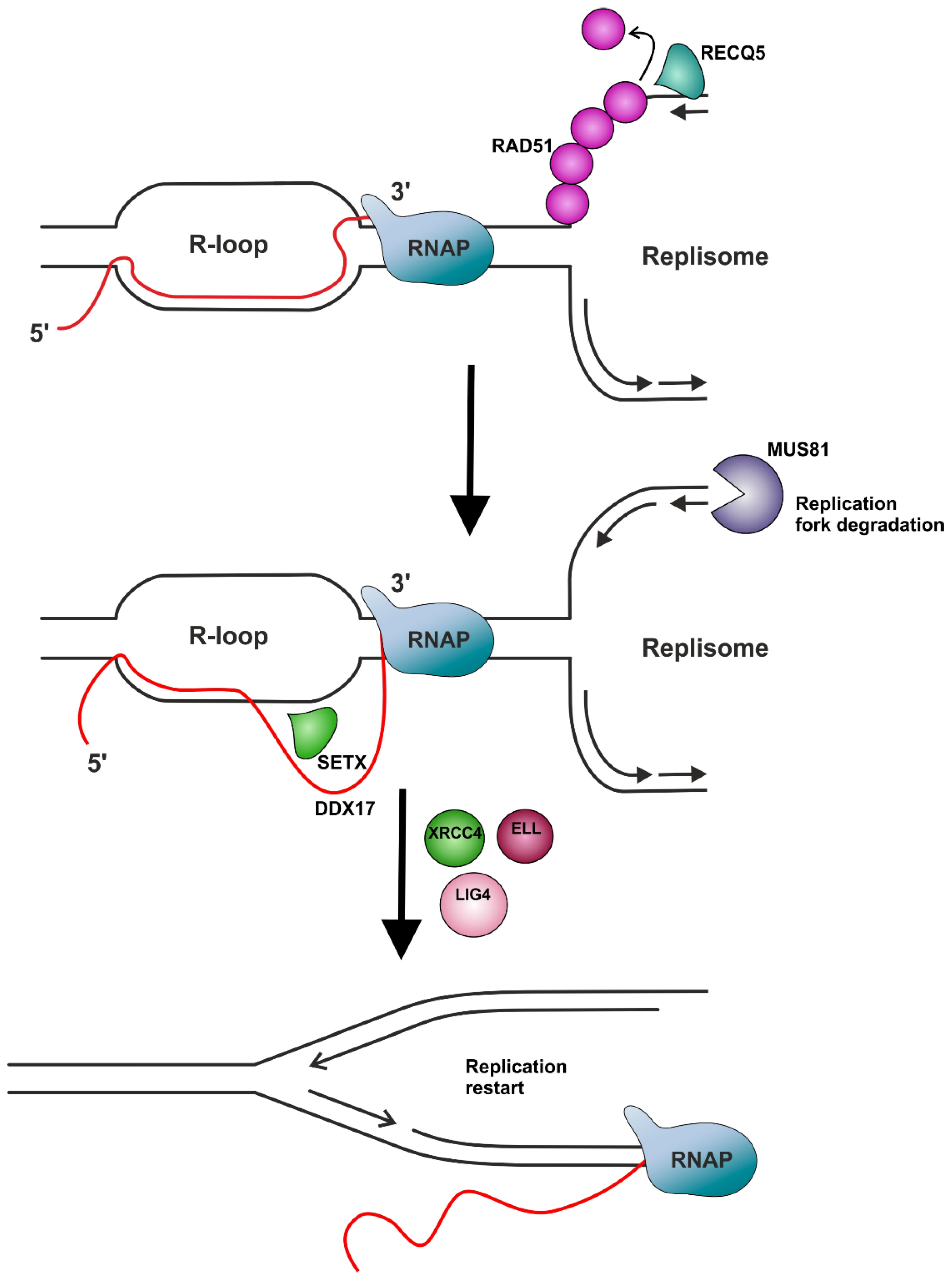
Factors Resolving R-Loops
- RNase H and others
- Helicases
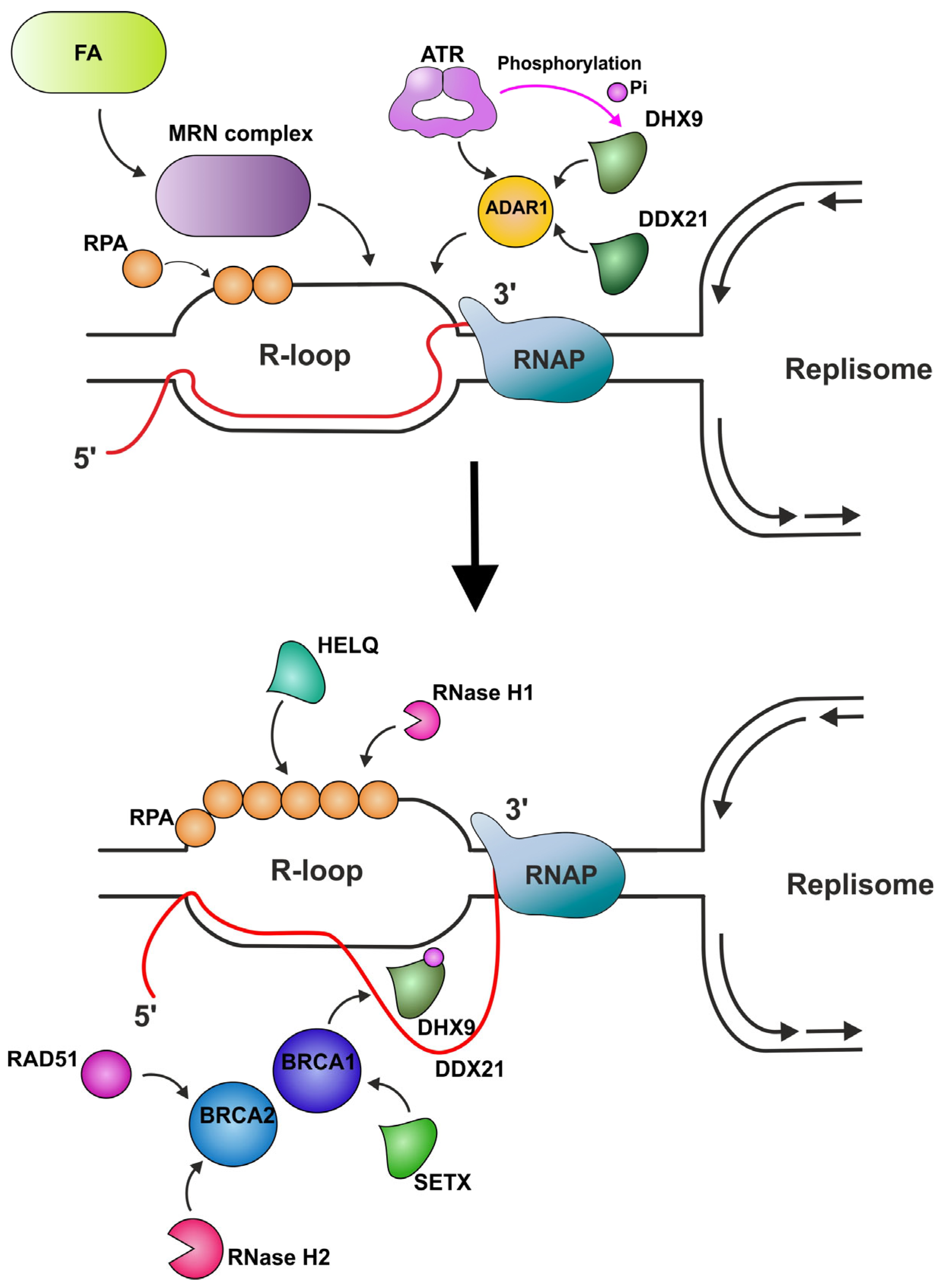
Factors Providing Indirect R-Loop Removal
- DNA repair complexes
- Transcription factors
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| hm5C | 5-hydroxymethylcytosine |
| m5C | 5-methylcytosine |
| AQR | Aquarius protein |
| ATR | Ataxia telangiectasia-mutated (ATM) and RAD3-related DDR kinase |
| BER | Base excision repair |
| BLM | Bloom syndrome protein |
| BRCA1/2 | Breast cancer type 1/2 susceptibility gene protein |
| CD-TRC | Co-directional TRC |
| DSBs | Double-strand breaks |
| dsDNA | Double-stranded DNA |
| dsRNA | Double-stranded RNA |
| GADD45A | DNA damage protein 45A |
| DDR | DNA damage response |
| LIG4 | DNA ligase IV |
| DNMT | DNA methyltransferase |
| Erα | Estrogen receptor alpha |
| FA | Fanconi anemia |
| FEN1 | Flap endonuclease 1 |
| H3K9me2 | H3K9 dimethylation |
| HO-TRC | Head-on TRC |
| HR | Homologous recombination |
| METTL3 | Methyltransferase-like 3 |
| MMR | Mismatch repair |
| m6A | N6-methyladenosine |
| NAT10 | N-acetyltransferase 10 |
| NER | Nucleotide excision repair |
| PARP1 | Poly [ADP-ribose] polymerase 1 |
| PRC2 | Polycomb repressive complex 2 |
| PCBP1 | Polycytosine (poly(C))-binding protein 1 |
| PGCs | Primordial germ cells |
| RHD | RecD helicase domain |
| RPA | Replication protein A |
| rDNA | Ribosomal DNA |
| RNAP | RNA polymerase |
| SETX | Senataxin |
| ssDNA | Single-stranded DNA |
| SSB | Single-stranded DNA-binding protein |
| SF2 | Superfamily 2 helicases |
| TET | Ten-eleven translocation DNA dioxygenase |
| TonEBP | Tonicity-responsive enhancer-binding protein |
| Top1 | Topoisomerase I |
| Top2 | Topoisomerase II |
| TOP3B | Topoisomerase III beta |
| TC-NER | Transcription-coupled nucleotide excision repair |
| TRCs | Transcription–replication collisions/conflicts |
| TLS | Trans-lesion synthesis |
| WRN | Werner syndrome helicase |
| WTAP | Wilms tumor 1-associated protein |
References
- Goehring, L.; Keegan, S.; Lahiri, S.; Xia, W.; Kong, M.; Jimenez-Sainz, J.; Gupta, D.; Drapkin, R.; Jensen, R.B.; Smith, D.J.; et al. Dormant Origin Firing Promotes Head-on Transcription-Replication Conflicts at Transcription Termination Sites in Response to BRCA2 Deficiency. Nat. Commun. 2024, 15, 4716. [Google Scholar] [CrossRef]
- Gaillard, H.; Aguilera, A. Transcription as a Threat to Genome Integrity. Annu. Rev. Biochem. 2016, 85, 291–317. [Google Scholar] [CrossRef]
- Aguilera, A.; García-Muse, T. Causes of Genome Instability. Annu. Rev. Genet. 2013, 47, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.M.; Newlon, C.S. DNA Replication Fork Pause Sites Dependent on Transcription. Science 1996, 272, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- French, S. Consequences of Replication Fork Movement through Transcription Units in Vivo. Science 1992, 258, 1362–1365. [Google Scholar] [CrossRef]
- Aguilera, A.; Gaillard, H. Transcription and Recombination: When RNA Meets DNA. Cold Spring Harb. Perspect. Biol. 2014, 6, a016543. [Google Scholar] [CrossRef]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA Template during Transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef]
- Isik, E.; Shukla, K.; Pospisilova, M.; König, C.; Andrs, M.; Rao, S.; Rosano, V.; Dobrovolna, J.; Krejci, L.; Janscak, P. MutSβ-MutLβ-FANCJ Axis Mediates the Restart of DNA Replication after Fork Stalling at Cotranscriptional G4/R-Loops. Sci. Adv. 2024, 10, eadk2685. [Google Scholar] [CrossRef]
- Brill, S.J.; Sternglanz, R. Transcription-Dependent DNA Supercoiling in Yeast DNA Topoisomerase Mutants. Cell 1988, 54, 403–411. [Google Scholar] [CrossRef]
- Sperling, A.S.; Jeong, K.S.; Kitada, T.; Grunstein, M. Topoisomerase II Binds Nucleosome-Free DNA and Acts Redundantly with Topoisomerase I to Enhance Recruitment of RNA Pol II in Budding Yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 12693–12698. [Google Scholar] [CrossRef]
- Christman, M.F.; Dietrich, F.S.; Fink, G.R. Mitotic Recombination in the rDNA of S. Cerevisiae Is Suppressed by the Combined Action of DNA Topoisomerases I and II. Cell 1988, 55, 413–425. [Google Scholar] [CrossRef]
- García-Rubio, M.L.; Aguilera, A. Topological Constraints Impair RNA Polymerase II Transcription and Causes Instability of Plasmid-Borne Convergent Genes. Nucleic Acids Res. 2012, 40, 1050–1064. [Google Scholar] [CrossRef]
- Duardo, R.C.; Guerra, F.; Pepe, S.; Capranico, G. Non-B DNA Structures as a Booster of Genome Instability. Biochimie 2023, 214, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Belotserkovskii, B.P.; Mirkin, S.M.; Hanawalt, P.C. DNA Sequences That Interfere with Transcription: Implications for Genome Function and Stability. Chem. Rev. 2013, 113, 8620–8637. [Google Scholar] [CrossRef] [PubMed]
- Hanawalt, P.C.; Spivak, G. Transcription-Coupled DNA Repair: Two Decades of Progress and Surprises. Nat. Rev. Mol. Cell Biol. 2008, 9, 958–970. [Google Scholar] [CrossRef]
- Hamperl, S.; Cimprich, K.A. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell 2016, 167, 1455–1467. [Google Scholar] [CrossRef]
- Gómez-González, B.; Aguilera, A. Transcription-Mediated Replication Hindrance: A Major Driver of Genome Instability. Genes Dev. 2019, 33, 1008–1026. [Google Scholar] [CrossRef]
- Breier, A.M.; Weier, H.U.G.; Cozzarelli, N.R. Independence of Replisomes in Escherichia Coli Chromosomal Replication. Proc. Natl. Acad. Sci. USA 2005, 102, 3942–3947. [Google Scholar] [CrossRef]
- Liu, B.; Wong, M.L.; Alberts, B. A Transcribing RNA Polymerase Molecule Survives DNA Replication without Aborting Its Growing RNA Chain. Proc. Natl. Acad. Sci. USA 1994, 91, 10660–10664. [Google Scholar] [CrossRef]
- Liu, B.; Lie Wong, M.; Tinker, R.L.; Geiduschek, E.P.; Alberts, B.M. The DNA Replication Fork Can Pass RNA Polymerase without Displacing the Nascent Transcript. Nature 1993, 366, 33–39. [Google Scholar] [CrossRef]
- Kumar, C.; Batra, S.; Griffith, J.D.; Remus, D. The Interplay of RNA:DNA Hybrid Structure and G-Quadruplexes Determines the Outcome of R-Loop-Replisome Collisions. Elife 2021, 10, e72286. [Google Scholar] [CrossRef]
- Stoy, H.; Zwicky, K.; Kuster, D.; Lang, K.S.; Krietsch, J.; Crossley, M.P.; Schmid, J.A.; Cimprich, K.A.; Merrikh, H.; Lopes, M. Direct Visualization of Transcription-Replication Conflicts Reveals Post-Replicative DNA:RNA Hybrids. Nat. Struct. Mol. Biol. 2023, 30, 348–359. [Google Scholar] [CrossRef]
- Liu, B.; Alberts, B.M. Head-on Collision between a DNA Replication Apparatus and RNA Polymerase Transcription Complex. Science 1995, 267, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Bedinger, P.; Hochstrasser, M.; Jongeneel, C.V.; Alberts, B.M. Properties of the T4 Bacteriophage DNA Replication Apparatus: The T4 Dda DNA Helicase Is Required to Pass a Bound RNA Polymerase Molecule. Cell 1983, 34, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Browning, K.R.; Merrikh, H. Replication-Transcription Conflicts: A Perpetual War on the Chromosome. Annu. Rev. Biochem. 2024, 93, 21–46. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, R.T.; O’Donnell, M. Direct Restart of a Replication Fork Stalled by a Head-on RNA Polymerase. Science 2010, 327, 590–592. [Google Scholar] [CrossRef]
- Pomerantz, R.T.; O’Donnell, M. The Replisome Uses mRNA as a Primer after Colliding with RNA Polymerase. Nature 2008, 456, 762–766. [Google Scholar] [CrossRef]
- Fan, J.; Leroux-Coyau, M.; Savery, N.J.; Strick, T.R. Reconstruction of Bacterial Transcription-Coupled Repair at Single-Molecule Resolution. Nature 2016, 536, 234–237. [Google Scholar] [CrossRef]
- Mellon, I.; Hanawalt, P.C. Induction of the Escherichia Coli Lactose Operon Selectively Increases Repair of Its Transcribed DNA Strand. Nature 1989, 342, 95–98. [Google Scholar] [CrossRef]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774–786.e19. [Google Scholar] [CrossRef]
- Rocha, E.P. Evolutionary Patterns in Prokaryotic Genomes. Curr. Opin. Microbiol. 2008, 11, 454–460. [Google Scholar] [CrossRef]
- Akerman, I.; Kasaai, B.; Bazarova, A.; Sang, P.B.; Peiffer, I.; Artufel, M.; Derelle, R.; Smith, G.; Rodriguez-Martinez, M.; Romano, M.; et al. A Predictable Conserved DNA Base Composition Signature Defines Human Core DNA Replication Origins. Nat. Commun. 2020, 11, 4826. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.H.; Nussenzweig, A. Replication Initiation and Genome Instability: A Crossroads for DNA and RNA Synthesis. Cell. Mol. Life Sci. 2014, 71, 4545–4559. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Keegan, S.; Kahli, M.; Tonzi, P.; Fenyö, D.; Huang, T.T.; Smith, D.J. Transcription Shapes DNA Replication Initiation and Termination in Human Cells. Nat. Struct. Mol. Biol. 2019, 26, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Guilbaud, G.; Murat, P.; Wilkes, H.S.; Lerner, L.K.; Sale, J.E.; Krude, T. Determination of Human DNA Replication Origin Position and Efficiency Reveals Principles of Initiation Zone Organisation. Nucleic Acids Res. 2022, 50, 7436–7450. [Google Scholar] [CrossRef]
- Prioleau, M.-N.; MacAlpine, D.M. DNA Replication Origins-Where Do We Begin? Genes Dev. 2016, 30, 1683–1697. [Google Scholar] [CrossRef]
- Koyanagi, E.; Kakimoto, Y.; Minamisawa, T.; Yoshifuji, F.; Natsume, T.; Higashitani, A.; Ogi, T.; Carr, A.M.; Kanemaki, M.T.; Daigaku, Y. Global Landscape of Replicative DNA Polymerase Usage in the Human Genome. Nat. Commun. 2022, 13, 7221. [Google Scholar] [CrossRef]
- Petryk, N.; Kahli, M.; d’Aubenton-Carafa, Y.; Jaszczyszyn, Y.; Shen, Y.; Silvain, M.; Thermes, C.; Chen, C.-L.; Hyrien, O. Replication Landscape of the Human Genome. Nat. Commun. 2016, 7, 10208. [Google Scholar] [CrossRef]
- Ide, S.; Miyazaki, T.; Maki, H.; Kobayashi, T. Abundance of Ribosomal RNA Gene Copies Maintains Genome Integrity. Science 2010, 327, 693–696. [Google Scholar] [CrossRef]
- Kobayashi, T.; Heck, D.J.; Nomura, M.; Horiuchi, T. Expansion and Contraction of Ribosomal DNA Repeats in Saccharomyces Cerevisiae: Requirement of Replication Fork Blocking (Fob1) Protein and the Role of RNA Polymerase I. Genes Dev. 1998, 12, 3821–3830. [Google Scholar] [CrossRef]
- Brewer, B.J.; Fangman, W.L. A Replication Fork Barrier at the 3’ End of Yeast Ribosomal RNA Genes. Cell 1988, 55, 637–643. [Google Scholar] [CrossRef]
- Defossez, P.-A.; Prusty, R.; Kaeberlein, M.; Lin, S.-J.; Ferrigno, P.; Silver, P.A.; Keil, R.L.; Guarente, L. Elimination of Replication Block Protein Fob1 Extends the Life Span of Yeast Mother Cells. Mol. Cell 1999, 3, 447–455. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Horiuchi, T.; Kobayashi, T. Transcription-Dependent Recombination and the Role of Fork Collision in Yeast rDNA. Genes Dev. 2003, 17, 1497–1506. [Google Scholar] [CrossRef]
- Merrikh, C.N.; Merrikh, H. Gene Inversion Potentiates Bacterial Evolvability and Virulence. Nat. Commun. 2018, 9, 4662. [Google Scholar] [CrossRef]
- Paul, S.; Million-Weaver, S.; Chattopadhyay, S.; Sokurenko, E.; Merrikh, H. Accelerated Gene Evolution through Replication-Transcription Conflicts. Nature 2013, 495, 512–515. [Google Scholar] [CrossRef]
- Dutta, D.; Shatalin, K.; Epshtein, V.; Gottesman, M.E.; Nudler, E. Linking RNA Polymerase Backtracking to Genome Instability in E. Coli. Cell 2011, 146, 533–543. [Google Scholar] [CrossRef]
- Merrikh, H.; Machón, C.; Grainger, W.H.; Grossman, A.D.; Soultanas, P. Co-Directional Replication-Transcription Conflicts Lead to Replication Restart. Nature 2011, 470, 554–557. [Google Scholar] [CrossRef]
- Prado, F.; Aguilera, A. Impairment of Replication Fork Progression Mediates RNA polII Transcription-Associated Recombination. EMBO J. 2005, 24, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Gottipati, P.; Cassel, T.N.; Savolainen, L.; Helleday, T. Transcription-Associated Recombination Is Dependent on Replication in Mammalian Cells. Mol. Cell. Biol. 2008, 28, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Tuduri, S.; Crabbé, L.; Conti, C.; Tourrière, H.; Holtgreve-Grez, H.; Jauch, A.; Pantesco, V.; De Vos, J.; Thomas, A.; Theillet, C.; et al. Topoisomerase I Suppresses Genomic Instability by Preventing Interference between Replication and Transcription. Nat. Cell Biol. 2009, 11, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, A.; Ballarino, M.; Tora, L. Collisions between Replication and Transcription Complexes Cause Common Fragile Site Instability at the Longest Human Genes. Mol. Cell 2011, 44, 966–977. [Google Scholar] [CrossRef]
- Le Tallec, B.; Koundrioukoff, S.; Wilhelm, T.; Letessier, A.; Brison, O.; Debatisse, M. Updating the Mechanisms of Common Fragile Site Instability: How to Reconcile the Different Views? Cell. Mol. Life Sci. 2014, 71, 4489–4494. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.S.; Merrikh, H. Topological Stress Is Responsible for the Detrimental Outcomes of Head-on Replication-Transcription Conflicts. Cell Rep. 2021, 34, 108797. [Google Scholar] [CrossRef] [PubMed]
- Brewer, B.J. When Polymerases Collide: Replication and the Transcriptional Organization of the E. coli Chromosome. Cell 1988, 53, 679–686. [Google Scholar] [CrossRef] [PubMed]
- García-Muse, T.; Aguilera, A. Transcription-Replication Conflicts: How They Occur and How They Are Resolved. Nat. Rev. Mol. Cell Biol. 2016, 17, 553–563. [Google Scholar] [CrossRef]
- Boubakri, H.; de Septenville, A.L.; Viguera, E.; Michel, B. The Helicases DinG, Rep and UvrD Cooperate to Promote Replication across Transcription Units in Vivo. EMBO J. 2010, 29, 145–157. [Google Scholar] [CrossRef]
- Merrikh, C.N.; Brewer, B.J.; Merrikh, H. The B. Subtilis Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units. PLoS Genet. 2015, 11, e1005289. [Google Scholar] [CrossRef]
- Azvolinsky, A.; Dunaway, S.; Torres, J.Z.; Bessler, J.B.; Zakian, V.A. The S. Cerevisiae Rrm3p DNA Helicase Moves with the Replication Fork and Affects Replication of All Yeast Chromosomes. Genes Dev. 2006, 20, 3104–3116. [Google Scholar] [CrossRef]
- Ivessa, A.S.; Lenzmeier, B.A.; Bessler, J.B.; Goudsouzian, L.K.; Schnakenberg, S.L.; Zakian, V.A. The Saccharomyces Cerevisiae Helicase Rrm3p Facilitates Replication Past Nonhistone Protein-DNA Complexes. Mol. Cell 2003, 12, 1525–1536. [Google Scholar] [CrossRef]
- Estep, K.N.; Butler, T.J.; Ding, J.; Brosh, R.M. G4-Interacting DNA Helicases and Polymerases: Potential Therapeutic Targets. Curr. Med. Chem. 2019, 26, 2881–2897. [Google Scholar] [CrossRef]
- Datta, A.; Pollock, K.J.; Kormuth, K.A.; Brosh, R.M. G-Quadruplex Assembly by Ribosomal DNA: Emerging Roles in Disease Pathogenesis and Cancer Biology. Cytogenet. Genome Res. 2021, 161, 285–296. [Google Scholar] [CrossRef]
- Kamath-Loeb, A.S.; Loeb, L.A.; Johansson, E.; Burgers, P.M.; Fry, M. Interactions between the Werner Syndrome Helicase and DNA Polymerase Delta Specifically Facilitate Copying of Tetraplex and Hairpin Structures of the d(CGG)n Trinucleotide Repeat Sequence. J. Biol. Chem. 2001, 276, 16439–16446. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-Quadruplex Structures Mark Human Regulatory Chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Hanakahi, L.A.; Sun, H.; Maizels, N. High Affinity Interactions of Nucleolin with G-G-Paired rDNA. J. Biol. Chem. 1999, 274, 15908–15912. [Google Scholar] [CrossRef] [PubMed]
- Hershman, S.G.; Chen, Q.; Lee, J.Y.; Kozak, M.L.; Yue, P.; Wang, L.-S.; Johnson, F.B. Genomic Distribution and Functional Analyses of Potential G-Quadruplex-Forming Sequences in Saccharomyces Cerevisiae. Nucleic Acids Res. 2008, 36, 144–156. [Google Scholar] [CrossRef]
- Bharti, S.K.; Awate, S.; Banerjee, T.; Brosh, R.M. Getting Ready for the Dance: FANCJ Irons Out DNA Wrinkles. Genes 2016, 7, 31. [Google Scholar] [CrossRef]
- Mendoza, O.; Bourdoncle, A.; Boulé, J.-B.; Brosh, R.M.; Mergny, J.-L. G-Quadruplexes and Helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef]
- Sauer, M.; Paeschke, K. G-Quadruplex Unwinding Helicases and Their Function in Vivo. Biochem. Soc. Trans. 2017, 45, 1173–1182. [Google Scholar] [CrossRef]
- Sabouri, N.; McDonald, K.R.; Webb, C.J.; Cristea, I.M.; Zakian, V.A. DNA Replication through Hard-to-Replicate Sites, Including Both Highly Transcribed RNA Pol II and Pol III Genes, Requires the S. Pombe Pfh1 Helicase. Genes Dev. 2012, 26, 581–593. [Google Scholar] [CrossRef]
- Steinacher, R.; Osman, F.; Dalgaard, J.Z.; Lorenz, A.; Whitby, M.C. The DNA Helicase Pfh1 Promotes Fork Merging at Replication Termination Sites to Ensure Genome Stability. Genes Dev. 2012, 26, 594–602. [Google Scholar] [CrossRef]
- Wallgren, M.; Mohammad, J.B.; Yan, K.-P.; Pourbozorgi-Langroudi, P.; Ebrahimi, M.; Sabouri, N. G-Rich Telomeric and Ribosomal DNA Sequences from the Fission Yeast Genome Form Stable G-Quadruplex DNA Structures in Vitro and Are Unwound by the Pfh1 DNA Helicase. Nucleic Acids Res. 2016, 44, 6213–6231. [Google Scholar] [CrossRef]
- Eddy, S.; Ketkar, A.; Zafar, M.K.; Maddukuri, L.; Choi, J.-Y.; Eoff, R.L. Human Rev1 Polymerase Disrupts G-Quadruplex DNA. Nucleic Acids Res. 2014, 42, 3272–3285. [Google Scholar] [CrossRef]
- Bétous, R.; Rey, L.; Wang, G.; Pillaire, M.-J.; Puget, N.; Selves, J.; Biard, D.S.F.; Shin-ya, K.; Vasquez, K.M.; Cazaux, C.; et al. Role of TLS DNA Polymerases Eta and Kappa in Processing Naturally Occurring Structured DNA in Human Cells. Mol. Carcinog. 2009, 48, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.; Maddukuri, L.; Ketkar, A.; Zafar, M.K.; Henninger, E.E.; Pursell, Z.F.; Eoff, R.L. Evidence for the Kinetic Partitioning of Polymerase Activity on G-Quadruplex DNA. Biochemistry 2015, 54, 3218–3230. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.; Tillman, M.; Maddukuri, L.; Ketkar, A.; Zafar, M.K.; Eoff, R.L. Human Translesion Polymerase κ Exhibits Enhanced Activity and Reduced Fidelity Two Nucleotides from G-Quadruplex DNA. Biochemistry 2016, 55, 5218–5229. [Google Scholar] [CrossRef]
- Koole, W.; van Schendel, R.; Karambelas, A.E.; van Heteren, J.T.; Okihara, K.L.; Tijsterman, M. A Polymerase Theta-Dependent Repair Pathway Suppresses Extensive Genomic Instability at Endogenous G4 DNA Sites. Nat. Commun. 2014, 5, 3216. [Google Scholar] [CrossRef]
- Huertas, P.; Aguilera, A. Cotranscriptionally Formed DNA:RNA Hybrids Mediate Transcription Elongation Impairment and Transcription-Associated Recombination. Mol. Cell 2003, 12, 711–721. [Google Scholar] [CrossRef]
- Piruat, J.I.; Aguilera, A. A Novel Yeast Gene, THO2, Is Involved in RNA Pol II Transcription and Provides New Evidence for Transcriptional Elongation-Associated Recombination. EMBO J. 1998, 17, 4859–4872. [Google Scholar] [CrossRef]
- Prado, F.; Piruat, J.I.; Aguilera, A. Recombination between DNA Repeats in Yeast Hpr1delta Cells Is Linked to Transcription Elongation. EMBO J. 1997, 16, 2826–2835. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef]
- Marnef, A.; Legube, G. R-Loops as Janus-Faced Modulators of DNA Repair. Nat. Cell Biol. 2021, 23, 305–313. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Proudfoot, N.J. A Double-Edged Sword: R Loops as Threats to Genome Integrity and Powerful Regulators of Gene Expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef]
- Sollier, J.; Cimprich, K.A. Breaking Bad: R-Loops and Genome Integrity. Trends Cell Biol. 2015, 25, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Drolet, M.; Bi, X.; Liu, L.F. Hypernegative Supercoiling of the DNA Template during Transcription Elongation in Vitro. J. Biol. Chem. 1994, 269, 2068–2074. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; García-Muse, T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Arab, K.; Karaulanov, E.; Musheev, M.; Trnka, P.; Schäfer, A.; Grummt, I.; Niehrs, C. GADD45A Binds R-Loops and Recruits TET1 to CpG Island Promoters. Nat. Genet. 2019, 51, 217–223. [Google Scholar] [CrossRef]
- Keskin, H.; Shen, Y.; Huang, F.; Patel, M.; Yang, T.; Ashley, K.; Mazin, A.V.; Storici, F. Transcript-RNA-Templated DNA Recombination and Repair. Nature 2014, 515, 436–439. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef]
- Castillo-Guzman, D.; Chédin, F. Defining R-Loop Classes and Their Contributions to Genome Instability. DNA Repair 2021, 106, 103182. [Google Scholar] [CrossRef]
- Beletskii, A.; Bhagwat, A.S. Transcription-Induced Mutations: Increase in C to T Mutations in the Nontranscribed Strand during Transcription in Escherichia Coli. Proc. Natl. Acad. Sci. USA 1996, 93, 13919–13924. [Google Scholar] [CrossRef]
- Sollier, J.; Stork, C.T.; García-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef]
- Gaillard, H.; Herrera-Moyano, E.; Aguilera, A. Transcription-Associated Genome Instability. Chem. Rev. 2013, 113, 8638–8661. [Google Scholar] [CrossRef]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-Loop-Mediated Genomic Instability Is Caused by Impairment of Replication Fork Progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef]
- Berti, M.; Cortez, D.; Lopes, M. The Plasticity of DNA Replication Forks in Response to Clinically Relevant Genotoxic Stress. Nat. Rev. Mol. Cell Biol. 2020, 21, 633–651. [Google Scholar] [CrossRef]
- Chappidi, N.; Nascakova, Z.; Boleslavska, B.; Zellweger, R.; Isik, E.; Andrs, M.; Menon, S.; Dobrovolna, J.; Balbo Pogliano, C.; Matos, J.; et al. Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-Transcriptional R-Loops. Mol. Cell 2020, 77, 528–541.e8. [Google Scholar] [CrossRef] [PubMed]
- Mijic, S.; Zellweger, R.; Chappidi, N.; Berti, M.; Jacobs, K.; Mutreja, K.; Ursich, S.; Ray Chaudhuri, A.; Nussenzweig, A.; Janscak, P.; et al. Replication Fork Reversal Triggers Fork Degradation in BRCA2-Defective Cells. Nat. Commun. 2017, 8, 859. [Google Scholar] [CrossRef] [PubMed]
- Adolph, M.B.; Cortez, D. Mechanisms and Regulation of Replication Fork Reversal. DNA Repair 2024, 141, 103731. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Zou, L. Hallmarks of DNA Replication Stress. Mol. Cell 2022, 82, 2298–2314. [Google Scholar] [CrossRef]
- Liao, H.; Ji, F.; Helleday, T.; Ying, S. Mechanisms for Stalled Replication Fork Stabilization: New Targets for Synthetic Lethality Strategies in Cancer Treatments. EMBO Rep. 2018, 19, e46263. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Cortez, D. ATR: An Essential Regulator of Genome Integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef]
- Zhang, B.; Li, Y.; Zhang, J.; Wang, Y.; Liang, C.; Lu, T.; Zhang, C.; Liu, L.; Qin, Y.; He, J.; et al. ADAR1 Links R-Loop Homeostasis to ATR Activation in Replication Stress Response. Nucleic Acids Res. 2023, 51, 11668–11687. [Google Scholar] [CrossRef] [PubMed]
- Hamperl, S.; Cimprich, K.A. The Contribution of Co-Transcriptional RNA:DNA Hybrid Structures to DNA Damage and Genome Instability. DNA Repair 2014, 19, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Wahba, L.; Gore, S.K.; Koshland, D. The Homologous Recombination Machinery Modulates the Formation of RNA-DNA Hybrids and Associated Chromosome Instability. Elife 2013, 2, e00505. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Clikeman, J.A.; Bates, D.B.; Kogoma, T. RecA Protein-Dependent R-Loop Formation in Vitro. Genes Dev. 2000, 14, 360–365. [Google Scholar] [CrossRef]
- Ginno, P.A.; Lott, P.L.; Christensen, H.C.; Korf, I.; Chédin, F. R-Loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters. Mol. Cell 2012, 45, 814–825. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Kamieniarz-Gdula, K.; Proudfoot, N.J. R-Loops Induce Repressive Chromatin Marks over Mammalian Gene Terminators. Nature 2014, 516, 436–439. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Proudfoot, N.J.; Gromak, N. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination. Mol. Cell 2011, 42, 794–805. [Google Scholar] [CrossRef]
- Ginno, P.A.; Lim, Y.W.; Lott, P.L.; Korf, I.; Chédin, F. GC Skew at the 5’ and 3’ Ends of Human Genes Links R-Loop Formation to Epigenetic Regulation and Transcription Termination. Genome Res. 2013, 23, 1590–1600. [Google Scholar] [CrossRef]
- Duquette, M.L.; Pham, P.; Goodman, M.F.; Maizels, N. AID Binds to Transcription-Induced Structures in c-MYC That Map to Regions Associated with Translocation and Hypermutation. Oncogene 2005, 24, 5791–5798. [Google Scholar] [CrossRef]
- Duquette, M.L.; Handa, P.; Vincent, J.A.; Taylor, A.F.; Maizels, N. Intracellular Transcription of G-Rich DNAs Induces Formation of G-Loops, Novel Structures Containing G4 DNA. Genes Dev. 2004, 18, 1618–1629. [Google Scholar] [CrossRef]
- Ribeiro de Almeida, C.; Dhir, S.; Dhir, A.; Moghaddam, A.E.; Sattentau, Q.; Meinhart, A.; Proudfoot, N.J. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol. Cell 2018, 70, 650–662.e8. [Google Scholar] [CrossRef]
- Elías-Arnanz, M.; Salas, M. Resolution of Head-on Collisions between the Transcription Machinery and Bacteriophage Φ29 DNA Polymerase Is Dependent on RNA Polymerase Translocation. EMBO J. 1999, 18, 5675–5682. [Google Scholar] [CrossRef]
- Elías-Arnanz, M.; Salas, M. Bacteriophage Φ29 DNA Replication Arrest Caused by Codirectional Collisions with the Transcription Machinery. EMBO J. 1997, 16, 5775–5783. [Google Scholar] [CrossRef]
- Belotserkovskii, B.P.; Shin, J.H.S.; Hanawalt, P.C. Strong Transcription Blockage Mediated by R-Loop Formation within a G-Rich Homopurine-Homopyrimidine Sequence Localized in the Vicinity of the Promoter. Nucleic Acids Res. 2017, 45, 6589–6599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, S.-B.; Yi, Z.; Qiao, Z.; Xu, B.; Geng, H.; Wang, H.; Yin, X.; Tang, M.; Ge, W.; et al. Global Coupling of R-Loop Dynamics with RNA Polymerase II Modulates Gene Expression and Early Development of Drosophila. Nucleic Acids Res. 2024, 52, 13110–13127. [Google Scholar] [CrossRef] [PubMed]
- Zardoni, L.; Nardini, E.; Brambati, A.; Lucca, C.; Choudhary, R.; Loperfido, F.; Sabbioneda, S.; Liberi, G. Elongating RNA Polymerase II and RNA:DNA Hybrids Hinder Fork Progression and Gene Expression at Sites of Head-on Replication-Transcription Collisions. Nucleic Acids Res. 2021, 49, 12769–12784. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.X.; Grunseich, C.; Fox, J.; Burdick, J.; Zhu, Z.; Ravazian, N.; Hafner, M.; Cheung, V.G. Human Proteins That Interact with RNA/DNA Hybrids. Genome Res. 2018, 28, 1405–1414. [Google Scholar] [CrossRef]
- Chakraborty, P.; Grosse, F. Human DHX9 Helicase Preferentially Unwinds RNA-Containing Displacement Loops (R-Loops) and G-Quadruplexes. DNA Repair 2011, 10, 654–665. [Google Scholar] [CrossRef]
- Cristini, A.; Groh, M.; Kristiansen, M.S.; Gromak, N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 2018, 23, 1891–1905. [Google Scholar] [CrossRef]
- Karam, J.A.Q.; Fréreux, C.; Mohanty, B.K.; Dalton, A.C.; Dincman, T.A.; Palanisamy, V.; Howley, B.V.; Howe, P.H. The RNA-Binding Protein PCBP1 Modulates Transcription by Recruiting the G-Quadruplex-Specific Helicase DHX9. J. Biol. Chem. 2024, 300, 107830. [Google Scholar] [CrossRef]
- Liu, M.-Y.; Lin, K.-R.; Chien, Y.-L.; Yang, B.-Z.; Tsui, L.-Y.; Chu, H.-P.C.; Wu, C.-S.P. ATR Phosphorylates DHX9 at Serine 321 to Suppress R-Loop Accumulation upon Genotoxic Stress. Nucleic Acids Res. 2024, 52, 204–222. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Kosarev, I.A.; Timofeyeva, N.A.; Novopashina, D.S.; Kuznetsov, N.A. Kinetic Features of Degradation of R-Loops by RNase H1 from Escherichia Coli. Int. J. Mol. Sci. 2024, 25, 12263. [Google Scholar] [CrossRef]
- Chakraborty, P.; Huang, J.T.J.; Hiom, K. DHX9 Helicase Promotes R-Loop Formation in Cells with Impaired RNA Splicing. Nat. Commun. 2018, 9, 4346. [Google Scholar] [CrossRef]
- Alecki, C.; Chiwara, V.; Sanz, L.A.; Grau, D.; Arias Pérez, O.; Boulier, E.L.; Armache, K.-J.; Chédin, F.; Francis, N.J. RNA-DNA Strand Exchange by the Drosophila Polycomb Complex PRC2. Nat. Commun. 2020, 11, 1781. [Google Scholar] [CrossRef] [PubMed]
- Mazina, O.M.; Somarowthu, S.; Kadyrova, L.Y.; Baranovskiy, A.G.; Tahirov, T.H.; Kadyrov, F.A.; Mazin, A.V. Replication Protein A Binds RNA and Promotes R-Loop Formation. J. Biol. Chem. 2020, 295, 14203–14213. [Google Scholar] [CrossRef] [PubMed]
- Sabino, J.C.; de Almeida, M.R.; Abreu, P.L.; Ferreira, A.M.; Caldas, P.; Domingues, M.M.; Santos, N.C.; Azzalin, C.M.; Grosso, A.R.; de Almeida, S.F. Epigenetic Reprogramming by TET Enzymes Impacts Co-Transcriptional R-Loops. Elife 2022, 11, e69476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Yuan, F.; Garcia-Martinez, L.; Lee, K.D.; Stransky, S.; Sidoli, S.; Verdun, R.E.; Zhang, Y.; Wang, Z.; et al. The Polycomb Protein RING1B Enables Estrogen-Mediated Gene Expression by Promoting Enhancer-Promoter Interaction and R-Loop Formation. Nucleic Acids Res. 2021, 49, 9768–9782. [Google Scholar] [CrossRef]
- Sanchez, A.; de Vivo, A.; Tonzi, P.; Kim, J.; Huang, T.T.; Kee, Y. Transcription-Replication Conflicts as a Source of Common Fragile Site Instability Caused by BMI1-RNF2 Deficiency. PLoS Genet. 2020, 16, e1008524. [Google Scholar] [CrossRef]
- Feng, S.; Manley, J.L. Replication Protein A Associates with Nucleolar R Loops and Regulates rRNA Transcription and Nucleolar Morphology. Genes Dev. 2021, 35, 1579–1594. [Google Scholar] [CrossRef]
- Davletgildeeva, A.T.; Kuznetsov, N.A. The Role of DNMT Methyltransferases and TET Dioxygenases in the Maintenance of the DNA Methylation Level. Biomolecules 2024, 14, 1117. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Pastor, W.A.; Aravind, L.; Rao, A. TETonic Shift: Biological Roles of TET Proteins in DNA Demethylation and Transcription. Nat. Rev. Mol. Cell Biol. 2013, 14, 341–356. [Google Scholar] [CrossRef]
- Hirasawa, R.; Chiba, H.; Kaneda, M.; Tajima, S.; Li, E.; Jaenisch, R.; Sasaki, H. Maternal and Zygotic Dnmt1 Are Necessary and Sufficient for the Maintenance of DNA Methylation Imprints during Preimplantation Development. Genes Dev. 2008, 22, 1607–1616. [Google Scholar] [CrossRef]
- Kaneda, M.; Okano, M.; Hata, K.; Sado, T.; Tsujimoto, N.; Li, E.; Sasaki, H. Essential Role for de Novo DNA Methyltransferase Dnmt3a in Paternal and Maternal Imprinting. Nature 2004, 429, 900–903. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Shih, H.-T.; Chen, W.-Y.; Wang, H.-Y.; Chao, T.; Huang, H.-D.; Chou, C.-H.; Chang, Z.-F. DNMT3b Protects Centromere Integrity by Restricting R-Loop-Mediated DNA Damage. Cell Death Dis. 2022, 13, 546. [Google Scholar] [CrossRef]
- Luna, R.; Gómez-González, B.; Aguilera, A. RNA Biogenesis and RNA Metabolism Factors as R-Loop Suppressors: A Hidden Role in Genome Integrity. Genes Dev. 2024, 38, 504–527. [Google Scholar] [CrossRef]
- Li, Y.; Liu, C.; Jia, X.; Bi, L.; Ren, Z.; Zhao, Y.; Zhang, X.; Guo, L.; Bao, Y.; Liu, C.; et al. RPA Transforms RNase H1 to a Bidirectional Exoribonuclease for Processive RNA-DNA Hybrid Cleavage. Nat. Commun. 2024, 15, 7464. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Yadav, T.; Giri, S.; Saez, B.; Graubert, T.A.; Zou, L. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol. Cell 2017, 65, 832–847.e4. [Google Scholar] [CrossRef]
- Pan, J.M.; Betts, H.; Cubbon, A.; He, L.; Bolt, E.L.; Soultanas, P. The Human HELQ Helicase and XRN2 Exoribonuclease Cooperate in R-Loop Resolution. Open Biol. 2025, 15, 240112. [Google Scholar] [CrossRef]
- Pan, H.; Jin, M.; Ghadiyaram, A.; Kaur, P.; Miller, H.E.; Ta, H.M.; Liu, M.; Fan, Y.; Mahn, C.; Gorthi, A.; et al. Cohesin SA1 and SA2 Are RNA Binding Proteins That Localize to RNA Containing Regions on DNA. Nucleic Acids Res. 2020, 48, 5639–5655. [Google Scholar] [CrossRef]
- Bayona-Feliu, A.; Herrera-Moyano, E.; Badra-Fajardo, N.; Galván-Femenía, I.; Soler-Oliva, M.E.; Aguilera, A. The Chromatin Network Helps Prevent Cancer-Associated Mutagenesis at Transcription-Replication Conflicts. Nat. Commun. 2023, 14, 6890. [Google Scholar] [CrossRef]
- Ahmad, M.; Xu, D.; Wang, W. Type IA Topoisomerases Can Be “Magicians” for Both DNA and RNA in All Domains of Life. RNA Biol. 2017, 14, 854–864. [Google Scholar] [CrossRef]
- El Hage, A.; French, S.L.; Beyer, A.L.; Tollervey, D. Loss of Topoisomerase I Leads to R-Loop-Mediated Transcriptional Blocks during Ribosomal RNA Synthesis. Genes Dev. 2010, 24, 1546–1558. [Google Scholar] [CrossRef]
- Yang, Y.; McBride, K.M.; Hensley, S.; Lu, Y.; Chedin, F.; Bedford, M.T. Arginine Methylation Facilitates the Recruitment of TOP3B to Chromatin to Prevent R Loop Accumulation. Mol. Cell 2014, 53, 484–497. [Google Scholar] [CrossRef]
- Zhang, T.; Wallis, M.; Petrovic, V.; Challis, J.; Kalitsis, P.; Hudson, D.F. Loss of TOP3B Leads to Increased R-Loop Formation and Genome Instability. Open Biol. 2019, 9, 190222. [Google Scholar] [CrossRef]
- Andrs, M.; Hasanova, Z.; Oravetzova, A.; Dobrovolna, J.; Janscak, P. RECQ5: A Mysterious Helicase at the Interface of DNA Replication and Transcription. Genes 2020, 11, 232. [Google Scholar] [CrossRef]
- Di Marco, S.; Hasanova, Z.; Kanagaraj, R.; Chappidi, N.; Altmannova, V.; Menon, S.; Sedlackova, H.; Langhoff, J.; Surendranath, K.; Hühn, D.; et al. RECQ5 Helicase Cooperates with MUS81 Endonuclease in Processing Stalled Replication Forks at Common Fragile Sites during Mitosis. Mol. Cell 2017, 66, 658–671.e8. [Google Scholar] [CrossRef]
- Hamadeh, Z.; Lansdorp, P. RECQL5 at the Intersection of Replication and Transcription. Front. Cell Dev. Biol. 2020, 8, 324. [Google Scholar] [CrossRef]
- Urban, V.; Dobrovolna, J.; Hühn, D.; Fryzelkova, J.; Bartek, J.; Janscak, P. RECQ5 Helicase Promotes Resolution of Conflicts between Replication and Transcription in Human Cells. J. Cell. Biol. 2016, 214, 401–415. [Google Scholar] [CrossRef]
- Šviković, S.; Crisp, A.; Tan-Wong, S.M.; Guilliam, T.A.; Doherty, A.J.; Proudfoot, N.J.; Guilbaud, G.; Sale, J.E. R-Loop Formation during S Phase Is Restricted by PrimPol-Mediated Repriming. EMBO J. 2019, 38, e99793. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Patidar, P.L.; Motea, E.A.; Dang, T.T.; Manley, J.L.; Boothman, D.A. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016, 12, e1006107. [Google Scholar] [CrossRef]
- Cornelio, D.A.; Sedam, H.N.C.; Ferrarezi, J.A.; Sampaio, N.M.V.; Argueso, J.L. Both R-Loop Removal and Ribonucleotide Excision Repair Activities of RNase H2 Contribute Substantially to Chromosome Stability. DNA Repair 2017, 52, 110–114. [Google Scholar] [CrossRef]
- Cristini, A.; Tellier, M.; Constantinescu, F.; Accalai, C.; Albulescu, L.O.; Heiringhoff, R.; Bery, N.; Sordet, O.; Murphy, S.; Gromak, N. RNase H2, Mutated in Aicardi-Goutières Syndrome, Resolves Co-Transcriptional R-Loops to Prevent DNA Breaks and Inflammation. Nat. Commun. 2022, 13, 2961. [Google Scholar] [CrossRef]
- Hiller, B.; Achleitner, M.; Glage, S.; Naumann, R.; Behrendt, R.; Roers, A. Mammalian RNase H2 Removes Ribonucleotides from DNA to Maintain Genome Integrity. J. Exp. Med. 2012, 209, 1419–1426. [Google Scholar] [CrossRef]
- Camino, L.P.; Dutta, A.; Barroso, S.; Pérez-Calero, C.; Katz, J.N.; García-Rubio, M.; Sung, P.; Gómez-González, B.; Aguilera, A. DICER Ribonuclease Removes Harmful R-Loops. Mol. Cell 2023, 83, 3707–3719.e5. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, S.Y.; Kim, S.; Kim, S.-H.; Lee, S.H.; Park, S.; Kim, J.J.; Kim, D.-W.; Kim, H. REXO5 Promotes Genomic Integrity through Regulating R-Loop Using Its Exonuclease Activity. Leukemia 2024, 38, 2150–2161. [Google Scholar] [CrossRef]
- Laverde, E.E.; Polyzos, A.A.; Tsegay, P.P.; Shaver, M.; Hutcheson, J.D.; Balakrishnan, L.; McMurray, C.T.; Liu, Y. Flap Endonuclease 1 Endonucleolytically Processes RNA to Resolve R-Loops through DNA Base Excision Repair. Genes 2022, 14, 98. [Google Scholar] [CrossRef]
- Laverde, E.E.; Lai, Y.; Leng, F.; Balakrishnan, L.; Freudenreich, C.H.; Liu, Y. R-Loops Promote Trinucleotide Repeat Deletion through DNA Base Excision Repair Enzymatic Activities. J. Biol. Chem. 2020, 295, 13902–13913. [Google Scholar] [CrossRef]
- Teasley, D.C.; Parajuli, S.; Nguyen, M.; Moore, H.R.; Alspach, E.; Lock, Y.J.; Honaker, Y.; Saharia, A.; Piwnica-Worms, H.; Stewart, S.A. Flap Endonuclease 1 Limits Telomere Fragility on the Leading Strand. J. Biol. Chem. 2015, 290, 15133–15145. [Google Scholar] [CrossRef]
- Osmundson, J.S.; Kumar, J.; Yeung, R.; Smith, D.J. Pif1-Family Helicases Cooperatively Suppress Widespread Replication-Fork Arrest at tRNA Genes. Nat. Struct. Mol. Biol. 2017, 24, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.L.T.; Pohl, T.J.; Chen, C.-F.; Chan, A.; Pott, S.; Zakian, V.A. PIF1 Family DNA Helicases Suppress R-Loop Mediated Genome Instability at tRNA Genes. Nat. Commun. 2017, 8, 15025. [Google Scholar] [CrossRef] [PubMed]
- Balajee, A.S.; Machwe, A.; May, A.; Gray, M.D.; Oshima, J.; Martin, G.M.; Nehlin, J.O.; Brosh, R.; Orren, D.K.; Bohr, V.A. The Werner Syndrome Protein Is Involved in RNA Polymerase II Transcription. Mol. Biol. Cell. 1999, 10, 2655–2668. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, E.Y.-C.; Novoa, C.A.; Aristizabal, M.J.; Coulombe, Y.; Segovia, R.; Chaturvedi, R.; Shen, Y.; Keong, C.; Tam, A.S.; Jones, S.J.M.; et al. RECQ-like Helicases Sgs1 and BLM Regulate R-Loop-Associated Genome Instability. J. Cell. Biol. 2017, 216, 3991–4005. [Google Scholar] [CrossRef]
- Gray, M.D.; Wang, L.; Youssoufian, H.; Martin, G.M.; Oshima, J. Werner Helicase Is Localized to Transcriptionally Active Nucleoli of Cycling Cells. Exp. Cell Res. 1998, 242, 487–494. [Google Scholar] [CrossRef]
- Marabitti, V.; Lillo, G.; Malacaria, E.; Palermo, V.; Sanchez, M.; Pichierri, P.; Franchitto, A. ATM Pathway Activation Limits R-Loop-Associated Genomic Instability in Werner Syndrome Cells. Nucleic Acids Res. 2019, 47, 3485–3502. [Google Scholar] [CrossRef]
- Shiratori, M.; Suzuki, T.; Itoh, C.; Goto, M.; Furuichi, Y.; Matsumoto, T. WRN Helicase Accelerates the Transcription of Ribosomal RNA as a Component of an RNA Polymerase I-Associated Complex. Oncogene 2002, 21, 2447–2454. [Google Scholar] [CrossRef]
- Hatchi, E.; Skourti-Stathaki, K.; Ventz, S.; Pinello, L.; Yen, A.; Kamieniarz-Gdula, K.; Dimitrov, S.; Pathania, S.; McKinney, K.M.; Eaton, M.L.; et al. BRCA1 Recruitment to Transcriptional Pause Sites Is Required for R-Loop-Driven DNA Damage Repair. Mol. Cell 2015, 57, 636–647. [Google Scholar] [CrossRef]
- Kanagaraj, R.; Mitter, R.; Kantidakis, T.; Edwards, M.M.; Benitez, A.; Chakravarty, P.; Fu, B.; Becherel, O.; Yang, F.; Lavin, M.F.; et al. Integrated Genome and Transcriptome Analyses Reveal the Mechanism of Genome Instability in Ataxia with Oculomotor Apraxia 2. Proc. Natl. Acad. Sci. USA 2022, 119, e2114314119. [Google Scholar] [CrossRef]
- Rao, S.; Andrs, M.; Shukla, K.; Isik, E.; König, C.; Schneider, S.; Bauer, M.; Rosano, V.; Prokes, J.; Müller, A.; et al. Senataxin RNA/DNA Helicase Promotes Replication Restart at Co-Transcriptional R-Loops to Prevent MUS81-Dependent Fork Degradation. Nucleic Acids Res. 2024, 52, 10355–10369. [Google Scholar] [CrossRef]
- Boleslavska, B.; Oravetzova, A.; Shukla, K.; Nascakova, Z.; Ibini, O.N.; Hasanova, Z.; Andrs, M.; Kanagaraj, R.; Dobrovolna, J.; Janscak, P. DDX17 Helicase Promotes Resolution of R-Loop-Mediated Transcription-Replication Conflicts in Human Cells. Nucleic Acids Res. 2022, 50, 12274–12290. [Google Scholar] [CrossRef] [PubMed]
- de Amorim, J.L.; Leung, S.W.; Haji-Seyed-Javadi, R.; Hou, Y.; Yu, D.S.; Ghalei, H.; Khoshnevis, S.; Yao, B.; Corbett, A.H. The Putative RNA Helicase DDX1 Associates with the Nuclear RNA Exosome and Modulates RNA/DNA Hybrids (R-Loops). J. Biol. Chem. 2024, 300, 105646. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Reyes, Y.; Fonseca-Rodríguez, C.; Freire, R.; Smits, V.A.J. DDX37 and DDX50 Maintain Genome Stability by Preventing Transcription-Dependent R-Loop Formation. J. Mol. Biol. 2025, 437, 169061. [Google Scholar] [CrossRef] [PubMed]
- Hodroj, D.; Recolin, B.; Serhal, K.; Martinez, S.; Tsanov, N.; Abou Merhi, R.; Maiorano, D. An ATR-Dependent Function for the Ddx19 RNA Helicase in Nuclear R-Loop Metabolism. EMBO J. 2017, 36, 1182–1198. [Google Scholar] [CrossRef]
- Kim, S.; Kang, N.; Park, S.H.; Wells, J.; Hwang, T.; Ryu, E.; Kim, B.-G.; Hwang, S.; Kim, S.-J.; Kang, S.; et al. ATAD5 Restricts R-Loop Formation through PCNA Unloading and RNA Helicase Maintenance at the Replication Fork. Nucleic Acids Res. 2020, 48, 7218–7238. [Google Scholar] [CrossRef]
- Li, L.; Germain, D.R.; Poon, H.-Y.; Hildebrandt, M.R.; Monckton, E.A.; McDonald, D.; Hendzel, M.J.; Godbout, R. DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks. Mol. Cell. Biol. 2016, 36, 2794–2810. [Google Scholar] [CrossRef]
- Mersaoui, S.Y.; Yu, Z.; Coulombe, Y.; Karam, M.; Busatto, F.F.; Masson, J.-Y.; Richard, S. Arginine Methylation of the DDX5 Helicase RGG/RG Motif by PRMT5 Regulates Resolution of RNA: DNA Hybrids. EMBO J. 2019, 38, e100986. [Google Scholar] [CrossRef]
- Song, C.; Hotz-Wagenblatt, A.; Voit, R.; Grummt, I. SIRT7 and the DEAD-Box Helicase DDX21 Cooperate to Resolve Genomic R Loops and Safeguard Genome Stability. Genes Dev. 2017, 31, 1370–1381. [Google Scholar] [CrossRef]
- Chimnaronk, S.; Suzuki, T.; Manita, T.; Ikeuchi, Y.; Yao, M.; Suzuki, T.; Tanaka, I. RNA Helicase Module in an Acetyltransferase That Modifies a Specific tRNA Anticodon. EMBO J. 2009, 28, 1362–1373. [Google Scholar] [CrossRef]
- Ito, S.; Horikawa, S.; Suzuki, T.; Kawauchi, H.; Tanaka, Y.; Suzuki, T.; Suzuki, T. Human NAT10 Is an ATP-Dependent RNA Acetyltransferase Responsible for N4-Acetylcytidine Formation in 18 S Ribosomal RNA (rRNA). J. Biol. Chem. 2014, 289, 35724–35730. [Google Scholar] [CrossRef]
- Kong, R.; Zhang, L.; Hu, L.; Peng, Q.; Han, W.; Du, X.; Ke, Y. hALP, a Novel Transcriptional U Three Protein (t-UTP), Activates RNA Polymerase I Transcription by Binding and Acetylating the Upstream Binding Factor (UBF). J. Biol. Chem. 2011, 286, 7139–7148. [Google Scholar] [CrossRef]
- Su, K.; Zhao, Z.; Wang, Y.; Sun, S.; Liu, X.; Zhang, C.; Jiang, Y.; Du, X. NAT10 Resolves Harmful Nucleolar R-Loops Depending on Its Helicase Domain and Acetylation of DDX21. Cell Commun. Signal. 2024, 22, 490. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Tibbetts, R.S.; Brumbaugh, K.M.; Fang, Y.; Richardson, D.A.; Ali, A.; Chen, S.M.; Abraham, R.T.; Wang, X.F. ATR/ATM-Mediated Phosphorylation of Human Rad17 Is Required for Genotoxic Stress Responses. Nature 2001, 411, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Hammond, E.M.; Denko, N.C.; Dorie, M.J.; Abraham, R.T.; Giaccia, A.J. Hypoxia Links ATR and P53 through Replication Arrest. Mol. Cell. Biol. 2002, 22, 1834–1843. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mazzanti, M.; Mistrik, M.; Kosar, M.; Beznoussenko, G.V.; Mironov, A.A.; Garrè, M.; Parazzoli, D.; Shivashankar, G.V.; Scita, G.; et al. ATR Mediates a Checkpoint at the Nuclear Envelope in Response to Mechanical Stress. Cell 2014, 158, 633–646. [Google Scholar] [CrossRef]
- Liu, S.; Shiotani, B.; Lahiri, M.; Maréchal, A.; Tse, A.; Leung, C.C.Y.; Glover, J.N.M.; Yang, X.H.; Zou, L. ATR Autophosphorylation as a Molecular Switch for Checkpoint Activation. Mol. Cell 2011, 43, 192–202. [Google Scholar] [CrossRef]
- Peterson, S.E.; Li, Y.; Wu-Baer, F.; Chait, B.T.; Baer, R.; Yan, H.; Gottesman, M.E.; Gautier, J. Activation of DSB Processing Requires Phosphorylation of CtIP by ATR. Mol. Cell 2013, 49, 657–667. [Google Scholar] [CrossRef]
- Tibbetts, R.S.; Cortez, D.; Brumbaugh, K.M.; Scully, R.; Livingston, D.; Elledge, S.J.; Abraham, R.T. Functional Interactions between BRCA1 and the Checkpoint Kinase ATR during Genotoxic Stress. Genes Dev. 2000, 14, 2989–3002. [Google Scholar] [CrossRef]
- Tibbetts, R.S.; Brumbaugh, K.M.; Williams, J.M.; Sarkaria, J.N.; Cliby, W.A.; Shieh, S.Y.; Taya, Y.; Prives, C.; Abraham, R.T. A Role for ATR in the DNA Damage-Induced Phosphorylation of P53. Genes Dev. 1999, 13, 152–157. [Google Scholar] [CrossRef]
- Chang, E.Y.-C.; Tsai, S.; Aristizabal, M.J.; Wells, J.P.; Coulombe, Y.; Busatto, F.F.; Chan, Y.A.; Kumar, A.; Dan Zhu, Y.; Wang, A.Y.-H.; et al. MRE11-RAD50-NBS1 Promotes Fanconi Anemia R-Loop Suppression at Transcription-Replication Conflicts. Nat. Commun. 2019, 10, 4265. [Google Scholar] [CrossRef]
- He, Y.J.; Meghani, K.; Caron, M.-C.; Yang, C.; Ronato, D.A.; Bian, J.; Sharma, A.; Moore, J.; Niraj, J.; Detappe, A.; et al. DYNLL1 Binds to MRE11 to Limit DNA End Resection in BRCA1-Deficient Cells. Nature 2018, 563, 522–526. [Google Scholar] [CrossRef]
- Paull, T.T. 20 Years of Mre11 Biology: No End in Sight. Mol. Cell 2018, 71, 419–427. [Google Scholar] [CrossRef]
- Shibata, A.; Moiani, D.; Arvai, A.S.; Perry, J.; Harding, S.M.; Genois, M.-M.; Maity, R.; van Rossum-Fikkert, S.; Kertokalio, A.; Romoli, F.; et al. DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities. Mol. Cell 2014, 53, 7–18. [Google Scholar] [CrossRef]
- Groh, M.; Albulescu, L.O.; Cristini, A.; Gromak, N. Senataxin: Genome Guardian at the Interface of Transcription and Neurodegeneration. J. Mol. Biol. 2017, 429, 3181–3195. [Google Scholar] [CrossRef]
- Herold, S.; Kalb, J.; Büchel, G.; Ade, C.P.; Baluapuri, A.; Xu, J.; Koster, J.; Solvie, D.; Carstensen, A.; Klotz, C.; et al. Recruitment of BRCA1 Limits MYCN-Driven Accumulation of Stalled RNA Polymerase. Nature 2019, 567, 545–549. [Google Scholar] [CrossRef]
- Hill, S.J.; Rolland, T.; Adelmant, G.; Xia, X.; Owen, M.S.; Dricot, A.; Zack, T.I.; Sahni, N.; Jacob, Y.; Hao, T.; et al. Systematic Screening Reveals a Role for BRCA1 in the Response to Transcription-Associated DNA Damage. Genes Dev. 2014, 28, 1957–1975. [Google Scholar] [CrossRef]
- Martin-Tumasz, S.; Brow, D.A. Saccharomyces Cerevisiae Sen1 Helicase Domain Exhibits 5’- to 3’-Helicase Activity with a Preference for Translocation on DNA Rather than RNA. J. Biol. Chem. 2015, 290, 22880–22889. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.N.; August, A.; Hanafusa, H. Evidence for a Transcriptional Activation Function of BRCA1 C-Terminal Region. Proc. Natl. Acad. Sci. USA 1996, 93, 13595–13599. [Google Scholar] [CrossRef] [PubMed]
- San Martin Alonso, M.; Noordermeer, S.M. Untangling the Crosstalk between BRCA1 and R-Loops during DNA Repair. Nucleic Acids Res. 2021, 49, 4848–4863. [Google Scholar] [CrossRef]
- Scully, R.; Anderson, S.F.; Chao, D.M.; Wei, W.; Ye, L.; Young, R.A.; Livingston, D.M.; Parvin, J.D. BRCA1 Is a Component of the RNA Polymerase II Holoenzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 5605–5610. [Google Scholar] [CrossRef]
- Bhatia, V.; Barroso, S.I.; García-Rubio, M.L.; Tumini, E.; Herrera-Moyano, E.; Aguilera, A. BRCA2 Prevents R-Loop Accumulation and Associates with TREX-2 mRNA Export Factor PCID2. Nature 2014, 511, 362–365. [Google Scholar] [CrossRef]
- D’Alessandro, G.; Whelan, D.R.; Howard, S.M.; Vitelli, V.; Renaudin, X.; Adamowicz, M.; Iannelli, F.; Jones-Weinert, C.W.; Lee, M.; Matti, V.; et al. BRCA2 Controls DNA:RNA Hybrid Level at DSBs by Mediating RNase H2 Recruitment. Nat. Commun. 2018, 9, 5376. [Google Scholar] [CrossRef] [PubMed]
- Shivji, M.K.K.; Renaudin, X.; Williams, Ç.H.; Venkitaraman, A.R. BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation. Cell Rep. 2018, 22, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Amon, J.D.; Koshland, D. RNase H Enables Efficient Repair of R-Loop Induced DNA Damage. Elife 2016, 5, e20533. [Google Scholar] [CrossRef] [PubMed]
- Jalan, M.; Sharma, A.; Pei, X.; Weinhold, N.; Buechelmaier, E.S.; Zhu, Y.; Ahmed-Seghir, S.; Ratnakumar, A.; Di Bona, M.; McDermott, N.; et al. RAD52 Resolves Transcription-Replication Conflicts to Mitigate R-Loop Induced Genome Instability. Nat. Commun. 2024, 15, 7776. [Google Scholar] [CrossRef]
- Laspata, N.; Kaur, P.; Mersaoui, S.Y.; Muoio, D.; Liu, Z.S.; Bannister, M.H.; Nguyen, H.D.; Curry, C.; Pascal, J.M.; Poirier, G.G.; et al. PARP1 Associates with R-Loops to Promote Their Resolution and Genome Stability. Nucleic Acids Res. 2023, 51, 2215–2237. [Google Scholar] [CrossRef]
- Abakir, A.; Giles, T.C.; Cristini, A.; Foster, J.M.; Dai, N.; Starczak, M.; Rubio-Roldan, A.; Li, M.; Eleftheriou, M.; Crutchley, J.; et al. N6-Methyladenosine Regulates the Stability of RNA:DNA Hybrids in Human Cells. Nat. Genet. 2020, 52, 48–55. [Google Scholar] [CrossRef]
- Hao, J.-D.; Liu, Q.-L.; Liu, M.-X.; Yang, X.; Wang, L.-M.; Su, S.-Y.; Xiao, W.; Zhang, M.-Q.; Zhang, Y.-C.; Zhang, L.; et al. DDX21 Mediates Co-Transcriptional RNA m6A Modification to Promote Transcription Termination and Genome Stability. Mol. Cell 2024, 84, 1711–1726.e11. [Google Scholar] [CrossRef]
- Horiuchi, K.; Kawamura, T.; Iwanari, H.; Ohashi, R.; Naito, M.; Kodama, T.; Hamakubo, T. Identification of Wilms’ Tumor 1-Associating Protein Complex and Its Role in Alternative Splicing and the Cell Cycle. J. Biol. Chem. 2013, 288, 33292–33302. [Google Scholar] [CrossRef]
- Kang, H.J.; Cheon, N.Y.; Park, H.; Jeong, G.W.; Ye, B.J.; Yoo, E.J.; Lee, J.H.; Hur, J.-H.; Lee, E.-A.; Kim, H.; et al. TonEBP Recognizes R-Loops and Initiates m6A RNA Methylation for R-Loop Resolution. Nucleic Acids Res. 2021, 49, 269–284. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Leela, J.K.; Syeda, A.H.; Anupama, K.; Gowrishankar, J. Rho-Dependent Transcription Termination Is Essential to Prevent Excessive Genome-Wide R-Loops in Escherichia Coli. Proc. Natl. Acad. Sci. USA 2013, 110, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Manley, J.L. Inactivation of the SR Protein Splicing Factor ASF/SF2 Results in Genomic Instability. Cell 2005, 122, 365–378. [Google Scholar] [CrossRef]
- García-Pichardo, D.; Cañas, J.C.; García-Rubio, M.L.; Gómez-González, B.; Rondón, A.G.; Aguilera, A. Histone Mutants Separate R Loop Formation from Genome Instability Induction. Mol. Cell 2017, 66, 597–609.e5. [Google Scholar] [CrossRef]
- Berger, S.L. The Complex Language of Chromatin Regulation during Transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef]
- Kubicek, S.; O’Sullivan, R.J.; August, E.M.; Hickey, E.R.; Zhang, Q.; Teodoro, M.L.; Rea, S.; Mechtler, K.; Kowalski, J.A.; Homon, C.A.; et al. Reversal of H3K9me2 by a Small-Molecule Inhibitor for the G9a Histone Methyltransferase. Mol. Cell 2007, 25, 473–481. [Google Scholar] [CrossRef]
- Zhou, H.; Li, L.; Wang, Q.; Hu, Y.; Zhao, W.; Gautam, M.; Li, L. H3K9 Demethylation-Induced R-Loop Accumulation Is Linked to Disorganized Nucleoli. Front. Genet. 2020, 11, 43. [Google Scholar] [CrossRef]
- Hao, S.; Wang, Y.; Zhao, Y.; Gao, W.; Cui, W.; Li, Y.; Cui, J.; Liu, Y.; Lin, L.; Xu, X.; et al. Dynamic Switching of Crotonylation to Ubiquitination of H2A at Lysine 119 Attenuates Transcription-Replication Conflicts Caused by Replication Stress. Nucleic Acids Res. 2022, 50, 9873–9892. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H2A Ubiquitination in Polycomb Silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Bayona-Feliu, A.; Casas-Lamesa, A.; Reina, O.; Bernués, J.; Azorín, F. Linker Histone H1 Prevents R-Loop Accumulation and Genome Instability in Heterochromatin. Nat. Commun. 2017, 8, 283. [Google Scholar] [CrossRef]
- Zeller, P.; Padeken, J.; van Schendel, R.; Kalck, V.; Tijsterman, M.; Gasser, S.M. Histone H3K9 Methylation Is Dispensable for Caenorhabditis Elegans Development but Suppresses RNA:DNA Hybrid-Associated Repeat Instability. Nat. Genet. 2016, 48, 1385–1395. [Google Scholar] [CrossRef]
- Phoenix, P.; Raymond, M.A.; Massé, E.; Drolet, M. Roles of DNA Topoisomerases in the Regulation of R-Loop Formation in Vitro. J. Biol. Chem. 1997, 272, 1473–1479. [Google Scholar] [CrossRef]
- Promonet, A.; Padioleau, I.; Liu, Y.; Sanz, L.; Biernacka, A.; Schmitz, A.-L.; Skrzypczak, M.; Sarrazin, A.; Mettling, C.; Rowicka, M.; et al. Topoisomerase 1 Prevents Replication Stress at R-Loop-Enriched Transcription Termination Sites. Nat. Commun. 2020, 11, 3940. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Yang, X.; Huang, S.-Y.N.; Agama, K.; Baechler, S.A.; Sun, Y.; Zhang, H.; Saha, L.K.; Su, S.; Jenkins, L.M.; et al. Resolution of R-Loops by Topoisomerase III-β (TOP3B) in Coordination with the DEAD-Box Helicase DDX5. Cell Rep. 2022, 40, 111067. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, E.V.; Mirkin, S.M. Replication Fork Stalling at Natural Impediments. Microbiol. Mol. Biol. Rev. 2007, 71, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, V.; Montermini, L.; Moltò, M.D.; Pianese, L.; Cossée, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A.; et al. Friedreich’s Ataxia: Autosomal Recessive Disease Caused by an Intronic GAA Triplet Repeat Expansion. Science 1996, 271, 1423–1427. [Google Scholar] [CrossRef]
- Frank-Kamenetskii, M.D.; Mirkin, S.M. Triplex DNA Structures. Annu. Rev. Biochem. 1995, 64, 65–95. [Google Scholar] [CrossRef]
- Groh, M.; Lufino, M.M.P.; Wade-Martins, R.; Gromak, N. R-Loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome. PLoS Genet. 2014, 10, e1004318. [Google Scholar] [CrossRef]
- Clark, R.M.; Bhaskar, S.S.; Miyahara, M.; Dalgliesh, G.L.; Bidichandani, S.I. Expansion of GAA Trinucleotide Repeats in Mammals. Genomics 2006, 87, 57–67. [Google Scholar] [CrossRef][Green Version]
- Willems, T.; Gymrek, M.; Highnam, G.; 1000 Genomes Project Consortium; Mittelman, D.; Erlich, Y. The Landscape of Human STR Variation. Genome Res. 2014, 24, 1894–1904. [Google Scholar] [CrossRef]
- Bianchi, J.; Rudd, S.G.; Jozwiakowski, S.K.; Bailey, L.J.; Soura, V.; Taylor, E.; Stevanovic, I.; Green, A.J.; Stracker, T.H.; Lindsay, H.D.; et al. PrimPol Bypasses UV Photoproducts during Eukaryotic Chromosomal DNA Replication. Mol. Cell 2013, 52, 566–573. [Google Scholar] [CrossRef]
- García-Gómez, S.; Reyes, A.; Martínez-Jiménez, M.I.; Chocrón, E.S.; Mourón, S.; Terrados, G.; Powell, C.; Salido, E.; Méndez, J.; Holt, I.J.; et al. PrimPol, an Archaic Primase/Polymerase Operating in Human Cells. Mol. Cell 2013, 52, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Lou, J.; Xia, Y.; Su, B.; Liu, T.; Cui, J.; Sun, Y.; Lou, H.; Huang, J. hPrimpol1/CCDC111 Is a Human DNA Primase-Polymerase Required for the Maintenance of Genome Integrity. EMBO Rep. 2013, 14, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- Schiavone, D.; Jozwiakowski, S.K.; Romanello, M.; Guilbaud, G.; Guilliam, T.A.; Bailey, L.J.; Sale, J.E.; Doherty, A.J. PrimPol Is Required for Replicative Tolerance of G Quadruplexes in Vertebrate Cells. Mol. Cell 2016, 61, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Costantino, L.; Koshland, D. The Yin and Yang of R-Loop Biology. Curr. Opin. Cell Biol. 2015, 34, 39–45. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The Enzymes in Eukaryotes. FEBS J. 2009, 276, 1494–1505. [Google Scholar] [CrossRef]
- Wahba, L.; Amon, J.D.; Koshland, D.; Vuica-Ross, M. RNase H and Multiple RNA Biogenesis Factors Cooperate to Prevent RNA:DNA Hybrids from Generating Genome Instability. Mol. Cell 2011, 44, 978–988. [Google Scholar] [CrossRef]
- Wolak, C.; Ma, H.J.; Soubry, N.; Sandler, S.J.; Reyes-Lamothe, R.; Keck, J.L. Interaction with Single-Stranded DNA-Binding Protein Localizes Ribonuclease HI to DNA Replication Forks and Facilitates R-Loop Removal. Mol. Microbiol. 2020, 114, 495–509. [Google Scholar] [CrossRef]
- Qiu, J.; Qian, Y.; Frank, P.; Wintersberger, U.; Shen, B. Saccharomyces Cerevisiae RNase H(35) Functions in RNA Primer Removal during Lagging-Strand DNA Synthesis, Most Efficiently in Cooperation with Rad27 Nuclease. Mol. Cell. Biol. 1999, 19, 8361–8371. [Google Scholar] [CrossRef]
- Holt, I.J. The Jekyll and Hyde Character of RNase H1 and Its Multiple Roles in Mitochondrial DNA Metabolism. DNA Repair 2019, 84, 102630. [Google Scholar] [CrossRef]
- Lima, W.F.; Murray, H.M.; Damle, S.S.; Hart, C.E.; Hung, G.; De Hoyos, C.L.; Liang, X.-H.; Crooke, S.T. Viable RNaseH1 Knockout Mice Show RNaseH1 Is Essential for R Loop Processing, Mitochondrial and Liver Function. Nucleic Acids Res. 2016, 44, 5299–5312. [Google Scholar] [CrossRef]
- Lockhart, A.; Pires, V.B.; Bento, F.; Kellner, V.; Luke-Glaser, S.; Yakoub, G.; Ulrich, H.D.; Luke, B. RNase H1 and H2 Are Differentially Regulated to Process RNA-DNA Hybrids. Cell Rep. 2019, 29, 2890–2900.e5. [Google Scholar] [CrossRef]
- Hyjek, M.; Figiel, M.; Nowotny, M. RNases H: Structure and Mechanism. DNA Repair 2019, 84, 102672. [Google Scholar] [CrossRef]
- Lang, K.S.; Hall, A.N.; Merrikh, C.N.; Ragheb, M.; Tabakh, H.; Pollock, A.J.; Woodward, J.J.; Dreifus, J.E.; Merrikh, H. Replication-Transcription Conflicts Generate R-Loops That Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 2017, 170, 787–799.e18. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.-H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m6A Methylation Regulates the Ultraviolet-Induced DNA Damage Response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a Bidentate Ribonuclease in the Initiation Step of RNA Interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Ciechanowska, K.; Pokornowska, M.; Kurzyńska-Kokorniak, A. Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases. Int. J. Mol. Sci. 2021, 22, 616. [Google Scholar] [CrossRef] [PubMed]
- Kurzynska-Kokorniak, A.; Koralewska, N.; Pokornowska, M.; Urbanowicz, A.; Tworak, A.; Mickiewicz, A.; Figlerowicz, M. The Many Faces of Dicer: The Complexity of the Mechanisms Regulating Dicer Gene Expression and Enzyme Activities. Nucleic Acids Res. 2015, 43, 4365–4380. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, L.; Bambara, R.A. Flap Endonuclease 1. Annu. Rev. Biochem. 2013, 82, 119–138. [Google Scholar] [CrossRef]
- Liu, Y.; Kao, H.-I.; Bambara, R.A. Flap Endonuclease 1: A Central Component of DNA Metabolism. Annu. Rev. Biochem. 2004, 73, 589–615. [Google Scholar] [CrossRef]
- Meng, F.; Li, T.; Singh, A.K.; Wang, Y.; Attiyeh, M.; Kohram, F.; Feng, Q.; Li, Y.R.; Shen, B.; Williams, T.; et al. Base-Excision Repair Pathway Regulates Transcription-Replication Conflicts in Pancreatic Ductal Adenocarcinoma. Cell Rep. 2024, 43, 114820. [Google Scholar] [CrossRef] [PubMed]
- Groslambert, J.; Prokhorova, E.; Ahel, I. ADP-Ribosylation of DNA and RNA. DNA Repair 2021, 105, 103144. [Google Scholar] [CrossRef] [PubMed]
- Matta, E.; Kiribayeva, A.; Khassenov, B.; Matkarimov, B.T.; Ishchenko, A.A. Insight into DNA Substrate Specificity of PARP1-Catalysed DNA Poly(ADP-Ribosyl)Ation. Sci. Rep. 2020, 10, 3699. [Google Scholar] [CrossRef]
- Ziegler, R.G.; Weinstein, S.J.; Fears, T.R. Nutritional and Genetic Inefficiencies in One-Carbon Metabolism and Cervical Cancer Risk. J. Nutr. 2002, 132, 2345S–2349S. [Google Scholar] [CrossRef]
- Pohl, T.J.; Zakian, V.A. Pif1 Family DNA Helicases: A Helpmate to RNase H? DNA Repair 2019, 84, 102633. [Google Scholar] [CrossRef] [PubMed]
- Fairman-Williams, M.E.; Guenther, U.-P.; Jankowsky, E. SF1 and SF2 Helicases: Family Matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef]
- Mischo, H.E.; Gómez-González, B.; Grzechnik, P.; Rondón, A.G.; Wei, W.; Steinmetz, L.; Aguilera, A.; Proudfoot, N.J. Yeast Sen1 Helicase Protects the Genome from Transcription-Associated Instability. Mol. Cell 2011, 41, 21–32. [Google Scholar] [CrossRef]
- Bochman, M.L.; Sabouri, N.; Zakian, V.A. Unwinding the Functions of the Pif1 Family Helicases. DNA Repair 2010, 9, 237–249. [Google Scholar] [CrossRef]
- Rivosecchi, J.; Larochelle, M.; Teste, C.; Grenier, F.; Malapert, A.; Ricci, E.P.; Bernard, P.; Bachand, F.; Vanoosthuyse, V. Senataxin Homologue Sen1 Is Required for Efficient Termination of RNA Polymerase III Transcription. EMBO J. 2019, 38, e101955. [Google Scholar] [CrossRef]
- Sberna, S.; Filipuzzi, M.; Bianchi, N.; Croci, O.; Fardella, F.; Soriani, C.; Rohban, S.; Carnevali, S.; Albertini, A.A.; Crosetto, N.; et al. Senataxin Prevents Replicative Stress Induced by the Myc Oncogene. Cell Death Dis. 2025, 16, 187. [Google Scholar] [CrossRef]
- Zhao, H.; Hartono, S.R.; de Vera, K.M.F.; Yu, Z.; Satchi, K.; Zhao, T.; Sciammas, R.; Sanz, L.; Chédin, F.; Barlow, J. Senataxin and RNase H2 Act Redundantly to Suppress Genome Instability during Class Switch Recombination. Elife 2022, 11, e78917. [Google Scholar] [CrossRef]
- Cobb, J.A.; Bjergbaek, L. RecQ Helicases: Lessons from Model Organisms. Nucleic Acids Res. 2006, 34, 4106–4114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Croteau, D.L.; Popuri, V.; Opresko, P.L.; Bohr, V.A. Human RecQ Helicases in DNA Repair, Recombination, and Replication. Annu. Rev. Biochem. 2014, 83, 519–552. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Davis, A.J. Human RecQ Helicases in DNA Double-Strand Break Repair. Front. Cell Dev. Biol. 2021, 9, 640755. [Google Scholar] [CrossRef] [PubMed]
- Budhathoki, J.B.; Maleki, P.; Roy, W.A.; Janscak, P.; Yodh, J.G.; Balci, H. A Comparative Study of G-Quadruplex Unfolding and DNA Reeling Activities of Human RECQ5 Helicase. Biophys J. 2016, 110, 2585–2596. [Google Scholar] [CrossRef]
- Popuri, V.; Bachrati, C.Z.; Muzzolini, L.; Mosedale, G.; Costantini, S.; Giacomini, E.; Hickson, I.D.; Vindigni, A. The Human RecQ Helicases, BLM and RECQ1, Display Distinct DNA Substrate Specificities. J. Biol. Chem. 2008, 283, 17766–17776. [Google Scholar] [CrossRef]
- Kang, M.-S.; Ryu, E.; Lee, S.-W.; Park, J.; Ha, N.Y.; Ra, J.S.; Kim, Y.J.; Kim, J.; Abdel-Rahman, M.; Park, S.H.; et al. Regulation of PCNA Cycling on Replicating DNA by RFC and RFC-like Complexes. Nat. Commun. 2019, 10, 2420. [Google Scholar] [CrossRef]
- Kubota, T.; Nishimura, K.; Kanemaki, M.T.; Donaldson, A.D. The Elg1 Replication Factor C-like Complex Functions in PCNA Unloading during DNA Replication. Mol. Cell 2013, 50, 273–280. [Google Scholar] [CrossRef]
- Lee, K.; Fu, H.; Aladjem, M.I.; Myung, K. ATAD5 Regulates the Lifespan of DNA Replication Factories by Modulating PCNA Level on the Chromatin. J. Cell. Biol. 2013, 200, 31–44. [Google Scholar] [CrossRef]
- Brambati, A.; Zardoni, L.; Achar, Y.J.; Piccini, D.; Galanti, L.; Colosio, A.; Foiani, M.; Liberi, G. Dormant Origins and Fork Protection Mechanisms Rescue Sister Forks Arrested by Transcription. Nucleic Acids Res. 2018, 46, 1227–1239. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, W.; Gao, F.; Wen, C.; Zhao, S.; Yu, Y.; Jiao, W.; Mi, X.; Qin, Y.; Chen, Z.-J.; et al. Transcription-Replication Conflicts in Primordial Germ Cells Necessitate the Fanconi Anemia Pathway to Safeguard Genome Stability. Proc. Natl. Acad. Sci. USA 2022, 119, e2203208119. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Zhang, J.; Nepal, M.; Han, B.; Fei, P. Multifaceted Fanconi Anemia Signaling. Trends Genet. 2018, 34, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Olazabal-Herrero, A.; He, B.; Kwon, Y.; Gupta, A.K.; Dutta, A.; Huang, Y.; Boddu, P.; Liang, Z.; Liang, F.; Teng, Y.; et al. The FANCI/FANCD2 Complex Links DNA Damage Response to R-Loop Regulation through SRSF1-Mediated mRNA Export. Cell Rep. 2024, 43, 113610. [Google Scholar] [CrossRef] [PubMed]
- Christou, C.M.; Kyriacou, K. BRCA1 and Its Network of Interacting Partners. Biology 2013, 2, 40–63. [Google Scholar] [CrossRef]
- Hall, J.M.; Lee, M.K.; Newman, B.; Morrow, J.E.; Anderson, L.A.; Huey, B.; King, M.C. Linkage of Early-Onset Familial Breast Cancer to Chromosome 17q21. Science 1990, 250, 1684–1689. [Google Scholar] [CrossRef]
- Smith, S.A.; Easton, D.F.; Evans, D.G.; Ponder, B.A. Allele Losses in the Region 17q12-21 in Familial Breast and Ovarian Cancer Involve the Wild-Type Chromosome. Nat. Genet. 1992, 2, 128–131. [Google Scholar] [CrossRef]
- Anderson, S.F.; Schlegel, B.P.; Nakajima, T.; Wolpin, E.S.; Parvin, J.D. BRCA1 Protein Is Linked to the RNA Polymerase II Holoenzyme Complex via RNA Helicase A. Nat. Genet. 1998, 19, 254–256. [Google Scholar] [CrossRef]
- Patel, P.S.; Abraham, K.J.; Guturi, K.K.N.; Halaby, M.-J.; Khan, Z.; Palomero, L.; Ho, B.; Duan, S.; St-Germain, J.; Algouneh, A.; et al. RNF168 Regulates R-Loop Resolution and Genomic Stability in BRCA1/2-Deficient Tumors. J. Clin. Investig. 2021, 131, e140105. [Google Scholar] [CrossRef]
- Yasuhara, T.; Kato, R.; Hagiwara, Y.; Shiotani, B.; Yamauchi, M.; Nakada, S.; Shibata, A.; Miyagawa, K. Human Rad52 Promotes XPG-Mediated R-Loop Processing to Initiate Transcription-Associated Homologous Recombination Repair. Cell 2018, 175, 558–570.e11. [Google Scholar] [CrossRef]
- Washburn, R.S.; Gottesman, M.E. Transcription Termination Maintains Chromosome Integrity. Proc. Natl. Acad. Sci. USA 2011, 108, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Krogan, N.J.; Vasiljeva, L.; Rando, O.J.; Nedea, E.; Greenblatt, J.F.; Buratowski, S. The Yeast Rat1 Exonuclease Promotes Transcription Termination by RNA Polymerase II. Nature 2004, 432, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Mérida-Cerro, J.A.; Maraver-Cárdenas, P.; Rondón, A.G.; Aguilera, A. Rat1 Promotes Premature Transcription Termination at R-Loops. Nucleic Acids Res. 2024, 52, 3623–3635. [Google Scholar] [CrossRef] [PubMed]
- Tehranchi, A.K.; Blankschien, M.D.; Zhang, Y.; Halliday, J.A.; Srivatsan, A.; Peng, J.; Herman, C.; Wang, J.D. The Transcription Factor DksA Prevents Conflicts between DNA Replication and Transcription Machinery. Cell 2010, 141, 595–605. [Google Scholar] [CrossRef]
- Trautinger, B.W.; Jaktaji, R.P.; Rusakova, E.; Lloyd, R.G. RNA Polymerase Modulators and DNA Repair Activities Resolve Conflicts between DNA Replication and Transcription. Mol. Cell 2005, 19, 247–258. [Google Scholar] [CrossRef]
- Jiang, C.; Hong, Z.; Liu, S.; Hong, Z.; Dai, B. Roles of CDK12 Mutations in PCa Development and Treatment. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2025, 1880, 189247. [Google Scholar] [CrossRef]
- Milano, L.; Gautam, A.; Caldecott, K.W. DNA Damage and Transcription Stress. Mol. Cell 2024, 84, 70–79. [Google Scholar] [CrossRef]
- Tien, J.C.-Y.; Luo, J.; Chang, Y.; Zhang, Y.; Cheng, Y.; Wang, X.; Yang, J.; Mannan, R.; Mahapatra, S.; Shah, P.; et al. CDK12 Loss Drives Prostate Cancer Progression, Transcription-Replication Conflicts, and Synthetic Lethality with Paralog CDK13. Cell Rep. Med. 2024, 5, 101758. [Google Scholar] [CrossRef]
- Gómez-González, B.; García-Rubio, M.; Bermejo, R.; Gaillard, H.; Shirahige, K.; Marín, A.; Foiani, M.; Aguilera, A. Genome-Wide Function of THO/TREX in Active Genes Prevents R-Loop-Dependent Replication Obstacles. EMBO J. 2011, 30, 3106–3119. [Google Scholar] [CrossRef]
- Yang, Z.; Li, M.; Sun, Q. RHON1 Co-Transcriptionally Resolves R-Loops for Arabidopsis Chloroplast Genome Maintenance. Cell Rep. 2020, 30, 243–256.e5. [Google Scholar] [CrossRef]
- Alzu, A.; Bermejo, R.; Begnis, M.; Lucca, C.; Piccini, D.; Carotenuto, W.; Saponaro, M.; Brambati, A.; Cocito, A.; Foiani, M.; et al. Senataxin Associates with Replication Forks to Protect Fork Integrity across RNA-Polymerase-II-Transcribed Genes. Cell 2012, 151, 835–846. [Google Scholar] [CrossRef]
- Chang, E.Y.-C.; Stirling, P.C. Replication Fork Protection Factors Controlling R-Loop Bypass and Suppression. Genes 2017, 8, 33. [Google Scholar] [CrossRef]
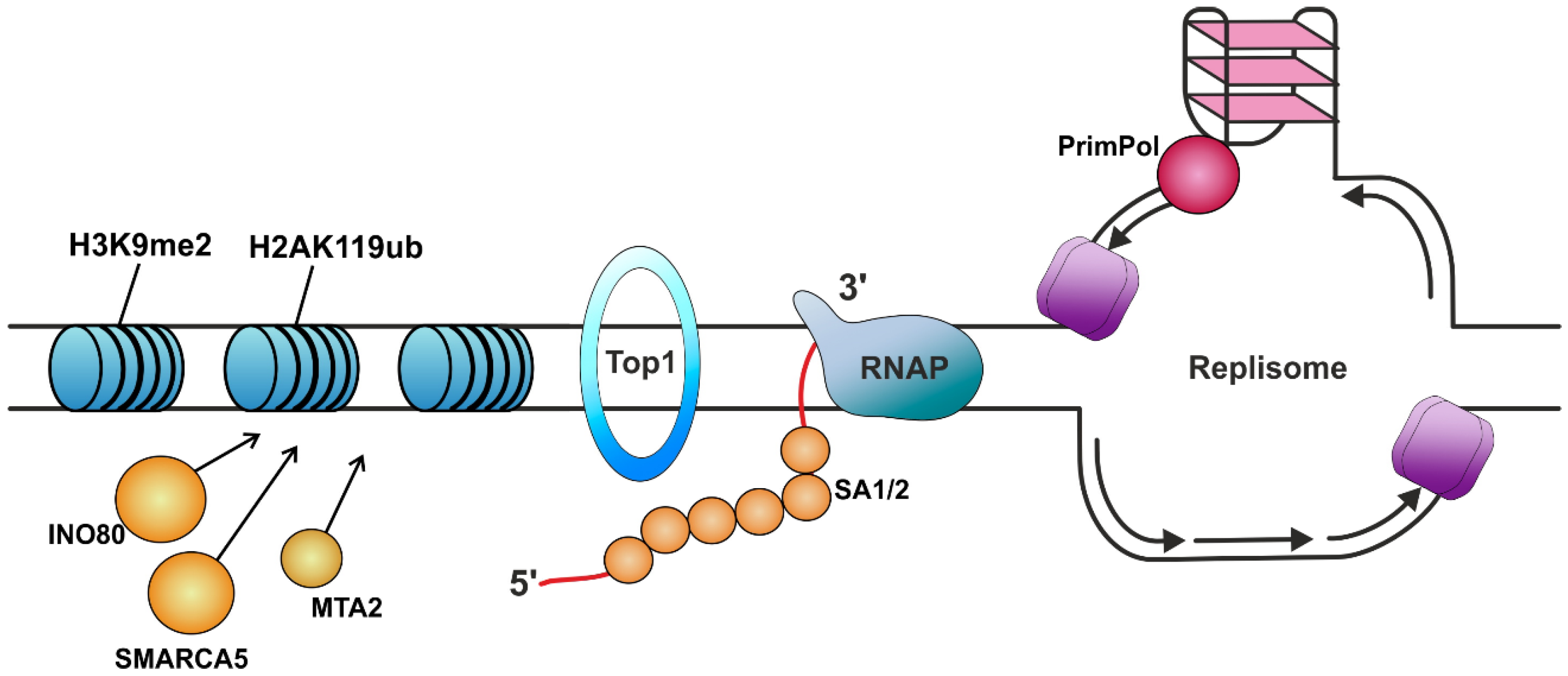
| Protein | Protein Function (Known Role in The Organism) | Role in R-Loop Suppression | Ref. |
|---|---|---|---|
| Prevention of R-loop formation | |||
| RPA | Binds and stabilizes ssDNA intermediates that form during DNA replication or following DNA stress. | Protects the nascent RNA by coating it. Allows human DNA polymerases to initiate DNA synthesis utilizing RPA-generated R-loops, thus reproducing replication restart in vivo. Prevents R-loop-induced DSB formation in SETX-deficient cells. Attracts RNase H1 to the regions of the formed R-loops and enhances its nuclease activity. Recruits HELQ helicase to R-loops. Signals ATR activation. | [100,125,129,138,139,140] |
| SA1, SA2 | Components of the cohesin complex that play an important role in 3D chromatin organization. Strongly bind to RNA, especially DNA–RNA hybrids. | Protect the nascent RNA by coating it. | [141] |
| INO80 | ATP-ase of the chromatin remodeling complex. | Suppression of TRCs due to heterochromatinization. | [142] |
| SMARCA5 | Helicase that possesses intrinsic ATP-dependent nucleosome-remodeling activity. | ||
| MTA2 | Acts as a component of the histone deacetylase NuRD complex, which participates in the remodeling of chromatin. | ||
| Fob1 | Nucleolar protein that binds to the rDNA replication fork barrier site. Required for replication fork blocking. | Suppression of TRCs due to replication blockage. | [43] |
| Topo IV *, DNA gyrase *, Top1 *, TOP1 TOP3B | Topoisomerases. | Prevention of topological stress accumulation in DNA. | [50,53,143,144,145,146] |
| T4 Dda *, Rep *, UvrD *, DinG *, PcrA *, Rrm3, Mfd * | Helicases. | Prevent TRCs from occurring by displacing proteins that may be an obstacle to the passage of the replication fork and RNAP from DNA. Mfd is an elongation factor that displaces RNAP from the replisome pathway. | [23,26,56,57,58,59] |
| RECQ5 | Helicase. | Plays an important role in the resolution of TRCs, particularly by removing RAD51 from the stalled replication fork to facilitate MUS81 endonuclease’s cleavage of the fork. | [95,147,148,149,150] |
| PrimPol | DNA polymerase called Primase-Polymerase. Plays a role in DNA damage tolerance in eukaryotes. | The participation of PrimPol in the replication process of sequences that are prone to R-loop formation reduces their formation. | [151] |
| XRN2 | 5’-3’ exoribonuclease is implicated in transcription termination. | Supposably prevents formation of R-loop-degrading downstream RNA containing a 5′ monophosphate as part of the termination process for most RNAP II transcripts. | [152] |
| Direct resolution of R-loops | |||
| RNase H | Endonucleases that specifically degrade RNA in DNA–RNA hybrids. | Direct nuclease digestion of R-loops. RNase H2 interacts with RNAP II. | [153,154,155] |
| DICER | Double-stranded RNA (dsRNA) endoribonuclease playing a central role in short dsRNA-mediated post-transcriptional gene silencing. | Specifically cleaving RNA within R-loops. | [156] |
| REXO5 | RNA exonuclease. | [157] | |
| FEN1 | Structure-specific nuclease with 5′-flap endonuclease and 5′-3′ exonuclease activities involved in DNA replication and repair. | [158,159,160] | |
| Pif1, Rrm3 | Specific helicases. | Directly resolve R-loops and/or G-quadruplexes. | [161,162] |
| BLM, WRN, RTEL1, PIF1 | [67,163,164,165,166,167] | ||
| FANCJ | Directly resolve R-loops through interacting with MutSβ and MLH1, the components of the mismatch repair complex MutLβ. | [8] | |
| SETX (Sen1) | RNA/DNA helicase involved in diverse aspects of RNA metabolism and genomic integrity. | The mechanism of resolution of TRCs via the helicase activity of SETX seems to be associated with the promotion of replication restart at R-loop formation sites through the MUS81–LIG4–ELL pathway, involving MUS81-mediated cleavage of the leading chain of the stalled fork, DNA ligase IV (LIG4)/XRCC4 complex-facilitated religation of the fork, and RNAP II passage provided by the elongation factor ELL. | [95,168,169,170] |
| DDX1, DDX5, DHX9, DDX17, DDX19, DDX21, DDX37, DDX50 | DEAD/DEXH-box RNA helicases. | Directly resolve R-loops. | [101,119,171,172,173,174,175,176,177,178] |
| HELQ | Helicase belonging to the superfamily 2 (SF2). | Unwinds R-loops in vitro as well as in cells. Interacts with nuclear 5’ to 3’ exoribonuclease, a transcription termination factor of Rat1/XRN2, and supposably coordinating the unwinding and degradation of R-loops. | [140] |
| NAT10 | RNA cytidine acetyltransferase that catalyzes the modification of N4-acetylcytidine on mRNAs, 18S rRNA, and tRNAs. | Resolves R-loops through its RecD helicase domain activity. Also acetylates DDX21 at K236 and K573, thus enhancing its helicase activity towards nucleolar R-loops. | [179,180,181,182] |
| Indirect impact on R-loop resolution | |||
| ATAD5 | Tumor suppressor. Functions as a PCNA (the eukaryotic sliding clamp for replicative polymerases) unloader. | Increases the abundance of DEAD/DEXH-box RNA helicases at replication fork sites, thus participating in R-loop resolution. | [119,172,173,174,175,176,177,178] |
| ATR | Serine/threonine protein kinase which activates checkpoint signaling upon genotoxic stresses such as ionizing radiation, ultraviolet light, or DNA replication stalling, thereby acting as a DNA damage sensor. | Activated in response to HO-TRCs. Phosphorylates BRCA1, CHEK1, MCM2, RAD17, RBBP8, RPA2, SMC1, DHX9, and p53/TP53, which collectively inhibit DNA replication and mitosis and promotes DNA repair, recombination, and apoptosis. The phosphorylation of DHX9 by ATR facilitates its interaction with BRCA1 and RPA, leading to its accumulation at R-loops. | [100,121,183,184,185,186,187,188,189] |
| ADAR1 | Catalyzes the hydrolytic deamination of adenosine to inosine in double-stranded RNA (dsRNA), referred to as A-to-I RNA editing. | Attracts ATR to R-loops through its interaction with TOPBP1 (scaffold protein that acts as a key protein–protein adapter in DNA replication and DNA repair and promotes the loading of RAD51). Is also suggested to attract DHX9 and DDX21 helicases to R-loops. | [101] |
| MRN complex (MRE11-RAD50-NBS1) | Plays an important role in early DNA damage signaling and the processing of DNA ends at DSBs. | Plays a role in suppressing R-loops through the recruitment of FA pathway complexes to these sites. | [190,191,192,193] |
| BRCA1 | Tumor suppressor, participates in a variety of cellular processes, including DNA damage repair, replication fork protection, transcription, and cell cycle regulation, through its interactions with different partners. Participates in the FA pathway. | Participates in transcription activation and R-loop resolution and is able to interact with RNAP II through DHX9. Attracts SETX to R-loops. Was demonstrated to participate in the activation of the persistent R-loop-induced homologue recombination pathway. | [194,195,196,197,198,199,200] |
| BRCA2 | Involved in double-strand break repair and homologous recombination. Participates in the FA pathway. | Acts by targeting RAD51 to ssDNA over dsDNA, enabling RAD51 to displace replication protein A (RPA) from ssDNA and stabilizing RAD51-ssDNA filaments by blocking ATP hydrolysis. Was also shown to recruit RNase H2. | [1,94,95,201,202,203] |
| RAD52 | Involved in double-strand break repair. Plays a central role in genetic recombination and DNA repair by promoting the annealing of complementary ssDNA and stimulating RAD51 recombinase. | Signaling of R-loop-associated damage and homologous recombination. | [204,205] |
| PARP1 | Poly-ADP-ribosyltransferase that mediates poly-ADP-ribosylation of proteins and plays a key role in DNA repair. | Exhibits a tendency to accumulate at R-loop sites in cells, possibly attracting other DNA repair enzymes. | [206] |
| XPF (ERCC1) and XPG (ERCC5) | NER endonucleases. | Attracted to R-loops and initiate DSBs at their ssDNA sites. | [136] |
| METTL3, METTL14 and Wilms tumor 1-associated protein (WTAP) | N6-methyltransferase complex that methylates adenosine residues at the N6 position of some RNAs and regulates various processes, such as the circadian clock, response to DNA damage, and primary miRNA processing. | Attracts RNase H1 through m6A methylation of RNA. Possibly attracted to R-loops by DDX21 helicase. | [207,208,209,210,211,212] |
| Rho * | Transcription termination factor. | Prevents R-loop formation, possibly through transcriptional arrest. | [213] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davletgildeeva, A.T.; Kuznetsov, N.A. Participants in Transcription–Replication Conflict and Their Role in Formation and Resolution of R-Loops. Int. J. Mol. Sci. 2025, 26, 6951. https://doi.org/10.3390/ijms26146951
Davletgildeeva AT, Kuznetsov NA. Participants in Transcription–Replication Conflict and Their Role in Formation and Resolution of R-Loops. International Journal of Molecular Sciences. 2025; 26(14):6951. https://doi.org/10.3390/ijms26146951
Chicago/Turabian StyleDavletgildeeva, Anastasiia T., and Nikita A. Kuznetsov. 2025. "Participants in Transcription–Replication Conflict and Their Role in Formation and Resolution of R-Loops" International Journal of Molecular Sciences 26, no. 14: 6951. https://doi.org/10.3390/ijms26146951
APA StyleDavletgildeeva, A. T., & Kuznetsov, N. A. (2025). Participants in Transcription–Replication Conflict and Their Role in Formation and Resolution of R-Loops. International Journal of Molecular Sciences, 26(14), 6951. https://doi.org/10.3390/ijms26146951







