Ethnic-Specific and UV-Independent Mutational Signatures of Basal Cell Carcinoma in Koreans
Abstract
1. Introduction
2. Results
2.1. Study Cohort
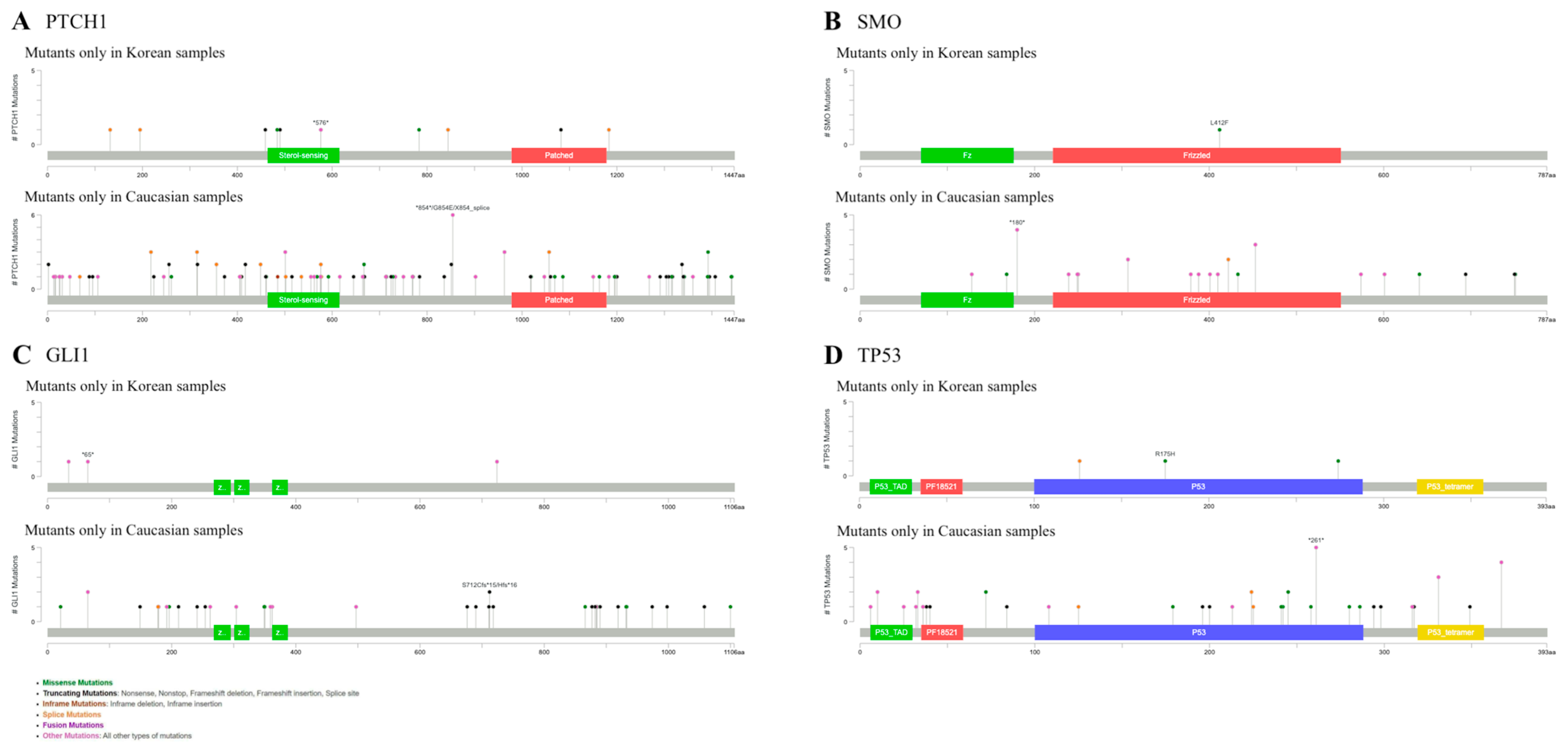
2.2. Mutations in Caucasians and Koreans Reveal Different Patterns in BCC
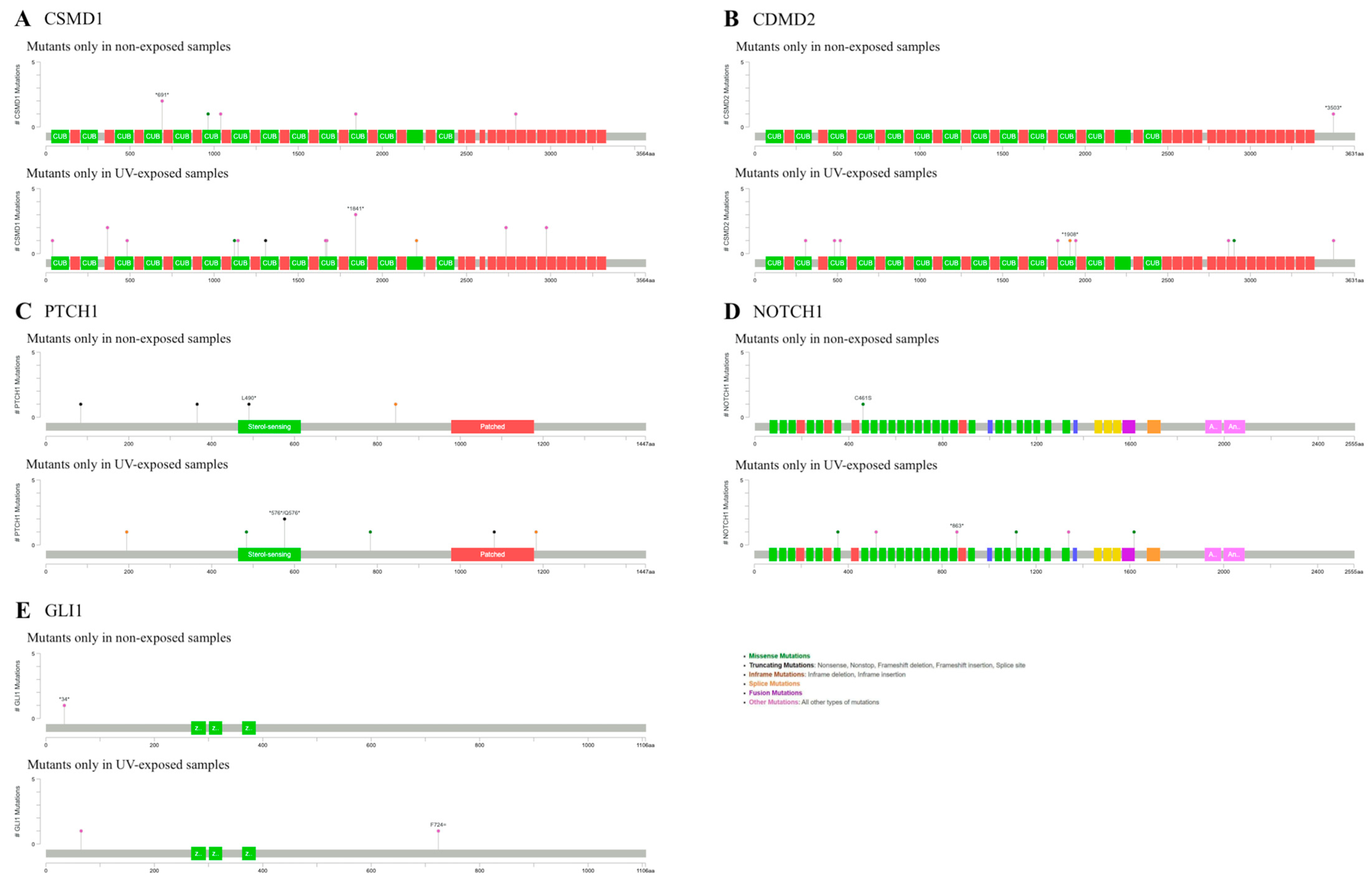
2.3. BCCs in Non-Exposure Areas Exhibited Significant Mutations in CSMD1, PTCH1 and NOTCH1
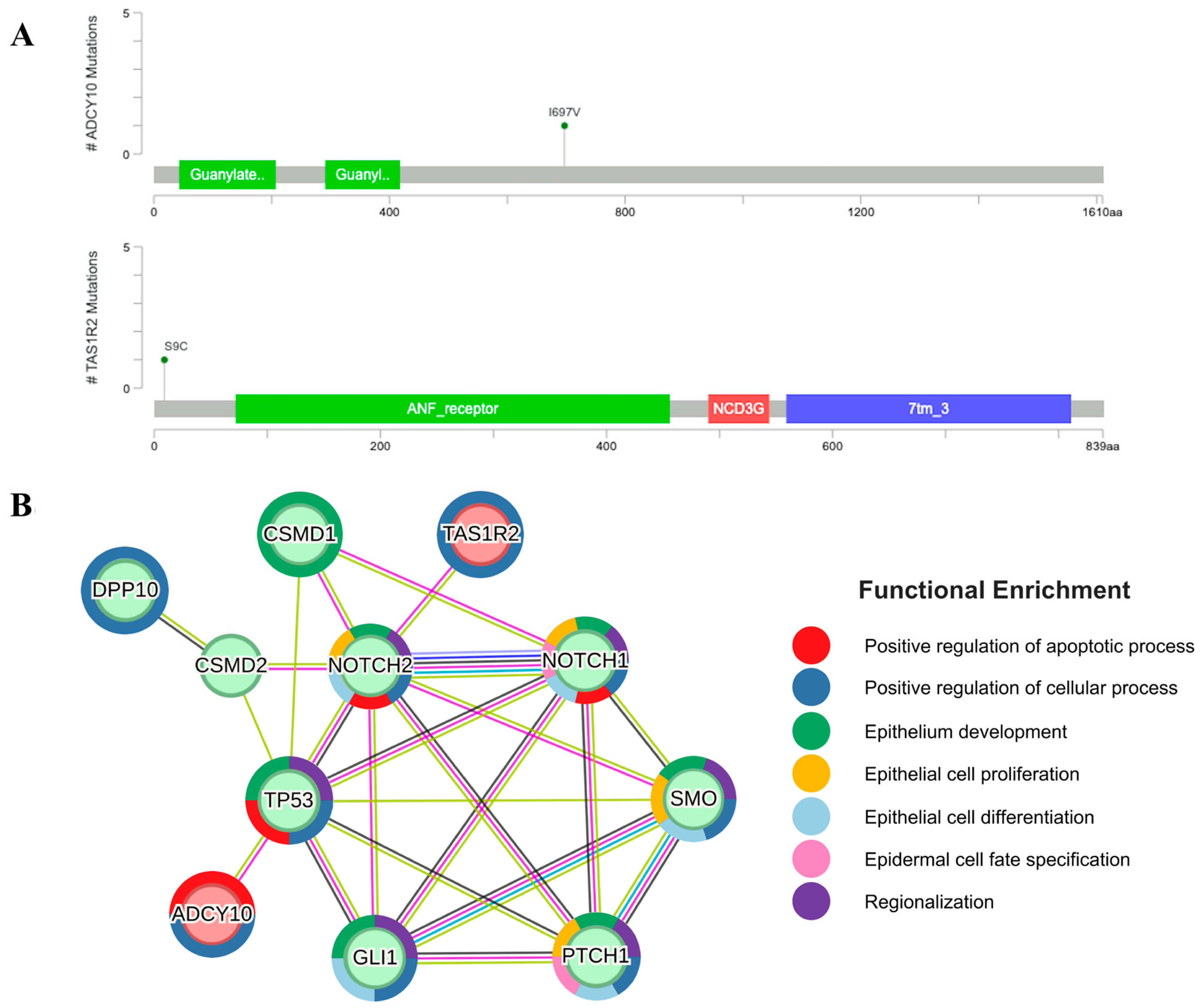
2.4. Mutations of ADCY10 and TAS1R2 Found in the Exposed Areas Interact with Ith BCC Marker Genes
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. DNA Extraction from Tumor Samples
4.3. Sequencing Data Processing
4.4. Variant Calling Analysis and Protein–Protein Interaction Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCC | Basal cancer cell |
| PPI | Protein–protein interaction |
| Hh | Hedgehog |
| FSPT | Fitzpatrick skin phototype |
| UV | Ultraviolet |
| PTCH1 | Patched 1 |
| SMO | Smoothened |
| GLI1 | Glioma-associated transcription factor 1 |
| TP53 | Tumor protein P53 |
| CSMD1 | CUB And Sushi Multiple Domains 1 |
| CSMD2 | CUB And Sushi Multiple Domains 2 |
| NOTCH1 | Notch Receptor 1 |
| NOTCH2 | Notch Receptor 2 |
| ITIH2 | Inter-Alpha-Trypsin Inhibitor Heavy Chain 2 |
| DPP10 | Dipeptidyl Peptidase Like 10 |
| STEAP4 | Six-Transmembrane Epithelial Antigen of Prostate 4 |
References
- Verkouteren, J.; Ramdas, K.; Wakkee, M.; Nijsten, T. Epidemiology of basal cell carcinoma: Scholarly review. Br. J. Dermatol. 2017, 177, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Choi, S.; Kim, J.S.; Kim, S.S.; Jue, M.; Seo, S.H.; Park, J.; Roh, M.R.; Mun, J.; Kim, J.Y.; et al. Incidence and survival rates of primary cutaneous malignancies in Korea, 1999–2019: A nationwide population-based study. J. Dermatol. 2024, 51, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, C.; Maturo, M.G.; Di Nardo, L.; Ciciarelli, V.; García-Rodrigo, C.G.; Fargnoli, M.C. Understanding the Molecular Genetics of Basal Cell Carcinoma. Int. J. Mol. Sci. 2017, 18, 2485. [Google Scholar] [CrossRef] [PubMed]
- Gailani, M.R.; Ståhle-Bäckdahl, M.; Leffell, D.J.; Glyn, M.; Zaphiropoulos, P.G.; Undén, A.B.; Dean, M.; Brash, D.E.; Bale, A.E.; Toftgård, R. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat. Genet. 1996, 14, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Park, H.J.; Baek, S.C.; Byun, D.G.; Houh, D. Mutations of the p53 and PTCH gene in basal cell carcinomas: UV mutation signature and strand bias. J. Dermatol. Sci. 2002, 29, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Bu, D.; Li, X.; Ma, Z.; Yang, Y.; Lin, Z.; Lu, F.; Tu, P.; Li, H. Unique features of PTCH1 mutation spectrum in Chinese sporadic basal cell carcinoma. J. Eur. Acad. Dermatol. Venereol. 2012, 27, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.S.; Rayhan, D.J.; Hazany, S.; Kolodney, M.S. Mutational Landscape of Basal Cell Carcinomas by Whole-Exome Sequencing. J. Investig. Dermatol. 2014, 134, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Teh, M.-T.; Blaydon, D.; Chaplin, T.; Foot, N.J.; Skoulakis, S.; Raghavan, M.; Harwood, C.A.; Proby, C.M.; Philpott, M.P.; Young, B.D.; et al. Genomewide Single Nucleotide Polymorphism Microarray Mapping in Basal Cell Carcinomas Unveils Uniparental Disomy as a Key Somatic Event. Cancer Res. 2005, 65, 8597–8603. [Google Scholar] [CrossRef] [PubMed]
- Reifenberger, J.; Wolter, M.; Knobbe, C.B.; Köhler, B.; Schönicke, A.; Scharwächter, C.; Kumar, K.; Blaschke, B.; Ruzicka, T.; Reifenberger, G. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br. J. Dermatol. 2005, 152, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, X.; Parmentier, L.; King, B.; Bezrukov, F.; Kaya, G.; Zoete, V.; Seplyarskiy, V.B.; Sharpe, H.J.; McKee, T.; Letourneau, A.; et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet. 2016, 48, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Murone, M.; Luoh, S.-M.; Ryan, A.; Gu, Q.; Zhang, C.; Bonifas, J.M.; Lam, C.-W.; Hynes, M.; Goddard, A.; et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 1998, 391, 90–92. [Google Scholar] [CrossRef] [PubMed]
- Rady, P.; Scinicariello, F.; Wagner, R.F., Jr.; Tyring, S.K. p53 mutations in basal cell carcinomas. Cancer Res. 1992, 52, 3804–3806. [Google Scholar] [PubMed]
- Gailani, M.R.; Leffell, D.J.; Ziegler, A.; Gross, E.G.; Brash, D.E.; Bale, A.E. Relationship Between Sunlight Exposure and a Key Genetic Alteration in Basal Cell Carcinoma. JNCI J. Natl. Cancer Inst. 1996, 88, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.; Leffell, D.J.; Kunala, S.; Sharma, H.W.; Gailani, M.; A Simon, J.; Halperin, A.J.; Baden, H.P.; E Shapiro, P.; E Bale, A. Mutation hotspots due to sunlight in the p53 gene of nonmelanoma skin cancers. Proc. Natl. Acad. Sci. USA 1993, 90, 4216–4220. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, B.S.; Phelps, R.G.; Weinstock, M.A.; Bernstein, J.L.; Gordon, M.L.; Rudikoff, D.; Kantor, I.; Shelton, R.; Lebwohl, M.G. p53 Mutations in Basal Cell Carcinomas Arising in Routine Users of Sunscreens. Photochem. Photobiol. 1999, 70, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Bolshakov, S.; Walker, C.M.; Strom, S.S.; Selvan, M.S.; Clayman, G.L.; El-Naggar, A.; Lippman, S.M.; Kripke, M.L.; Ananthaswamy, H.N. p53 mutations in human aggressive and nonaggressive basal and squamous cell carcinomas. Clin. Cancer Res. 2003, 9, 228–234. [Google Scholar] [PubMed]
- Wang, Y.M.; Huang, Y.S.; Ma, Z.H.; Bu, D.F.; Tu, P.; Li, H. Frequency and features of TP53 mutation in 30 Chinese patients with sporadic basal cell carcinoma. Clin. Exp. Dermatol. 2014, 39, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Wang, J.; Mancianti, M.L.; Epstein, E.H., Jr. Basal cell carcinomas arise from hair follicle stem cells in Ptch1+/− mice. Cancer Cell 2011, 19, 114–124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ikehata, H.; Ono, T. The mechanisms of UV mutagenesis. J. Radiat. Res. 2011, 52, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, J.; Plaza, P.; Brettel, K. Repair of (6-4) Lesions in DNA by (6-4) Photolyase: 20 Years of Quest for the Photoreaction Mechanism. Photochem. Photobiol. 2017, 93, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Scrivener, Y.; Grosshans, E.; Cribier, B. Variations of basal cell carcinomas according to gender, age, location and histopathological subtype. Br. J. Dermatol. 2002, 147, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Betti, R.; Bruscagin, C.; Inselvini, E.; Crosti, C. Basal cell carcinomas of covered and unusual sites of the body. Int. J. Dermatol. 1997, 36, 503–505. [Google Scholar] [CrossRef] [PubMed]
- Roh, S.G.; Park, J.; Song, K.H.; Nam, K.H.; Yun, S.K.; Kim, H.U. Clinical and histopathological characteristics of extra-facial basal cell carcinoma: Analysis of 35 patients at the Chonbuk National University Hospital in Korea. Australas. J. Dermatol. 2014, 55, e65–e68. [Google Scholar] [CrossRef] [PubMed]
- Di Nardo, L.; Pellegrini, C.; Di Stefani, A.; Ricci, F.; Fossati, B.; Del Regno, L.; Carbone, C.; Piro, G.; Corbo, V.; Delfino, P.; et al. Molecular alterations in basal cell carcinoma subtypes. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lobl, M.; Hass, B.; Clarey, D.; Higgins, S.; Sutton, A.; Wysong, A. Basal Cell Carcinoma Gene Mutations Differ Between Asian, Hispanic, and Caucasian Patients: A Pilot Study. J. Drugs Dermatol. 2021, 20, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Oren, M. Regulation of the p53 Tumor Suppressor Protein. J. Biol. Chem. 1999, 274, 36031–36034. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Jung, A.R.; Eun, Y.-G.; Lee, Y.C.; Noh, J.K.; Kwon, K.H. Clinical Significance of CUB and Sushi Multiple Domains 1 Inactivation in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2018, 19, 3996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, T.; Ren, X.; Fang, X.; Chen, X.; Wei, H.; Sun, W.; Wang, Y. Integrated pan-cancer analysis of CSMD2 as a potential prognostic, diagnostic, and immune biomarker. Front. Genet. 2022, 13, 918486. [Google Scholar] [CrossRef] [PubMed]
- Eberl, M.; Mangelberger, D.; Swanson, J.B.; Verhaegen, M.E.; Harms, P.W.; Frohm, M.L.; Dlugosz, A.A.; Wong, S.Y. Tumor Architecture and Notch Signaling Modulate Drug Response in Basal Cell Carcinoma. Cancer Cell 2018, 33, 229–243.e4. [Google Scholar] [CrossRef] [PubMed]
- Griewank, K.G.; Murali, R.; Schilling, B.; Schimming, T.; Möller, I.; Moll, I.; Schwamborn, M.; Sucker, A.; Zimmer, L.; Schadendorf, D.; et al. TERT Promoter Mutations Are Frequent in Cutaneous Basal Cell Carcinoma and Squamous Cell Carcinoma. PLoS ONE 2013, 8, e80354. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.A.; Laughlin, T.S.; Rothberg, P.G. Mutations of the TERT promoter are common in basal cell carcinoma and squamous cell carcinoma. Mod. Pathol. 2014, 27, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Pópulo, H.; Boaventura, P.; Vinagre, J.; Batista, R.; Mendes, A.; Caldas, R.; Pardal, J.; Azevedo, F.; Honavar, M.; Guimarães, I.; et al. TERT Promoter Mutations in Skin Cancer: The Effects of Sun Exposure and X-Irradiation. J. Investig. Dermatol. 2014, 134, 2251–2257. [Google Scholar] [CrossRef] [PubMed]
- Denisova, E.; Heidenreich, B.; Nagore, E.; Rachakonda, P.S.; Hosen, I.; Akrap, I.; Traves, V.; García-Casado, Z.; López-Guerrero, J.A.; Requena, C.; et al. Frequent DPH3 promoter mutations in skin cancers. Oncotarget 2015, 6, 35922–35930. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.B.; Jin, G.; Lee, S.Y.; Park, J.Y.; Kim, M.J.; Choi, J.E.; Jeon, H.S.; Cha, S.I.; Cho, S.; Kim, C.H.; et al. TP53 Mutations in Korean Patients with Non-small Cell Lung Cancer. J. Korean Med. Sci. 2010, 25, 698–705. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, Y.; Park, G.S.; Chung, Y.; Lee, J.H. Whole-exome sequencing of secondary tumors arising from nevus sebaceous revealed additional genomic alterations besides RAS mutations. J. Dermatol. 2023, 50, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Thélu, J.; Rossio, P.; Favier, B. Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing. BMC Dermatol. 2002, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Nawrocka, P.M.; Galka-Marciniak, P.; Urbanek-Trzeciak, M.O.; M-Thirusenthilarasan, I.; Szostak, N.; Philips, A.; Susok, L.; Sand, M.; Kozlowski, P. Profile of Basal Cell Carcinoma Mutations and Copy Number Alterations—Focus on Gene-Associated Noncoding Variants. Front. Oncol. 2021, 11, 752579. [Google Scholar] [CrossRef] [PubMed]
- Karampinis, E.; Koumaki, D.; Sgouros, D.; Nechalioti, P.-M.; Toli, O.; Pappa, G.; Papadakis, M.; Georgopoulou, K.-E.; Schulze-Roussaki, A.-V.; Kouretas, D. Non-Melanoma Skin Cancer: Assessing the Systemic Burden of the Disease. Cancers 2025, 17, 703. [Google Scholar] [CrossRef] [PubMed]
- Draper, E.; Li, Y.Y.; Mahadevan, N.R.; Laga, A.C.M.; Hanna, J.; Russell-Goldman, E. Clinicopathologic and Molecular Characterization of Basal Cell Carcinoma Arising at Sun-protected Sites. Am. J. Surg. Pathol. 2025, 49, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, M.; Pazzaglia, S.; Tanori, M.; Hahn, H.; Merola, P.; Rebessi, S.; Atkinson, M.J.; Di Majo, V.; Covelli, V.; Saran, A. Basal cell carcinoma and its development: Insights from radiation-induced tumors in Ptch1-deficient mice. Cancer Res. 2004, 64, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Carey, R.M.; Kim, T.; Cohen, N.A.; Lee, R.J.; Nead, K.T. Impact of sweet, umami, and bitter taste receptor (TAS1R and TAS2R) genomic and expression alterations in solid tumors on survival. Sci. Rep. 2022, 12, 8937. [Google Scholar] [CrossRef] [PubMed]
- Shaji, C.S.; Saraswathy, R. Taste receptors influencing effective modalities in human health–A cutting edge update on TAS1R and TAS2R receptor polymorphisms in taste perception and disease risk. Nutr. Health 2023. [Google Scholar] [CrossRef] [PubMed]
- González-Vela, M.d.C.; Curiel-Olmo, S.; Derdak, S.; Beltran, S.; Santibañez, M.; Martínez, N.; Castillo-Trujillo, A.; Gut, M.; Sánchez-Pacheco, R.; Almaraz, C.; et al. Shared Oncogenic Pathways Implicated in Both Virus-Positive and UV-Induced Merkel Cell Carcinomas. J. Investig. Dermatol. 2017, 137, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Mamoor, S. Adenylyl Cyclase 10 Is Differentially Expressed in the Lymph Nodes of Patients with Metastatic Breast Cancer. OSF Preprints 2021. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, K.; Ma, L.; Qian, Z.; Tian, X.; Miao, Y.; Niu, Y.; Xu, X.; Guo, S.; Yang, Y.; et al. Endogenous glutamate determines ferroptosis sensitivity via ADCY10-dependent YAP suppression in lung adenocarcinoma. Theranostics 2021, 11, 5650–5674. [Google Scholar] [CrossRef] [PubMed]
- Zippin, J.H.; Chadwick, P.A.; Levin, L.R.; Buck, J.; Magro, C.M. Soluble Adenylyl Cyclase Defines a Nuclear cAMP Microdomain in Keratinocyte Hyperproliferative Skin Diseases. J. Investig. Dermatol. 2010, 130, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
| Exposed (n = 8) | Non-Exposed (n = 6) | All (n = 14) | |
|---|---|---|---|
| Age (mean ± SD) | 65.38 ± 8.42 | 70.33 ± 7.26 | 67.50 ± 8.06 |
| Sex, n (%) | |||
| Male | 6 (75.0) | 3 (50.0) | 9 (64.3) |
| Female | 2 (25.0) | 3 (50.0) | 5 (35.7) |
| Ethnicity | |||
| Korean | 8 (100) | 6 (100) | 15 (100) |
| Skin type, n (%) | |||
| III | 6 (75.0) | 5 (83.3) | 11 (78.6) |
| IV | 2 (25.0) | 1 (16.7) | 3 (21.4) |
| History of other NMSC, n (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| History of sunburn, n (%) | 0 (0.0) | 1 (14.3) | 1 (6.3) |
| Immunosuppression, n (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Anatomical site, n (%) | |||
| Cheek | 4 (50.0) | 0 (0.0) | 4 (28.6) |
| Nose | 2 (25.0) | 0 (0.0) | 2 (14.3) |
| Eyelid | 1 (12.5) | 0 (0.0) | 1 (7.1) |
| Neck | 1 (12.5) | 0 (0.0) | 1 (7.1) |
| Back | 0 (0.0) | 2 (33.3) | 2 (14.3) |
| Abdomen | 0 (0.0) | 2 (33.3) | 2 (14.3) |
| Scalp | 0 (0.0) | 1 (16.7) | 1 (7.1) |
| Calf | 0 (0.0) | 1 (16.7) | 1 (7.1) |
| Histopathological subtype, n (%) | |||
| Nodular | 6 (75.0) | 3 (50.0) | 9 (64.3) |
| Superficial | 2 (25.0) | 3 (50.0) | 5 (35.7) |
| Pigmentation, n (%) | 8 (100) | 6 (100) | 14 (100) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-A.; Myung, S.; Choi, Y.; Kim, J.; Lee, Y.; Lee, K.; Lew, B.-L.; Kim, M.S.; Kwon, S.-H. Ethnic-Specific and UV-Independent Mutational Signatures of Basal Cell Carcinoma in Koreans. Int. J. Mol. Sci. 2025, 26, 6941. https://doi.org/10.3390/ijms26146941
Kim Y-A, Myung S, Choi Y, Kim J, Lee Y, Lee K, Lew B-L, Kim MS, Kwon S-H. Ethnic-Specific and UV-Independent Mutational Signatures of Basal Cell Carcinoma in Koreans. International Journal of Molecular Sciences. 2025; 26(14):6941. https://doi.org/10.3390/ijms26146941
Chicago/Turabian StyleKim, Ye-Ah, Seokho Myung, Yueun Choi, Junghyun Kim, Yoonsung Lee, Kiwon Lee, Bark-Lynn Lew, Man S. Kim, and Soon-Hyo Kwon. 2025. "Ethnic-Specific and UV-Independent Mutational Signatures of Basal Cell Carcinoma in Koreans" International Journal of Molecular Sciences 26, no. 14: 6941. https://doi.org/10.3390/ijms26146941
APA StyleKim, Y.-A., Myung, S., Choi, Y., Kim, J., Lee, Y., Lee, K., Lew, B.-L., Kim, M. S., & Kwon, S.-H. (2025). Ethnic-Specific and UV-Independent Mutational Signatures of Basal Cell Carcinoma in Koreans. International Journal of Molecular Sciences, 26(14), 6941. https://doi.org/10.3390/ijms26146941







