Role of Individual Amino Acid Residues Directly Involved in Damage Recognition in Active Demethylation by ABH2 Dioxygenase
Abstract
1. Introduction
2. Results and Discussion
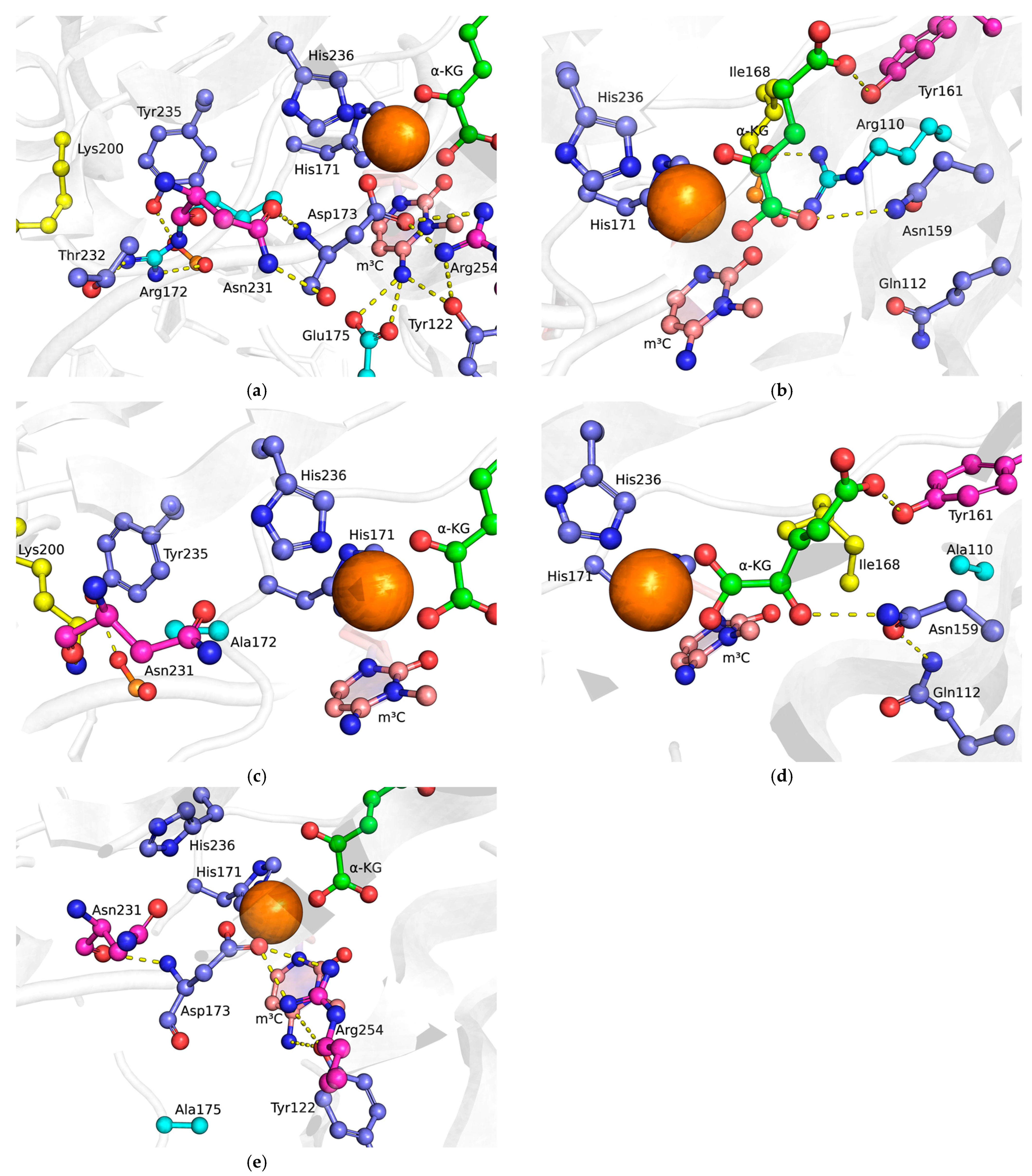
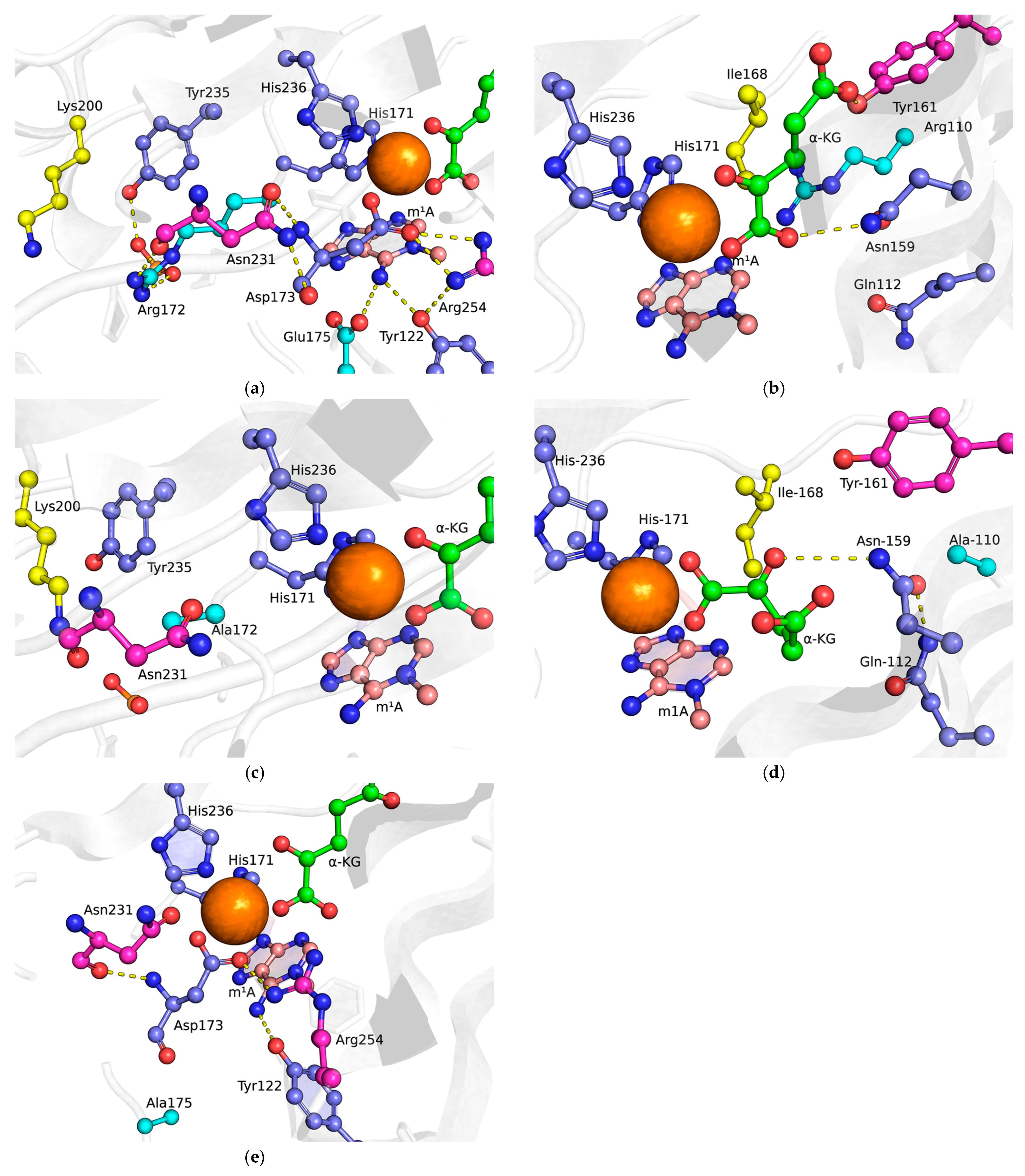
2.1. Choosing Potentially Important Amino Acid Residues Forming Specific Contacts with Damaged Bases
2.2. MD Simulations of Mutant Forms of the Enzyme
2.3. Circular Dichroism (CD) Spectra
2.4. The Melting Temperature
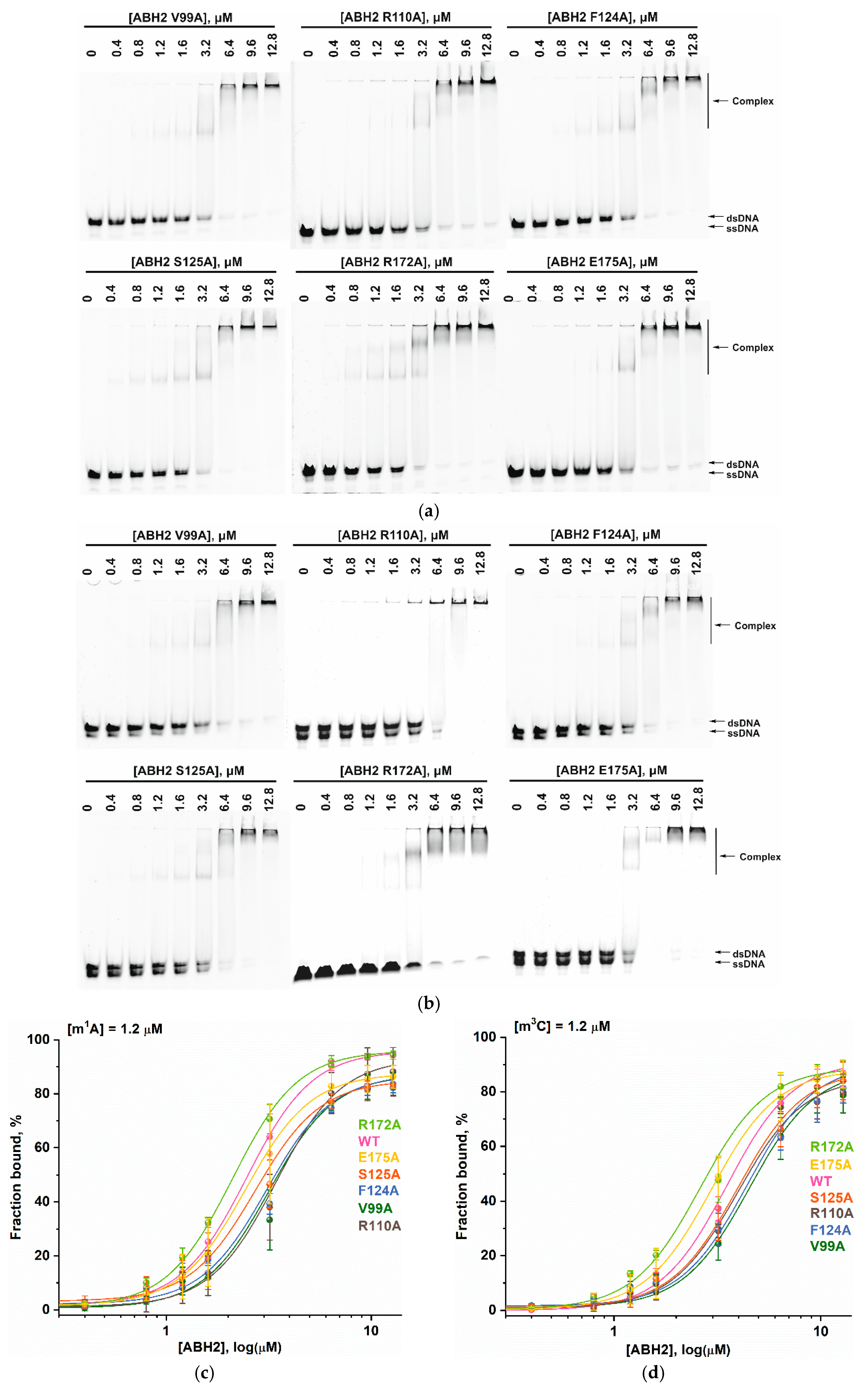
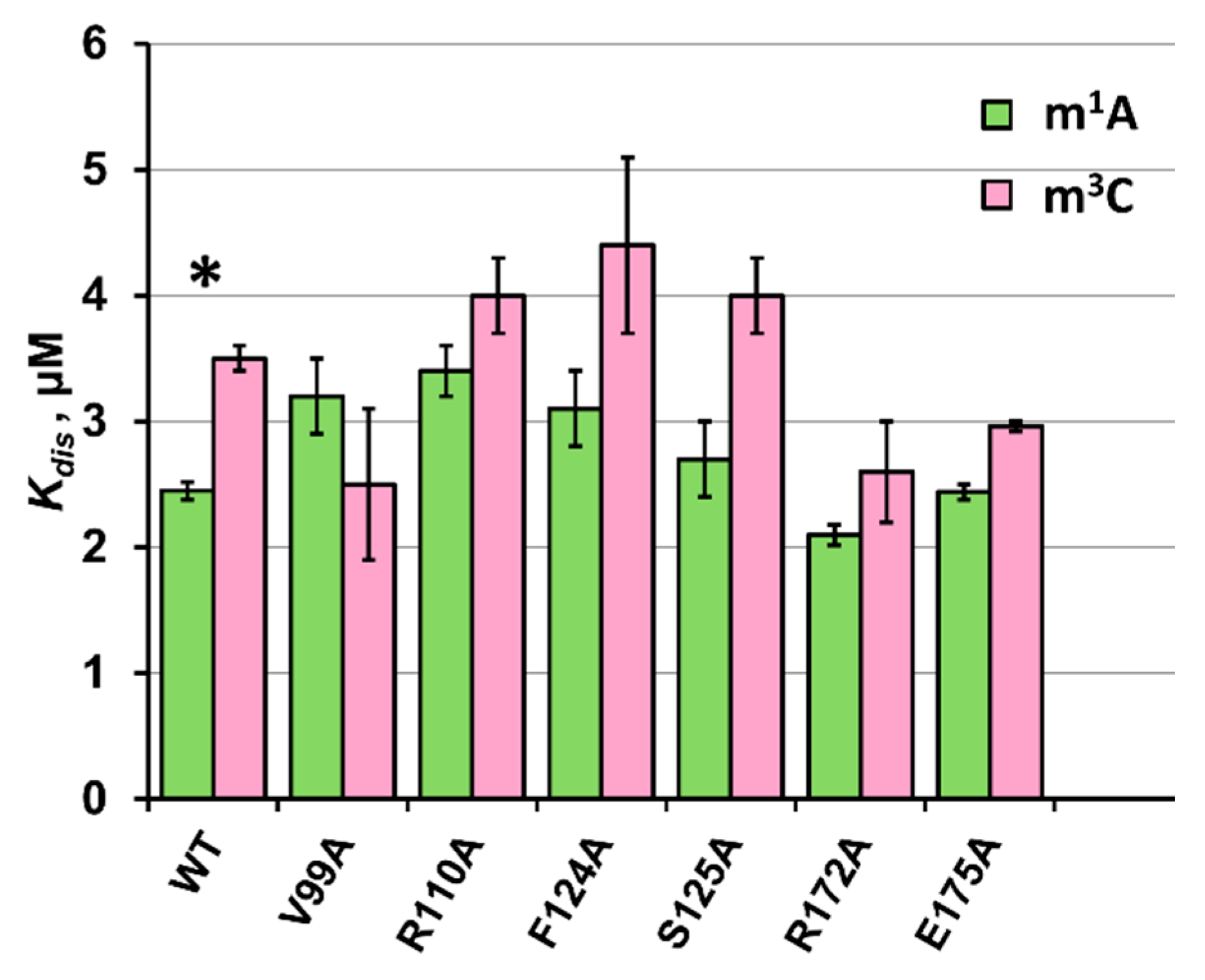
2.5. Equilibrium Binding of ABH2 Mutant Forms to Methylated DNA
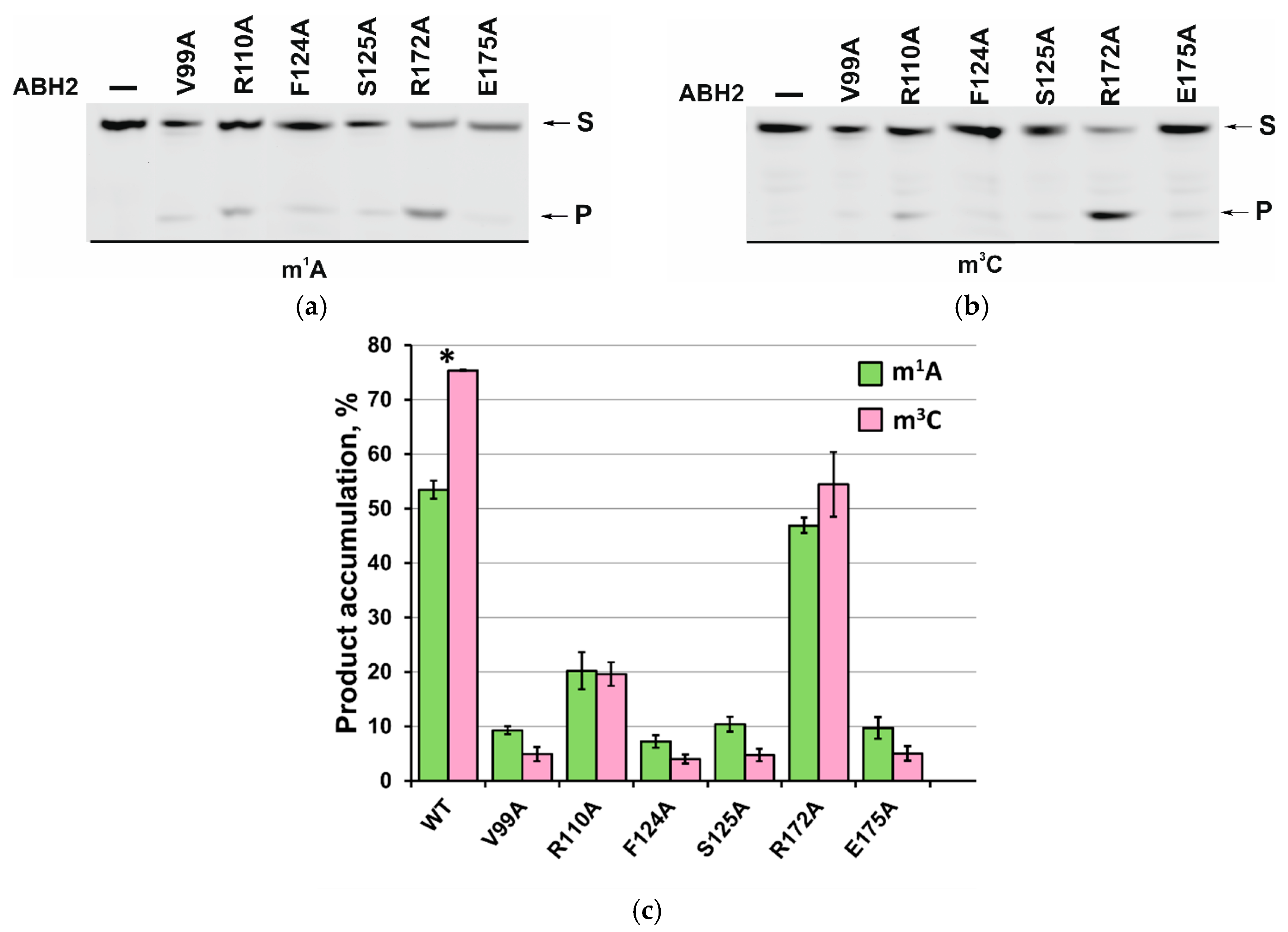
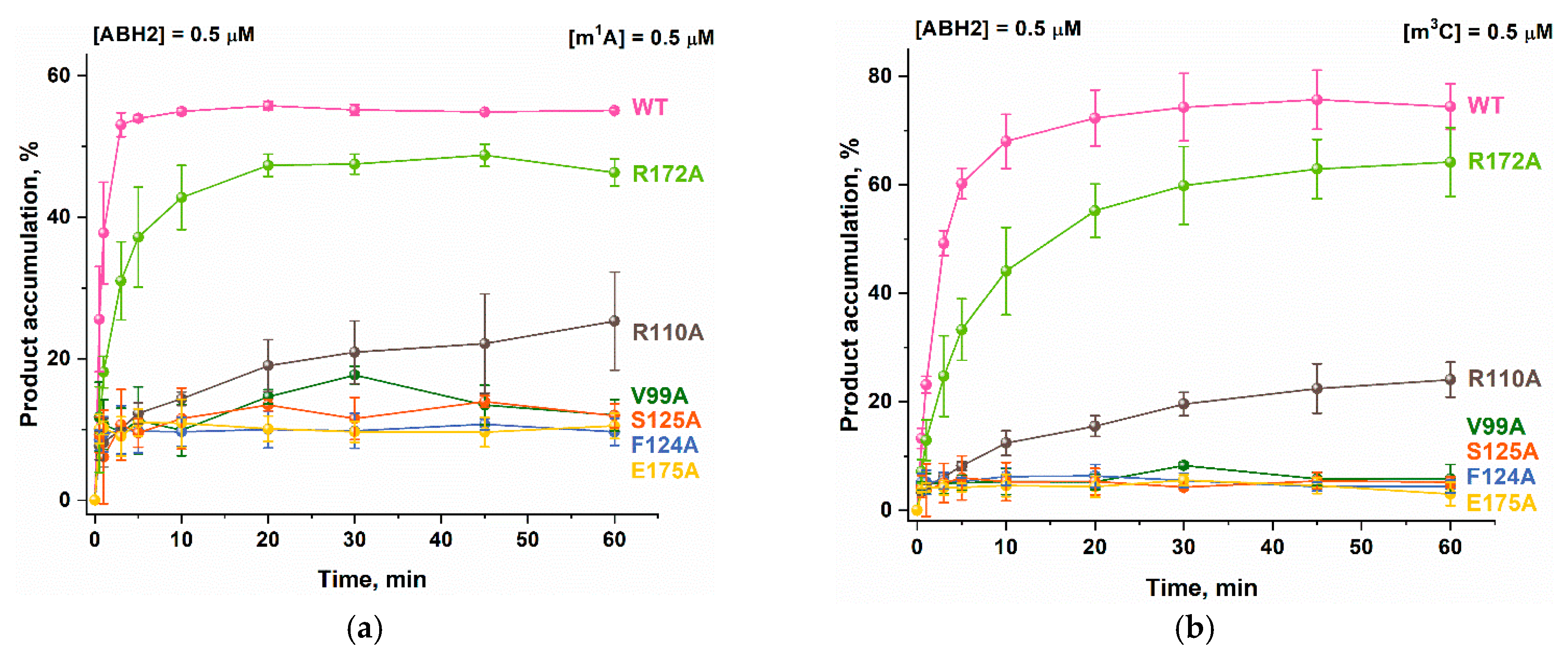
2.6. Activity of ABH2 Mutant Forms Toward DNA Containing m1A or m3C
3. Materials and Methods
3.1. Oligodeoxyribonucleotides
3.2. Site-Directed Mutagenesis
3.3. Enzyme Purification
3.4. CD Measurements
3.5. Fluorescence Thermal Shift Assay
3.6. Gel Mobility Shift Assay for the DNA Substrate Binding by ABH2 Mutant Forms
3.7. Methylated DNA Repair Activity Assays
3.8. MD Simulations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schofield, C.J.; Zhang, Z. Structural and Mechanistic Studies on 2-Oxoglutarate-Dependent Oxygenases and Related Enzymes. Curr. Opin. Struct. Biol. 1999, 9, 722–731. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, B. Repairing DNA-Methylation Damage. Nat. Rev. Mol. Cell Biol. 2004, 5, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Mishina, Y.; Duguid, E.M.; He, C. Direct Reversal of DNA Alkylation Damage. Chem. Rev. 2006, 106, 215–232. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.S. DNA Adduct Formation from Tobacco-Specific N-Nitrosamines. Mutat. Res. 1999, 424, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB Family of Fe(II)/α-Ketoglutarate-Dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Kanazhevskaya, L.Y.; Fedorova, O.S. DNA Demethylation in the Processes of Repair and Epigenetic Regulation Performed by 2-Ketoglutarate-Dependent DNA Dioxygenases. Int. J. Mol. Sci. 2021, 22, 10540. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Sedgwick, B.; Sekiguchi, M.; Nakabeppu, Y. Regulation and Expression of the Adaptive Response to Alkylating Agents. Annu. Rev. Biochem. 1988, 57, 133–157. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. The DNA-Repair Protein AlkB, EGL-9, and Leprecan Define New Families of 2-Oxoglutarate- and Iron-Dependent Dioxygenases. Genome Biol. 2001, 2, research0007.1. [Google Scholar] [CrossRef] [PubMed]
- Falnes, P.Ø.; Johansen, R.F.; Seeberg, E. AlkB-Mediated Oxidative Demethylation Reverses DNA Damage in Escherichia coli. Nature 2002, 419, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Trewick, S.C.; Henshaw, T.F.; Hausinger, R.P.; Lindahl, T.; Sedgwick, B. Oxidative Demethylation by Escherichia coli AlkB Directly Reverts DNA Base Damage. Nature 2002, 419, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Ougland, R.; Rognes, T.; Klungland, A.; Larsen, E. Non-Homologous Functions of the AlkB Homologs. J. Mol. Cell Biol. 2015, 7, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; He, C. DNA Repair by Reversal of DNA Damage. Cold Spring Harb. Perspect. Biol. 2013, 5, a012575. [Google Scholar] [CrossRef] [PubMed]
- Duncan, T.; Trewick, S.C.; Koivisto, P.; Bates, P.A.; Lindahl, T.; Sedgwick, B. Reversal of DNA Alkylation Damage by Two Human Dioxygenases. Proc. Natl. Acad. Sci. USA 2002, 99, 16660–16665. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.; Girard, C.A.; Tung, Y.-C.L.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.H.; McDonough, M.A.; Cunliffe, S.; McNeill, L.A.; et al. The Obesity-Associated FTO Gene Encodes a 2-Oxoglutarate-Dependent Nucleic Acid Demethylase. Science 2007, 318, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Kurowski, M.A.; Bhagwat, A.S.; Papaj, G.; Bujnicki, J.M. Phylogenomic Identification of Five New Human Homologs of the DNA Repair Enzyme AlkB. BMC Genomics 2003, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.F.; Carter, K.C.; Wang, R.P.; Shell, B.K. Molecular Cloning and Functional Analysis of a Human cDNA Encoding an Escherichia coli AlkB Homolog, a Protein Involved in DNA Alkylation Damage Repair. Nucleic Acids Res. 1996, 24, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhu, Q. The Role of Demethylase AlkB Homologs in Cancer. Front. Oncol. 2023, 13, 1153463. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Chen, B.; Qi, B.; Zhang, W.; Jia, G.; Zhang, L.; Li, C.J.; Dinner, A.R.; Yang, C.-G.; He, C. Duplex Interrogation by a Direct DNA Repair Protein in Search of Base Damage. Nat. Struct. Mol. Biol. 2012, 19, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.G.; Yi, C.; Duguid, E.M.; Sullivan, C.T.; Jian, X.; Rice, P.A.; He, C. Crystal Structures of DNA/RNA Repair Enzymes AlkB and ABH2 Bound to dsDNA. Nature 2008, 452, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Fu, Y.; He, C. Nucleic Acid Oxidation in DNA Damage Repair and Epigenetics. Chem. Rev. 2014, 114, 4602–4620. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yi, C. Switching Demethylation Activities between AlkB Family RNA/DNA Demethylases through Exchange of Active-Site Residues. Angew. Chem. Int. Ed. Engl. 2014, 53, 3659–3662. [Google Scholar] [CrossRef] [PubMed]
- Hausinger, R.P. FeII/Alpha-Ketoglutarate-Dependent Hydroxylases and Related Enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 21–68. [Google Scholar] [CrossRef] [PubMed]
- Clifton, I.J.; McDonough, M.A.; Ehrismann, D.; Kershaw, N.J.; Granatino, N.; Schofield, C.J. Structural Studies on 2-Oxoglutarate Oxygenases and Related Double-Stranded Beta-Helix Fold Proteins. J. Inorg. Biochem. 2006, 100, 644–669. [Google Scholar] [CrossRef] [PubMed]
- Waheed, S.O.; Ramanan, R.; Chaturvedi, S.S.; Lehnert, N.; Schofield, C.J.; Christov, C.Z.; Karabencheva-Christova, T.G. Role of Structural Dynamics in Selectivity and Mechanism of Non-Heme Fe(II) and 2-Oxoglutarate-Dependent Oxygenases Involved in DNA Repair. ACS Cent. Sci. 2020, 6, 795–814. [Google Scholar] [CrossRef] [PubMed]
- Davletgildeeva, A.T.; Tyugashev, T.E.; Zhao, M.; Kuznetsov, N.A.; Ishchenko, A.A.; Saparbaev, M.; Kuznetsova, A.A. Individual Contributions of Amido Acid Residues Tyr122, Ile168, and Asp173 to the Activity and Substrate Specificity of Human DNA Dioxygenase ABH2. Cells 2023, 12, 1839. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liu, H.; Sun, X.; Yang, C.-G. Mechanistic Insight into the Recognition of Single-Stranded and Double-Stranded DNA Substrates by ABH2 and ABH3. Mol. Biosyst. 2010, 6, 2143–2149. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Jin, S.G.; Cai, S.; Chen, Y.; Pfeifer, G.P.; O’Connor, T.R. Repair of Methylation Damage in DNA and RNA by Mammalian AlkB Homologues. J. Biol. Chem. 2005, 280, 39448–39459. [Google Scholar] [CrossRef] [PubMed]
- Monsen, V.T.; Sundheim, O.; Aas, P.A.; Westbye, M.P.; Sousa, M.M.L.; Slupphaug, G.; Krokan, H.E. Divergent β-Hairpins Determine Double-Strand versus Single-Strand Substrate Recognition of Human AlkB-Homologues 2 and 3. Nucleic Acids Res. 2010, 38, 6447–6455. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pang, X.; Han, K.; Cui, Q. A Simple but Effective Modeling Strategy for Structural Properties of Non-Heme Fe(II) Sites in Proteins: Test of Force Field Models and Application to Proteins in the AlkB Family. J. Comput. Chem. 2013, 34, 1620–1635. [Google Scholar] [CrossRef] [PubMed]
- Bian, K.; Lenz, S.A.P.; Tang, Q.; Chen, F.; Qi, R.; Jost, M.; Drennan, C.L.; Essigmann, J.M.; Wetmore, S.D.; Li, D. DNA Repair Enzymes ALKBH2, ALKBH3, and AlkB Oxidize 5-Methylcytosine to 5-Hydroxymethylcytosine, 5-Formylcytosine and 5-Carboxylcytosine in Vitro. Nucleic Acids Res. 2019, 47, 5522–5529. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, H.; Tan, T. Rational Design of Methodology-Independent Metal Parameters Using a Nonbonded Dummy Model. J. Chem. Theory Comput. 2016, 12, 3250–3260. [Google Scholar] [CrossRef] [PubMed]
- Lenz, S.A.P.; Li, D.; Wetmore, S.D. Insights into the Direct Oxidative Repair of Etheno Lesions: MD and QM/MM Study on the Substrate Scope of ALKBH2 and AlkB. DNA Repair 2020, 96, 102944. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Merz, K.M. Metal Ion Modeling Using Classical Mechanics. Chem. Rev. 2017, 117, 1564–1686. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Liu, D.; Wang, Z.; Tian, R.; Zuo, Y. Multi-Substrate Selectivity Based on Key Loops and Non-Homologous Domains: New Insight into ALKBH Family. Cell. Mol. Life Sci. 2021, 78, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Abriata, L.A. A Simple Spreadsheet Program to Simulate and Analyze the Far-UV Circular Dichroism Spectra of Proteins. J. Chem. Educ. 2011, 88, 1268–1273. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. In Protein Structure Prediction; Methods Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2014; Volume 1137. [Google Scholar]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197, Erratum in J. Am. Chem. Soc. 1996, 118, 2309. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Zgarbová, M.; Šponer, J.; Otyepka, M.; Cheatham, T.E.; Galindo-Murillo, R.; Jurečka, P. Refinement of the Sugar-Phosphate Backbone Torsion Beta for AMBER Force Fields Improves the Description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. [Google Scholar] [CrossRef] [PubMed]
- Zgarbová, M.; Otyepka, M.; Sponer, J.; Mládek, A.; Banáš, P.; Cheatham, T.E.; Jurečka, P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Joung, I.S.; Cheatham, T.E. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef] [PubMed]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges: The RESP Model—ScienceOpen. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Meagher, K.L.; Redman, L.T.; Carlson, H.A. Development of Polyphosphate Parameters for Use with the AMBER Force Field. J. Comput. Chem. 2003, 24, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.-Y. R.E.D. Server: A Web Service for Deriving RESP and ESP Charges and Building Force Field Libraries for New Molecules and Molecular Fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic Atom Type and Bond Type Perception in Molecular Mechanical Calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Sousa da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Caston, R.A.; Gampala, S.; Armstrong, L.; Messmann, R.A.; Fishel, M.L.; Kelley, M.R. The Multifunctional APE1 DNA Repair–Redox Signaling Protein as a Drug Target in Human Disease. Drug Discov. Today 2021, 26, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Wennberg, C.L.; Murtola, T.; Hess, B.; Lindahl, E. Lennard-Jones Lattice Summation in Bilayer Simulations Has Critical Effects on Surface Tension and Lipid Properties. J. Chem. Theory Comput. 2013, 9, 3527–3537. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.; Fraaije, J. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Popov, A.V.; Vorobjev, Y.N.; Zharkov, D.O. MDTRA: A Molecular Dynamics Trajectory Analyzer with a Graphical User Interface. J. Comput. Chem. 2013, 34, 319–325. [Google Scholar] [CrossRef] [PubMed]

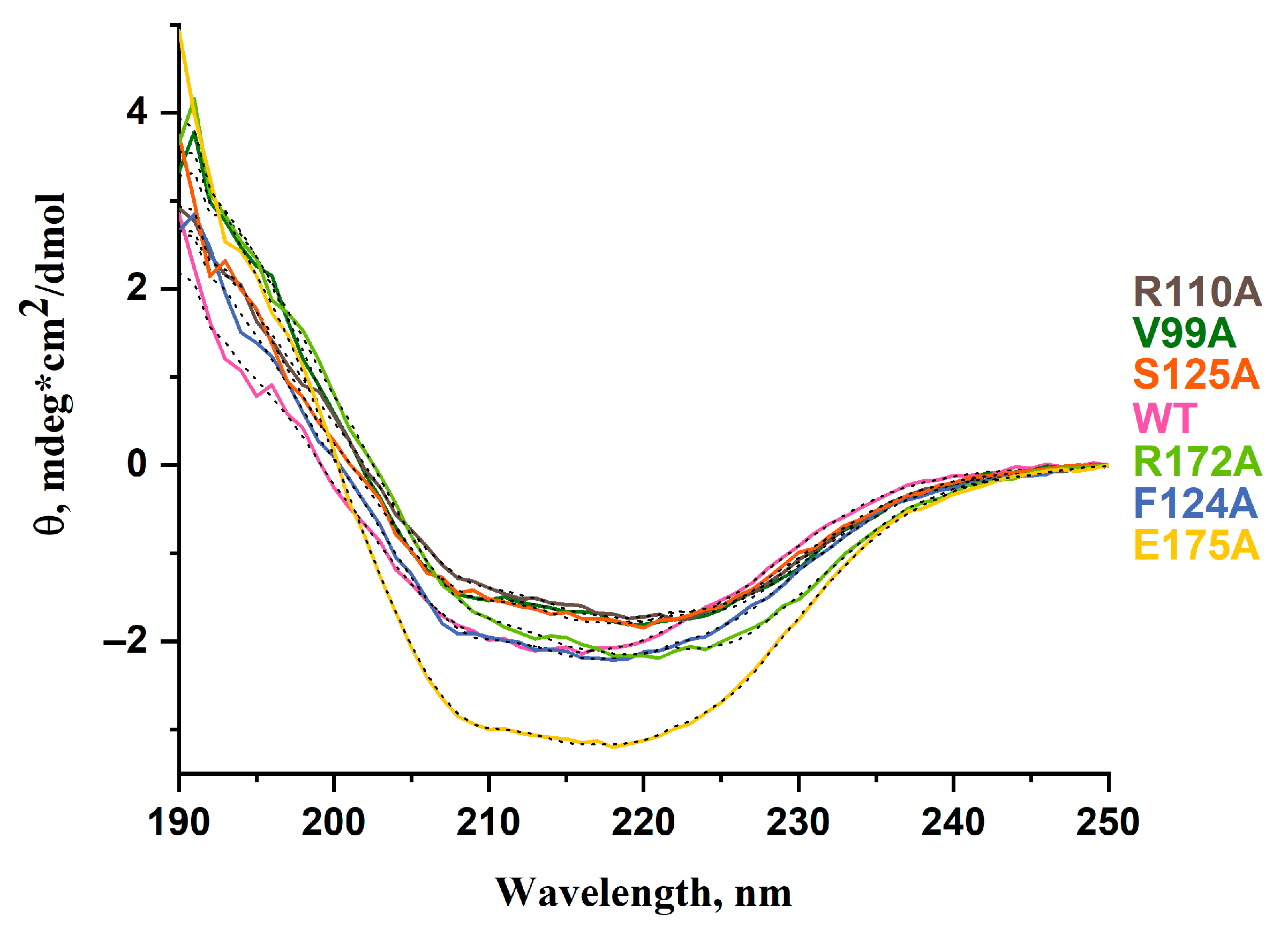



| α-Helix (%) | β-Sheet (%) | β-Turn (%) | Unordered (%) | |
|---|---|---|---|---|
| WT | 13 ± 3 | 34 ± 7 | 12 ± 2 | 42 ± 8 |
| V99A | 11 ± 2 | 34 ± 7 | 12 ± 2 | 43 ± 9 |
| R110A | 8 ± 2 | 36 ± 7 | 12 ± 2 | 44 ± 9 |
| F124A | 11 ± 2 | 33 ± 7 | 12 ± 2 | 44 ± 9 |
| S125A | 9 ± 2 | 36 ± 7 | 12 ± 2 | 43 ± 9 |
| R172A | 12 ± 2 | 35 ± 7 | 11 ± 2 | 42 ± 8 |
| E175A | 18 ± 4 | 28 ± 6 | 11 ± 2 | 42 ± 8 |
| WT | V99A | R110A | F124A | S125A | R172A | E175A | |
|---|---|---|---|---|---|---|---|
| Tm, °C | 46.7 ± 0.7 | 50.4 ± 0.4 | 49.1 ± 0.7 | 46 ± 1 | 49.1 ± 0.3 | 47.6 ± 0.2 | 47.7 ± 0.5 |
| Kd, µM | kobs, s−1 | ||
|---|---|---|---|
| WT * | 2.45 ± 0.07 | 4.1 ± 0.4 | m1A |
| V99A | 3.2 ± 0.3 | - | |
| R110A | 3.4 ± 0.2 | 1.0 ± 0.1 | |
| F124A | 3.1 ± 0.3 | - | |
| S125A | 2.7 ± 0.3 | - | |
| R172A | 2.10 ± 0.08 | 2.2 ± 0.2 | |
| E175A | 2.44 ± 0.06 | - | |
| WT * | 3.5 ± 0.1 | 4.3 ± 0.4 | m3C |
| V99A | 2.5 ± 0.6 | - | |
| R110A | 4.0 ± 0.3 | 0.63 ± 0.06 | |
| F124A | 4.4 ± 0.7 | - | |
| S125A | 4.0 ± 0.3 | - | |
| R172A | 2.6 ± 0.4 | 1.8 ± 0.2 | |
| E175A | 2.96 ± 0.04 | - |
| Shorthand | Sequence |
|---|---|
| m1A | 5′-FAM-AGTTCAATG-m1A-TCTTCAT-3′ 3′-TCAAGTTAC T AGAAGTA-5′ |
| m3C | 5′-FAM-AGTTCAATGAT-m3C-TTCAT-3′ 3′-TCAAGTTACTA G AAGTA-5′ |
| GATC | 5′-FAM-AGTTCAATGATCTTCAT-3′ 3′-TCAAGTTACTAGAAGTA-5′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davletgildeeva, A.T.; Tyugashev, T.E.; Zhao, M.; Ishchenko, A.A.; Saparbaev, M.; Kuznetsov, N.A. Role of Individual Amino Acid Residues Directly Involved in Damage Recognition in Active Demethylation by ABH2 Dioxygenase. Int. J. Mol. Sci. 2025, 26, 6912. https://doi.org/10.3390/ijms26146912
Davletgildeeva AT, Tyugashev TE, Zhao M, Ishchenko AA, Saparbaev M, Kuznetsov NA. Role of Individual Amino Acid Residues Directly Involved in Damage Recognition in Active Demethylation by ABH2 Dioxygenase. International Journal of Molecular Sciences. 2025; 26(14):6912. https://doi.org/10.3390/ijms26146912
Chicago/Turabian StyleDavletgildeeva, Anastasiia T., Timofey E. Tyugashev, Mingxing Zhao, Alexander A. Ishchenko, Murat Saparbaev, and Nikita A. Kuznetsov. 2025. "Role of Individual Amino Acid Residues Directly Involved in Damage Recognition in Active Demethylation by ABH2 Dioxygenase" International Journal of Molecular Sciences 26, no. 14: 6912. https://doi.org/10.3390/ijms26146912
APA StyleDavletgildeeva, A. T., Tyugashev, T. E., Zhao, M., Ishchenko, A. A., Saparbaev, M., & Kuznetsov, N. A. (2025). Role of Individual Amino Acid Residues Directly Involved in Damage Recognition in Active Demethylation by ABH2 Dioxygenase. International Journal of Molecular Sciences, 26(14), 6912. https://doi.org/10.3390/ijms26146912







