Transmembrane Protein 43: Molecular and Pathogenetic Implications in Arrhythmogenic Cardiomyopathy and Various Other Diseases
Abstract
1. Introduction
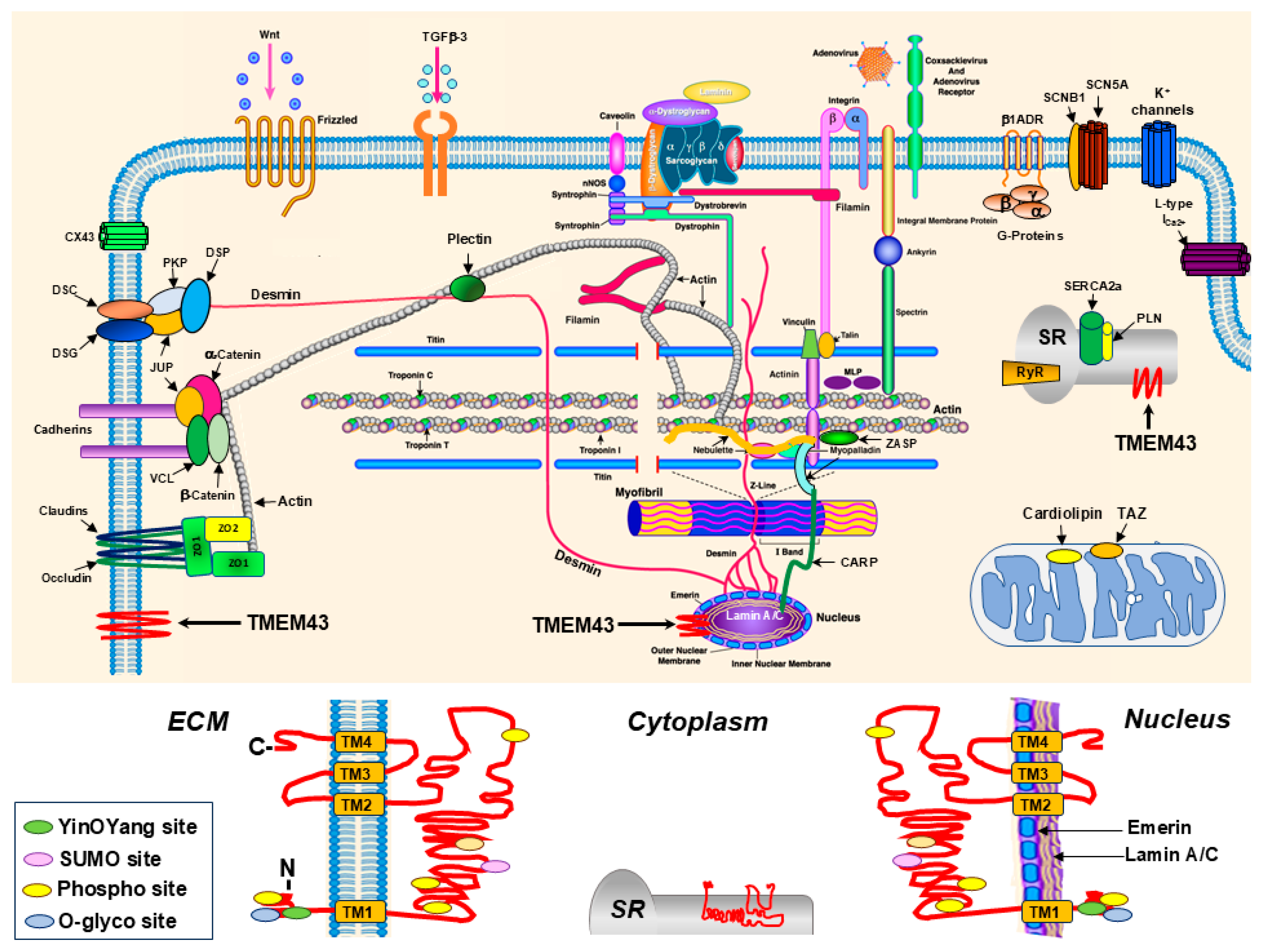
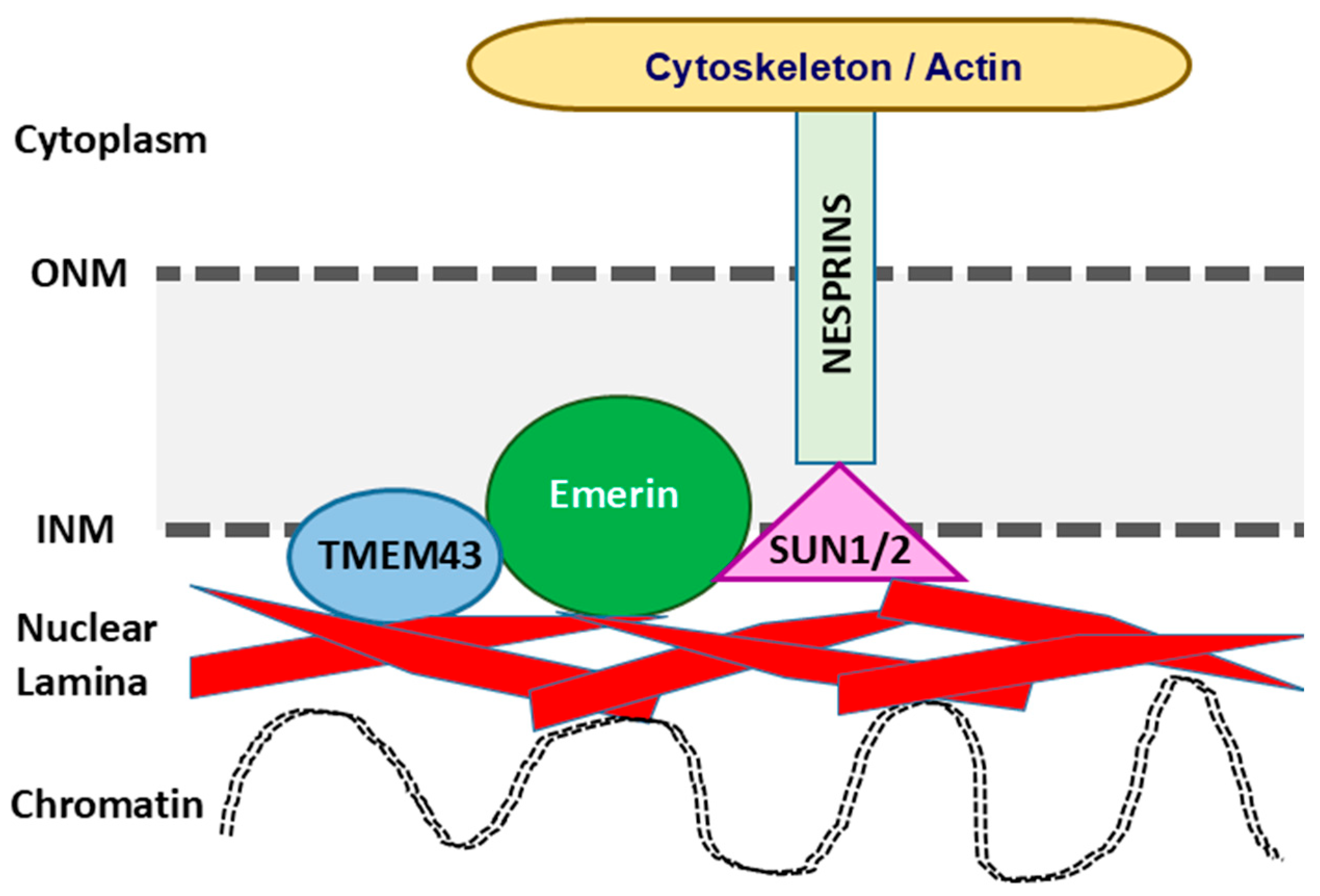
2. Biological and Molecular Functions of TMEM43
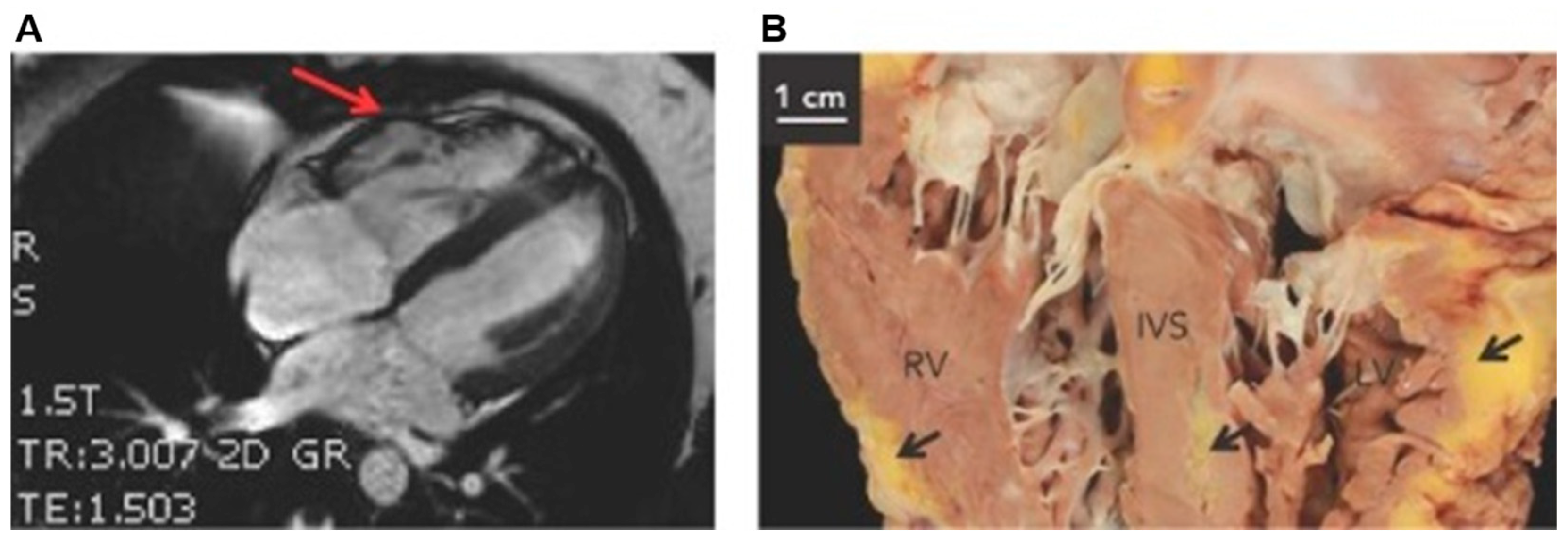
3. Arrhythmogenic Cardiomyopathies
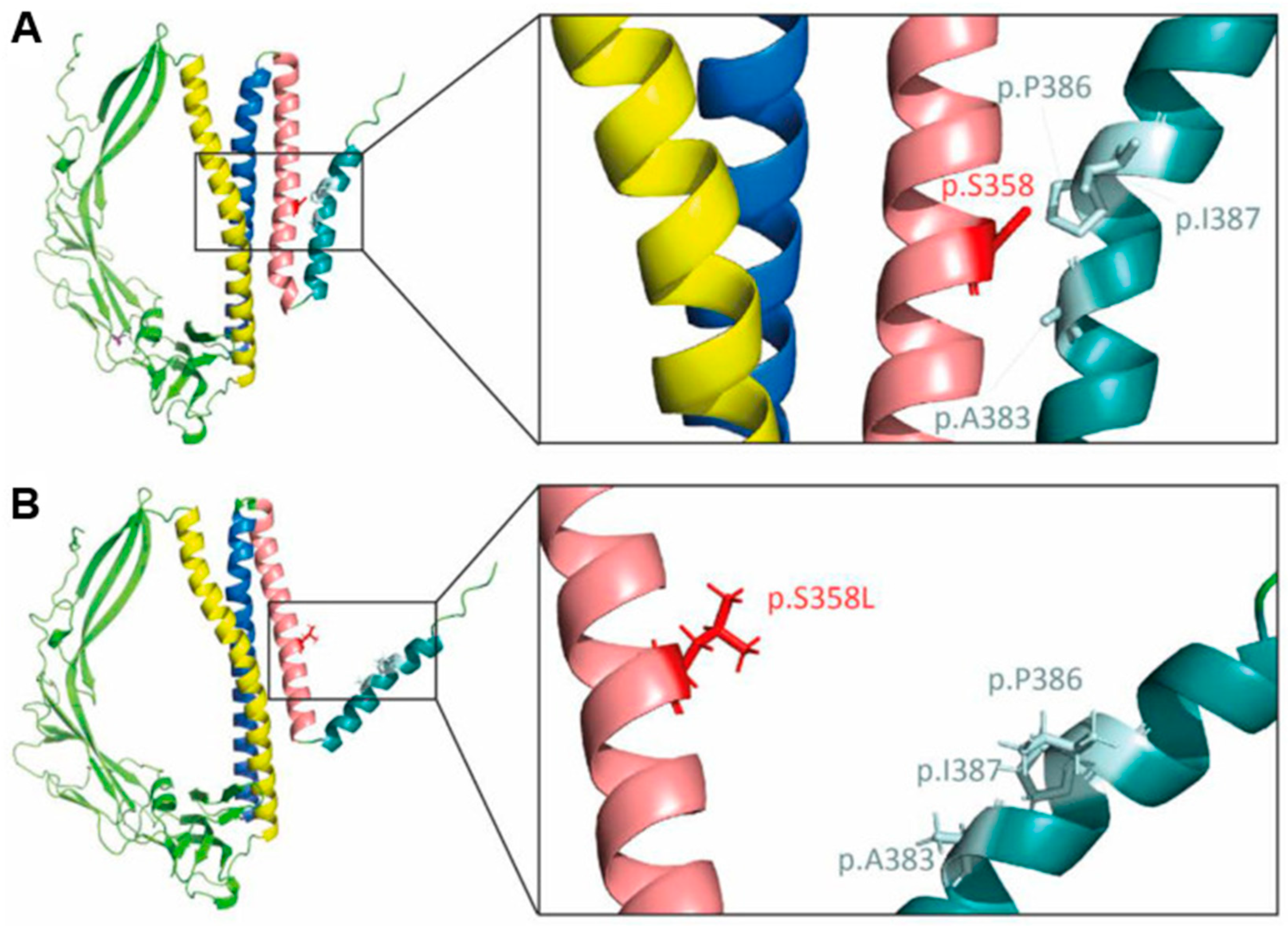
4. TMEM43 Mutations Causal for Arrhythmogenic Cardiomyopathies
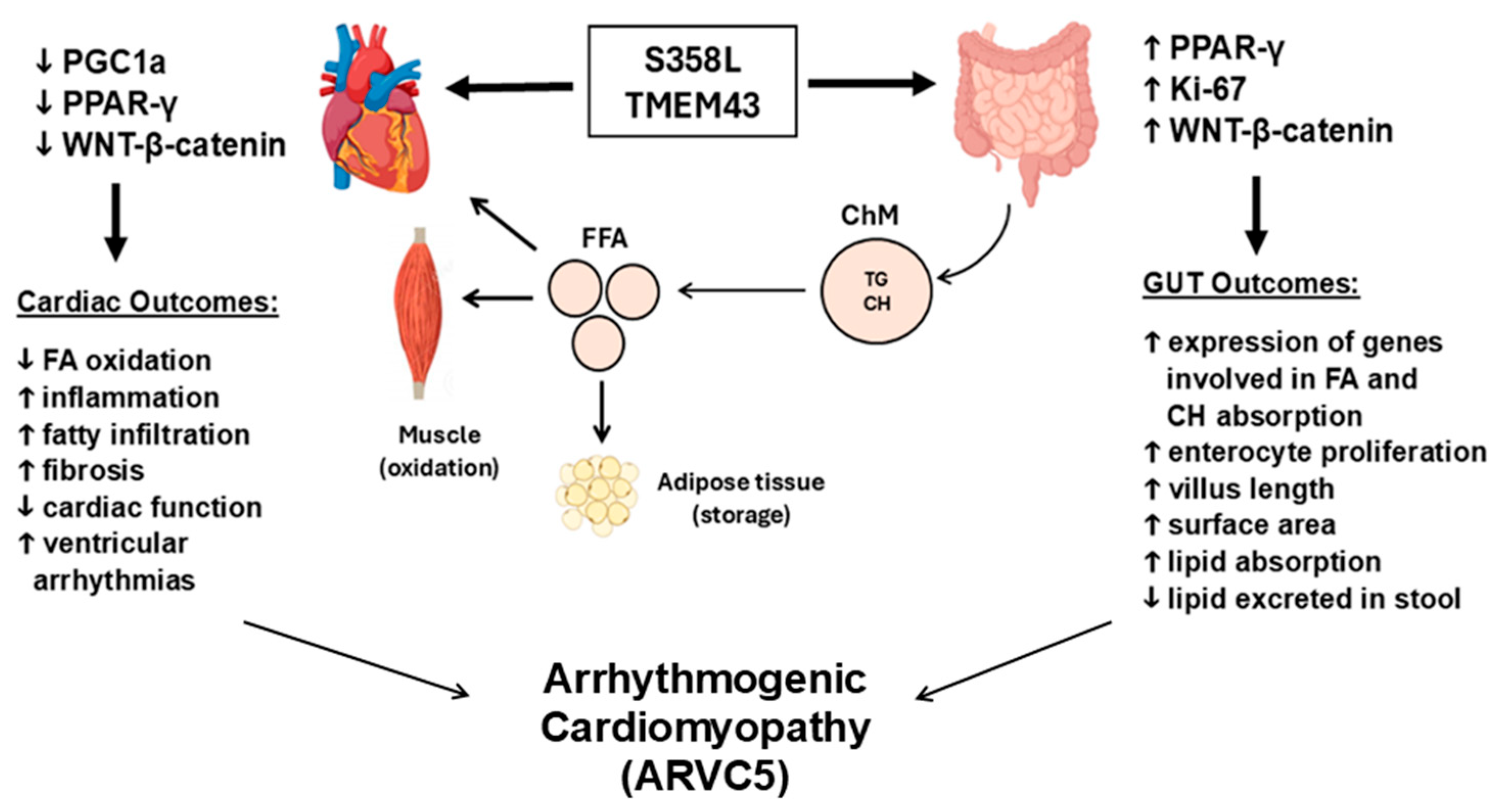
5. Mechanisms of Fibro-Fatty Replacement of the Myocardium in Tmem43-Associated Arrhythmogenic Cardiomyopathies
6. TMEM43 and Exercise in Arrhythmogenic Cardiomyopathies
7. Animal and In Vitro Modeling of the TMEM43 S358L Mutation
8. TMEM43 and Muscular Dystrophy
9. TMEM43 and Cancer Progression
10. TMEM43 and Auditory Neuropathy Spectrum Disorders
11. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | Adeno-associated virus |
| ACM | Arrhythmogenic cardiomyopathy |
| AKT | V-akt murine thymoma viral oncogene homolog |
| ANSD | auditory neuropathy spectrum disorders |
| ARVD/C | Arrhythmogenic right ventricular dysplasia/cardiomyopathy |
| CMR | Cardiac magnetic resonance |
| CX | Connexin |
| DDR | DNA damage response |
| DNA | Deoxyribonucleic acid |
| DSC | Desmocollin |
| DSG | Desmoglein |
| DSP | Desmoplakin |
| EAT | Epicardial adipose tissue |
| EDMD | Emery–Dreifuss Muscular Dystrophy |
| ER | Endoplasmic reticulum |
| GLSs | Glia-like supporting cells |
| HCC | Hepatocellular carcinoma |
| HF | Heart failure |
| ICD | Implantable cardioverter defibrillator |
| IGF-1 | Insulin-like growth factor 1 |
| INM | Inner nuclear membrane |
| JUP | Junctional plakoglobin |
| KASH | Klarsicht, ANC1, Syne Homology |
| LINC | Nucleoskeleton and cytoskeleton complex |
| mRNA | Messenger ribonucleic acid |
| NF-κB | Nuclear factor kappa B |
| ONM | Outer nuclear membrane |
| SUN | Sad1 and UNC-84 |
| SUMO | Small ubiquitin-like modifier |
| PKP | plakophilin |
| PPREs | Peroxisome proliferator response elements |
| PPAR | Peroxisome proliferator-activated receptor |
| PNS | Perinuclear space |
| SCD | Sudden cardiac death |
| SR | Sarcoplasmic reticulum |
| TGF | Transforming growth factor |
| TMEM | Transmembrane protein |
| VTA | Ventricular tachyarrhythmia |
| WNT | Wingless and Int |
| ZO-1 | Zonula occludens-1 |
References
- Bengtsson, L.; Otto, H. LUMA interacts with emerin and influences its distribution at the inner nuclear membrane. J. Cell Sci. 2008, 121 Pt 4, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Dreger, M.; Bengtsson, L.; Schoneberg, T.; Otto, H.; Hucho, F. Nuclear envelope proteomics: Novel integral membrane proteins of the inner nuclear membrane. Proc. Natl. Acad. Sci. USA 2001, 98, 11943–11948. [Google Scholar] [CrossRef] [PubMed]
- Fenech, E.J.; Lari, F.; Charles, P.D.; Fischer, R.; Laetitia-Thezenas, M.; Bagola, K.; Paton, A.W.; Paton, J.C.; Gyrd-Hansen, M.; Kessler, B.M.; et al. Interaction mapping of endoplasmic reticulum ubiquitin ligases identifies modulators of innate immune signalling. eLife 2020, 9, e57306. [Google Scholar] [CrossRef] [PubMed]
- Franke, W.W.; Dorflinger, Y.; Kuhn, C.; Zimbelmann, R.; Winter-Simanowski, S.; Frey, N.; Heid, H. Protein LUMA is a cytoplasmic plaque constituent of various epithelial adherens junctions and composite junctions of myocardial intercalated disks: A unifying finding for cell biology and cardiology. Cell Tissue Res. 2014, 357, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.W.; Kim, T.Y.; Sharma, K.; Kwon, J.; Yi, E.; Lee, C.J. A Deafness Associated Protein TMEM43 Interacts with KCNK3 (TASK-1) Two-pore Domain K(+) (K2P) Channel in the Cochlea. Exp. Neurobiol. 2021, 30, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Merner, N.D.; Hodgkinson, K.A.; Haywood, A.F.; Connors, S.; French, V.M.; Drenckhahn, J.D.; Kupprion, C.; Ramadanova, K.; Thierfelder, L.; McKenna, W.; et al. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am. J. Hum. Genet. 2008, 82, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.H.; Andersen, C.B.; Tybjaerg-Hansen, A.; Haunso, S.; Svendsen, J.H. Mutation analysis and evaluation of the cardiac localization of TMEM43 in arrhythmogenic right ventricular cardiomyopathy. Clin. Genet. 2011, 80, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.C.; Mitsuhashi, H.; Keduka, E.; Nonaka, I.; Noguchi, S.; Nishino, I.; Hayashi, Y.K. TMEM43 mutations in Emery-Dreifuss muscular dystrophy-related myopathy. Ann. Neurol. 2011, 69, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Mori-Yoshimura, M.; Nishikawa, A.; Hokkoku, K.; Sonoo, M.; Nishino, I.; Takahashi, Y. Emery-Dreifuss muscular dystrophy-related myopathy with TMEM43 mutations. Muscle Nerve 2019, 59, E5–E7. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.W.; Oh, D.Y.; Yi, E.; Liu, X.; Ling, J.; Kim, N.; Sharma, K.; Kim, T.Y.; Lee, S.; Kim, A.R.; et al. A nonsense TMEM43 variant leads to disruption of connexin-linked function and autosomal dominant auditory neuropathy spectrum disorder. Proc. Natl. Acad. Sci. USA 2021, 118, e2019681118. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.-C.; Roche, M.V.; Um, S.Y.; Gosstola, N.C.; Kim, M.Y.; Choi, B.Y.; Dykxhoorn, D.M.; Liu, X. Characterization of an induced pluripotent stem cell line (UMi040-A) bearing an auditory neuropathy spectrum disorder-associated variant in TMEM43. Stem Cell Res. 2022, 61, 102758. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Zhu, Y.; Zhou, Z.; Gumin, J.; Bengtsson, L.; Wu, W.; Songyang, Z.; Lang, F.F.; Lin, X. TMEM43/LUMA is a key signaling component mediating EGFR-induced NF-kappaB activation and tumor progression. Oncogene 2017, 36, 2813–2823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, F.; Yang, X.; Wang, Q.; Chang, R.; Zhu, L.; Feitelson, M.A.; Chen, Z. TMEM43 promotes the development of hepatocellular carcinoma by activating VDAC1 through USP7 deubiquitination. Transl. Gastroenterol. Hepatol. 2024, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, Y.; Zhang, C.; Wang, R.; Hua, L.; Guo, Y.; Gan, D.; Zhu, L.; Li, S.; Ma, P.; et al. TMEM43 promotes pancreatic cancer progression by stabilizing PRPF3 and regulating RAP2B/ERK axis. Cell Mol. Biol. Lett. 2022, 27, 24. [Google Scholar] [CrossRef] [PubMed]
- Rouhi, L.; Cheedipudi, S.M.; Chen, S.N.; Fan, S.; Lombardi, R.; Chen, X.; Coarfa, C.; Robertson, M.J.; Gurha, P.; Marian, A.J. Haplo-insufficiency of Tmem43 in cardiac myocytes activates the DNA damage response pathway leading to a Late-Onset Senescence-Associated pro-fibrotic cardiomyopathy. Cardiovasc. Res. 2020, 117, 2377–2394. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Yao, Y.R.; Ding, Y.; Zhang, X.W. Reduced expression of transmembrane protein 43 during cardiac hypertrophy leads to worsening heart failure in mice. Exp. Biol. Med. 2023, 248, 1437–1445. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cao, Z.; Gui, F.; Zhang, M.; Wu, X.; Peng, H.; Yu, B.; Li, W.; Ai, F.; Zhang, J. TMEM43 Protects against Sepsis-Induced Cardiac Injury via Inhibiting Ferroptosis in Mice. Cells 2022, 11, 2992. [Google Scholar] [CrossRef] [PubMed]
- Towbin, J.A. Inherited cardiomyopathies. Circ. J. 2014, 78, 2347–2356. [Google Scholar] [CrossRef] [PubMed]
- Susa, K.J.; Kruse, A.C.; Blacklow, S.C. Tetraspanins: Structure, dynamics, and principles of partner-protein recognition. Trends Cell Biol. 2024, 34, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Klinke, N.; Meyer, H.; Ratnavadivel, S.; Reinhardt, M.; Heinisch, J.J.; Malmendal, A.; Milting, H.; Paululat, A. A Drosophila melanogaster model for TMEM43-related arrhythmogenic right ventricular cardiomyopathy type 5. Cell. Mol. Life Sci. 2022, 79, 444. [Google Scholar] [CrossRef] [PubMed]
- Shinomiya, H.; Kato, H.; Kuramoto, Y.; Watanabe, N.; Tsuruda, T.; Arimura, T.; Miyashita, Y.; Miyasaka, Y.; Mashimo, T.; Takuwa, A.; et al. Aberrant accumulation of TMEM43 accompanied by perturbed transmural gene expression in arrhythmogenic cardiomyopathy. FASEB J. 2021, 35, e21994. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.; Kutay, U. The Diverse Cellular Functions of Inner Nuclear Membrane Proteins. Cold Spring Harb. Perspect. Biol. 2021, 13, a040477. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, E.C.; Florens, L.; Guan, T.; Yates, J.R., 3rd; Gerace, L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 2003, 301, 1380–1382. [Google Scholar] [CrossRef] [PubMed]
- Gunn, A.L.; Yashchenko, A.I.; Dubrulle, J.; Johnson, J.; Hatch, E.M. A high-content screen reveals new regulators of nuclear membrane stability. Sci. Rep. 2024, 14, 6013. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmsen, K.; Ketema, M.; Truong, H.; Sonnenberg, A. KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 2006, 119 Pt 24, 5021–5029. [Google Scholar] [CrossRef] [PubMed]
- Mejat, A.; Misteli, T. LINC complexes in health and disease. Nucleus 2010, 1, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Mellad, J.A.; Warren, D.T.; Shanahan, C.M. Nesprins LINC the nucleus and cytoskeleton. Curr. Opin. Cell Biol. 2011, 23, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.L.; Lammerding, J. Keeping the LINC: The importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem. Soc. Trans. 2011, 39, 1729–1734. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, K.H.; Kennedy, B.K. When lamins go bad: Nuclear structure and disease. Cell 2013, 152, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Stroud, M.J.; Banerjee, I.; Veevers, J.; Chen, J. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure, function, and disease. Circ. Res. 2014, 114, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Shimojima, M.; Yuasa, S.; Motoda, C.; Yozu, G.; Nagai, T.; Ito, S.; Lachmann, M.; Kashimura, S.; Takei, M.; Kusumoto, D.; et al. Emerin plays a crucial role in nuclear invagination and in the nuclear calcium transient. Sci. Rep. 2017, 7, 44312. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.A.; Warley, A.; Roberts, R.G.; Ehler, E.; Ellis, J.A. Identification of an emerin-beta-catenin complex in the heart important for intercalated disc architecture and beta-catenin localisation. Cell. Mol. Life Sci. 2010, 67, 781–796. [Google Scholar] [CrossRef] [PubMed]
- Siragam, V.; Cui, X.; Masse, S.; Ackerley, C.; Aafaqi, S.; Strandberg, L.; Tropak, M.; Fridman, M.D.; Nanthakumar, K.; Liu, J.; et al. TMEM43 mutation p.S358L alters intercalated disc protein expression and reduces conduction velocity in arrhythmogenic right ventricular cardiomyopathy. PLoS ONE 2014, 9, e109128. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, R.; Sembrat, J.C.; McDonough, B.; Seidman, C.E.; Ahmad, F. Functional effects of the TMEM43 Ser358Leu mutation in the pathogenesis of arrhythmogenic right ventricular cardiomyopathy. BMC Med. Genet. 2012, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Lemay, D.G.; Hwang, D.H. Genome-wide identification of peroxisome proliferator response elements using integrated computational genomics. J. Lipid Res. 2006, 47, 1583–1587. [Google Scholar] [CrossRef] [PubMed]
- Zink, M.; Seewald, A.; Rohrbach, M.; Brodehl, A.; Liedtke, D.; Williams, T.; Childs, S.J.; Gerull, B. Altered Expression of TMEM43 Causes Abnormal Cardiac Structure and Function in Zebrafish. Int. J. Mol. Sci. 2022, 23, 9530. [Google Scholar] [CrossRef] [PubMed]
- Freedman, A.N.; Eaves, L.A.; Rager, J.E.; Gavino-Lopez, N.; Smeester, L.; Bangma, J.; Santos, H.P.; Joseph, R.M.; Kuban, K.C.; O’Shea, T.M.; et al. The placenta epigenome-brain axis: Placental epigenomic and transcriptomic responses that preprogram cognitive impairment. Epigenomics 2022, 14, 897–911. [Google Scholar] [CrossRef] [PubMed]
- Marcus, F.; Towbin, J.A.; Zareba, W.; Moss, A.; Calkins, H.; Brown, M.; Gear, K.; Investigators, A.C. Arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C): A multidisciplinary study: Design and protocol. Circulation 2003, 107, 2975–2978. [Google Scholar] [CrossRef] [PubMed]
- Spezzacatene, A.; Sinagra, G.; Merlo, M.; Barbati, G.; Graw, S.L.; Brun, F.; Slavov, D.; Di Lenarda, A.; Salcedo, E.E.; Towbin, J.A.; et al. Arrhythmogenic Phenotype in Dilated Cardiomyopathy: Natural History and Predictors of Life-Threatening Arrhythmias. J. Am. Heart Assoc. 2015, 4, e002149. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Trummel, M.; Meyners, W. Prevalence of right ventricular dysplasia-cardiomyopathy in a non-referral hospital. Int. J. Cardiol. 2004, 97, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Groeneweg, J.A.; Bhonsale, A.; James, C.A.; te Riele, A.S.; Dooijes, D.; Tichnell, C.; Murray, B.; Wiesfeld, A.C.; Sawant, A.C.; Kassamali, B.; et al. Clinical Presentation, Long-Term Follow-Up, and Outcomes of 1001 Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Patients and Family Members. Circ. Cardiovasc. Genet. 2015, 8, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Dalla Volta, S.; Battaglia, G.; Zerbini, E. “Auricularization” of right ventricular pressure curve. Am. Heart J. 1961, 61, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Marcus, F.I.; Fontaine, G.H.; Guiraudon, G.; Frank, R.; Laurenceau, J.L.; Malergue, C.; Grosgogeat, Y. Right ventricular dysplasia: A report of 24 adult cases. Circulation 1982, 65, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Towbin, J.A.; Thiene, G.; Antzelevitch, C.; Corrado, D.; Arnett, D.; Moss, A.J.; Seidman, C.E.; Young, J.B.; American Heart, A.; et al. Prevention, Contemporary definitions and classification of the cardiomyopathies: An American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 2006, 113, 1807–1816. [Google Scholar] [PubMed]
- Sen-Chowdhry, S.; Syrris, P.; Ward, D.; Asimaki, A.; Sevdalis, E.; McKenna, W.J. Clinical and genetic characterization of families with arrhythmogenic right ventricular dysplasia/cardiomyopathy provides novel insights into patterns of disease expression. Circulation 2007, 115, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Capulzini, L.; Brugada, P.; Brugada, J.; Brugada, R. Arrhythmia and right heart disease: From genetic basis to clinical practice. Rev. Esp. Cardiol. 2010, 63, 963–983. [Google Scholar] [CrossRef] [PubMed]
- Towbin, J.A.; McKenna, W.J.; Abrams, D.J.; Ackerman, M.J.; Calkins, H.; Darrieux, F.C.C.; Daubert, J.P.; de Chillou, C.; DePasquale, E.C.; Desai, M.Y.; et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm 2019, 16, e301–e372. [Google Scholar] [CrossRef] [PubMed]
- Marcus, F.I.; Zareba, W.; Calkins, H.; Towbin, J.A.; Basso, C.; Bluemke, D.A.; Estes, N.A., 3rd; Picard, M.H.; Sanborn, D.; Thiene, G.; et al. Arrhythmogenic right ventricular cardiomyopathy/dysplasia clinical presentation and diagnostic evaluation: Results from the North American Multidisciplinary Study. Heart Rhythm 2009, 6, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Thiene, G.; Nava, A.; Corrado, D.; Rossi, L.; Pennelli, N. Right ventricular cardiomyopathy and sudden death in young people. N. Engl. J. Med. 1988, 318, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Marcus, F.I.; McKenna, W.J.; Sherrill, D.; Basso, C.; Bauce, B.; Bluemke, D.A.; Calkins, H.; Corrado, D.; Cox, M.G.; Daubert, J.P.; et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: Proposed modification of the task force criteria. Circulation 2010, 121, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, F.; Zorio, E.; Jimenez-Jaimez, J.; Salguero-Bodes, R.; Zwart, R.; Gonzalez-Lopez, E.; Molina, P.; Bermudez-Jimenez, F.; Delgado, J.F.; Braza-Boils, A.; et al. Clinical characteristics and determinants of the phenotype in TMEM43 arrhythmogenic right ventricular cardiomyopathy type 5. Heart Rhythm 2020, 17, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Drezner, J.; Basso, C.; Pelliccia, A.; Thiene, G. Strategies for the prevention of sudden cardiac death during sports. Eur. J. Prev. Cardiol. 2011, 18, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Chaitman, B.R.; Ackerman, M.J.; Bayes de Luna, A.; Corrado, D.; Crosson, J.E.; Deal, B.J.; Driscoll, D.J.; Estes, N.A., 3rd; Araujo, C.G.; et al. Recommendations for physical activity and recreational sports participation for young patients with genetic cardiovascular diseases. Circulation 2004, 109, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- James, C.A.; Bhonsale, A.; Tichnell, C.; Murray, B.; Russell, S.D.; Tandri, H.; Tedford, R.J.; Judge, D.P.; Calkins, H. Exercise increases age-related penetrance and arrhythmic risk in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. J. Am. Coll. Cardiol. 2013, 62, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Te Riele, A.; James, C.A.; Calkins, H.; Tsatsopoulou, A. Arrhythmogenic Right Ventricular Cardiomyopathy in Pediatric Patients: An Important but Underrecognized Clinical Entity. Front. Pediatr. 2021, 9, 750916. [Google Scholar] [CrossRef] [PubMed]
- Rootwelt-Norberg, C.; Lie, O.H.; Chivulescu, M.; Castrini, A.I.; Sarvari, S.I.; Lyseggen, E.; Almaas, V.M.; Bogsrud, M.P.; Edvardsen, T.; Haugaa, K.H. Sex differences in disease progression and arrhythmic risk in patients with arrhythmogenic cardiomyopathy. Europace 2021, 23, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Akdis, D.; Saguner, A.M.; Shah, K.; Wei, C.; Medeiros-Domingo, A.; von Eckardstein, A.; Luscher, T.F.; Brunckhorst, C.; Chen, H.S.V.; Duru, F. Sex hormones affect outcome in arrhythmogenic right ventricular cardiomyopathy/dysplasia: From a stem cell derived cardiomyocyte-based model to clinical biomarkers of disease outcome. Eur. Heart J. 2017, 38, 1498–1508. [Google Scholar] [CrossRef] [PubMed]
- Sen-Chowdhry, S.; Syrris, P.; McKenna, W.J. Genetics of right ventricular cardiomyopathy. J. Cardiovasc. Electrophysiol. 2005, 16, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.; Calkins, H. Risk Stratification in Arrhythmogenic Right Ventricular Cardiomyopathy. Arrhythmia Electrophysiol. Rev. 2021, 10, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Dalal, D.; James, C.; Devanagondi, R.; Tichnell, C.; Tucker, A.; Prakasa, K.; Spevak, P.J.; Bluemke, D.A.; Abraham, T.; Russell, S.D.; et al. Penetrance of mutations in plakophilin-2 among families with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J. Am. Coll. Cardiol. 2006, 48, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Vasireddi, S.K.; Sattayaprasert, P.; Yang, D.; Dennis, A.T.; Bektik, E.; Fu, J.D.; Mackall, J.A.; Laurita, K.R. Adipogenic Signaling Promotes Arrhythmia Substrates before Structural Abnormalities in TMEM43 ARVC. J. Pers. Med. 2022, 12, 1680. [Google Scholar] [CrossRef] [PubMed]
- Basharat, S.A.; Hsiung, I.; Garg, J.; Alsaid, A. Arrhythmogenic Cardiomyopathy: Evolving Diagnostic Criteria and Insight from Cardiac Magnetic Resonance Imaging. Heart Fail. Clin. 2023, 19, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Zorzi, A.; Cipriani, A.; Bauce, B.; Bariani, R.; Beffagna, G.; De Lazzari, M.; Migliore, F.; Pilichou, K.; Rampazzo, A.; et al. Evolving Diagnostic Criteria for Arrhythmogenic Cardiomyopathy. J. Am. Heart Assoc. 2021, 10, e021987. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, C.G.; Heddle, W.F. Left sided arrhythmogenic ventricular dysplasia in siblings. Heart 2001, 86, 128–130. [Google Scholar] [PubMed]
- Ruperto, C.; Mina, C.; Brun, F.; Liotta, R.; Pyxaras, S.; Clemenza, F.; Sinagra, G. Arrhythmogenic cardiomyopathy with biventricular involvement and noncompaction. J. Cardiovasc. Med. 2016, 17 (Suppl. S2), e244–e246. [Google Scholar] [CrossRef] [PubMed]
- Horimoto, M.; Akino, M.; Takenaka, T.; Igarashi, K.; Inoue, H.; Kawakami, Y. Evolution of left ventricular involvement in arrhythmogenic right ventricular cardiomyopathy. Cardiology 2000, 93, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Perazzolo Marra, M.; Zorzi, A.; Beffagna, G.; Cipriani, A.; Lazzari, M.; Migliore, F.; Pilichou, K.; Rampazzo, A.; Rigato, I.; et al. Diagnosis of arrhythmogenic cardiomyopathy: The Padua criteria. Int. J. Cardiol. 2020, 319, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Graziano, F.; Zorzi, A.; Cipriani, A.; De Lazzari, M.; Bauce, B.; Rigato, I.; Brunetti, G.; Pilichou, K.; Basso, C.; Perazzolo Marra, M.; et al. The 2020 “Padua Criteria” for Diagnosis and Phenotype Characterization of Arrhythmogenic Cardiomyopathy in Clinical Practice. J. Clin. Med. 2022, 11, 279. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Anastasakis, A.; Basso, C.; Bauce, B.; Blomstrom-Lundqvist, C.; Bucciarelli-Ducci, C.; Cipriani, A.; De Asmundis, C.; Gandjbakhch, E.; Jimenez-Jaimez, J.; et al. Proposed diagnostic criteria for arrhythmogenic cardiomyopathy: European Task Force consensus report. Int. J. Cardiol. 2024, 395, 131447. [Google Scholar] [CrossRef] [PubMed]
- Bhonsale, A.; Groeneweg, J.A.; James, C.A.; Dooijes, D.; Tichnell, C.; Jongbloed, J.D.; Murray, B.; te Riele, A.S.; van den Berg, M.P.; Bikker, H.; et al. Impact of genotype on clinical course in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated mutation carriers. Eur. Heart J. 2015, 36, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.A.; Gasperetti, A.; Bosman, L.P.; Schmidt, A.F.; Baas, A.F.; Amin, A.S.; Houweling, A.C.; Wilde, A.A.M.; Compagnucci, P.; Targetti, M.; et al. Individualized Family Screening for Arrhythmogenic Right Ventricular Cardiomyopathy. J. Am. Coll. Cardiol. 2023, 82, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Norman, M.; Simpson, M.; Mogensen, J.; Shaw, A.; Hughes, S.; Syrris, P.; Sen-Chowdhry, S.; Rowland, E.; Crosby, A.; McKenna, W.J. Novel mutation in desmoplakin causes arrhythmogenic left ventricular cardiomyopathy. Circulation 2005, 112, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Vatta, M.; Marcus, F.; Towbin, J.A. Arrhythmogenic right ventricular cardiomyopathy: A ‘final common pathway’ that defines clinical phenotype. Eur. Heart J. 2007, 28, 529–530. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Yang, Z.; Vatta, M.; Rampazzo, A.; Beffagna, G.; Pilichou, K.; Scherer, S.E.; Saffitz, J.; Kravitz, J.; Zareba, W.; et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J. Am. Coll. Cardiol. 2010, 55, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Marcus, F.I.; Edson, S.; Towbin, J.A. Genetics of arrhythmogenic right ventricular cardiomyopathy: A practical guide for physicians. J. Am. Coll. Cardiol. 2013, 61, 1945–1948. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Lindenfeld, J.; Mestroni, L.; Seidman, C.E.; Taylor, M.R.; Towbin, J.A. Genetic evaluation of cardiomyopathy—A Heart Failure Society of America practice guideline. J. Card. Fail. 2009, 15, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Gerull, B.; Heuser, A.; Wichter, T.; Paul, M.; Basson, C.T.; McDermott, D.A.; Lerman, B.B.; Markowitz, S.M.; Ellinor, P.T.; MacRae, C.A.; et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet. 2004, 36, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Asimaki, A.; Syrris, P.; Wichter, T.; Matthias, P.; Saffitz, J.E.; McKenna, W.J. A novel dominant mutation in plakoglobin causes arrhythmogenic right ventricular cardiomyopathy. Am. J. Hum. Genet. 2007, 81, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Genga, M.F.; Cuenca, S.; Dal Ferro, M.; Zorio, E.; Salgado-Aranda, R.; Climent, V.; Padron-Barthe, L.; Duro-Aguado, I.; Jimenez-Jaimez, J.; Hidalgo-Olivares, V.M.; et al. Truncating FLNC Mutations Are Associated With High-Risk Dilated and Arrhythmogenic Cardiomyopathies. J. Am. Coll. Cardiol. 2016, 68, 2440–2451. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Graw, S.; Sinagra, G.; Barnes, C.; Slavov, D.; Brun, F.; Pinamonti, B.; Salcedo, E.E.; Sauer, W.; Pyxaras, S.; et al. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy-overlap syndromes. Circulation 2011, 124, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Moncayo-Arlandi, J.; Brugada, R. Unmasking the molecular link between arrhythmogenic cardiomyopathy and Brugada syndrome. Nat. Rev. Cardiol. 2017, 14, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Cowan, J.; Morales, A.; Siegfried, J.D. Progress with genetic cardiomyopathies: Screening, counseling, and testing in dilated, hypertrophic, and arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ. Heart Fail. 2009, 2, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Basso, C.; Corrado, D.; Marcus, F.I.; Nava, A.; Thiene, G. Arrhythmogenic right ventricular cardiomyopathy. Lancet 2009, 373, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Rigato, I.; Bauce, B.; Rampazzo, A.; Zorzi, A.; Pilichou, K.; Mazzotti, E.; Migliore, F.; Marra, M.P.; Lorenzon, A.; De Bortoli, M.; et al. Compound and digenic heterozygosity predicts lifetime arrhythmic outcome and sudden cardiac death in desmosomal gene-related arrhythmogenic right ventricular cardiomyopathy. Circ. Cardiovasc. Genet. 2013, 6, 533–542. [Google Scholar] [CrossRef] [PubMed]
- De Bortoli, M.; Meraviglia, V.; Mackova, K.; Frommelt, L.S.; Konig, E.; Rainer, J.; Volani, C.; Benzoni, P.; Schlittler, M.; Cattelan, G.; et al. Modeling incomplete penetrance in arrhythmogenic cardiomyopathy by human induced pluripotent stem cell derived cardiomyocytes. Comput. Struct. Biotechnol. J. 2023, 21, 1759–1773. [Google Scholar] [CrossRef] [PubMed]
- Camors, E.M.; Purevjav, E.; Jefferies, J.L.; Saffitz, J.E.; Gong, N.; Ryan, T.D.; Lucky, A.W.; Taylor, M.D.; Sullivan, L.M.; Mestroni, L.; et al. Early Lethality Due to a Novel Desmoplakin Variant Causing Infantile Epidermolysis Bullosa Simplex With Fragile Skin, Aplasia Cutis Congenita, and Arrhythmogenic Cardiomyopathy. Circ. Genom. Precis. Med. 2020, 13, e002800. [Google Scholar] [CrossRef] [PubMed]
- Harvey, P.A.; Leinwand, L.A. The cell biology of disease: Cellular mechanisms of cardiomyopathy. J. Cell Biol. 2011, 194, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Akdis, D.; Brunckhorst, C.; Duru, F.; Saguner, A.M. Arrhythmogenic Cardiomyopathy: Electrical and Structural Phenotypes. Arrhythmia Electrophysiol. Rev. 2016, 5, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Tabib, A.; Loire, R.; Chalabreysse, L.; Meyronnet, D.; Miras, A.; Malicier, D.; Thivolet, F.; Chevalier, P.; Bouvagnet, P. Circumstances of death and gross and microscopic observations in a series of 200 cases of sudden death associated with arrhythmogenic right ventricular cardiomyopathy and/or dysplasia. Circulation 2003, 108, 3000–3005. [Google Scholar] [CrossRef] [PubMed]
- Lazzarini, E.; Jongbloed, J.D.; Pilichou, K.; Thiene, G.; Basso, C.; Bikker, H.; Charbon, B.; Swertz, M.; van Tintelen, J.P.; van der Zwaag, P.A. The ARVD/C genetic variants database: 2014 update. Hum. Mutat. 2015, 36, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Asimaki, A.; Kleber, A.G.; Saffitz, J.E. Pathogenesis of Arrhythmogenic Cardiomyopathy. Can. J. Cardiol. 2015, 31, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Sen-Chowdhry, S.; Syrris, P.; Prasad, S.K.; Hughes, S.E.; Merrifield, R.; Ward, D.; Pennell, D.J.; McKenna, W.J. Left-dominant arrhythmogenic cardiomyopathy: An under-recognized clinical entity. J. Am. Coll. Cardiol. 2008, 52, 2175–2187. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gras, E.; Lombardi, R.; Giocondo, M.J.; Willerson, J.T.; Schneider, M.D.; Khoury, D.S.; Marian, A.J. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J. Clin. Investig. 2006, 116, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Djouadi, F.; Lecarpentier, Y.; Hebert, J.L.; Charron, P.; Bastin, J.; Coirault, C. A potential link between peroxisome proliferator-activated receptor signalling and the pathogenesis of arrhythmogenic right ventricular cardiomyopathy. Cardiovasc. Res. 2009, 84, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.N.; Gurha, P.; Lombardi, R.; Ruggiero, A.; Willerson, J.T.; Marian, A.J. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ. Res. 2014, 114, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Delva, E.; Tucker, D.K.; Kowalczyk, A.P. The desmosome. Cold Spring Harb. Perspect. Biol. 2009, 1, a002543. [Google Scholar] [CrossRef] [PubMed]
- Hatzfeld, M.; Keil, R.; Magin, T.M. Desmosomes and Intermediate Filaments: Their Consequences for Tissue Mechanics. Cold Spring Harb. Perspect. Biol. 2017, 9, a029157. [Google Scholar] [CrossRef] [PubMed]
- Bowles, N.E.; Bowles, K.R.; Towbin, J.A. The “final common pathway” hypothesis and inherited cardiovascular disease. The role of cytoskeletal proteins in dilated cardiomyopathy. Herz 2000, 25, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Martherus, R.; Jain, R.; Takagi, K.; Mendsaikhan, U.; Turdi, S.; Osinska, H.; James, J.F.; Kramer, K.; Purevjav, E.; Towbin, J.A. Accelerated cardiac remodeling in desmoplakin transgenic mice in response to endurance exercise is associated with perturbed Wnt/beta-catenin signaling. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H174–H187. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, K.A.; Connors, S.P.; Merner, N.; Haywood, A.; Young, T.L.; McKenna, W.J.; Gallagher, B.; Curtis, F.; Bassett, A.S.; Parfrey, P.S. The natural history of a genetic subtype of arrhythmogenic right ventricular cardiomyopathy caused by a p.S358L mutation in TMEM43. Clin. Genet. 2013, 83, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, K.A.; Howes, A.J.; Boland, P.; Shen, X.S.; Stuckless, S.; Young, T.L.; Curtis, F.; Collier, A.; Parfrey, P.S.; Connors, S.P. Long-Term Clinical Outcome of Arrhythmogenic Right Ventricular Cardiomyopathy in Individuals With a p.S358L Mutation in TMEM43 Following Implantable Cardioverter Defibrillator Therapy. Circ. Arrhythmia Electrophysiol. 2016, 9, e003589. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Wong, J.; Wen, J.; Wang, S.; Wang, C.; Spiering, S.; Kan, N.G.; Forcales, S.; Puri, P.L.; Leone, T.C.; et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 2013, 494, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Baskin, B.; Skinner, J.R.; Sanatani, S.; Terespolsky, D.; Krahn, A.D.; Ray, P.N.; Scherer, S.W.; Hamilton, R.M. TMEM43 mutations associated with arrhythmogenic right ventricular cardiomyopathy in non-Newfoundland populations. Hum. Genet. 2013, 132, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Haywood, A.F.; Merner, N.D.; Hodgkinson, K.A.; Houston, J.; Syrris, P.; Booth, V.; Connors, S.; Pantazis, A.; Quarta, G.; Elliott, P.; et al. Recurrent missense mutations in TMEM43 (ARVD5) due to founder effects cause arrhythmogenic cardiomyopathies in the UK and Canada. Eur. Heart J. 2013, 34, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Aalbaek Kjaergaard, K.; Kristensen, J.; Molgaard, H.; Cosedis Nielsen, J.; Jensen, H.K. Failure of ICD therapy in lethal arrhythmogenic right ventricular cardiomyopathy type 5 caused by the TMEM43 p.Ser358Leu mutation. Hear. Case Rep. 2016, 2, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Paulin, F.L.; Hodgkinson, K.A.; MacLaughlan, S.; Stuckless, S.N.; Templeton, C.; Shah, S.; Bremner, H.; Roberts, J.D.; Young, T.L.; Parfrey, P.S.; et al. Exercise and arrhythmic risk in TMEM43 p.S358L arrhythmogenic right ventricular cardiomyopathy. Heart Rhythm 2020, 17, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Milting, H.; Klauke, B.; Christensen, A.H.; Musebeck, J.; Walhorn, V.; Grannemann, S.; Munnich, T.; Saric, T.; Rasmussen, T.B.; Jensen, H.K.; et al. The TMEM43 Newfoundland mutation p.S358L causing ARVC-5 was imported from Europe and increases the stiffness of the cell nucleus. Eur. Heart J. 2015, 36, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Orgil, B.O.; Munkhsaikhan, U.; Pierre, J.F.; Li, N.; Xu, F.; Alberson, N.R.; Johnson, J.N.; Wetzel, G.T.; Boukens, B.J.D.; Lu, L.; et al. The TMEM43 S358L mutation affects cardiac, small intestine, and metabolic homeostasis in a knock-in mouse model. Am. J. Physiol. Heart Circ. Physiol. 2023, 324, H866–H880. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Jiang, C.; Li, Y.; Yang, D.; Ma, Y.; Zhang, B.; Li, X.; Zhang, P.; Hu, X.; Zhao, X.; et al. TMEM43-S358L mutation enhances NF-kappaB-TGFbeta signal cascade in arrhythmogenic right ventricular dysplasia/cardiomyopathy. Protein Cell 2018, 10, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Padron-Barthe, L.; Villalba-Orero, M.; Gomez-Salinero, J.M.; Dominguez, F.; Roman, M.; Larrasa-Alonso, J.; Ortiz-Sanchez, P.; Martinez, F.; Lopez-Olaneta, M.; Bonzon-Kulichenko, E.; et al. Severe Cardiac Dysfunction and Death Caused by Arrhythmogenic Right Ventricular Cardiomyopathy Type 5 Are Improved by Inhibition of Glycogen Synthase Kinase-3beta. Circulation 2019, 140, 1188–1204. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Kanai, Y.; Ohno, S.; Ando, H.; Honda, M.; Niwano, S.; Ishii, M. Fetal arrhythmogenic right ventricular cardiomyopathy with double mutations in TMEM43. Pediatr. Int. 2016, 58, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Matos, J.; Helle, E.; Care, M.; Moayedi, Y.; Gollob, M.H.; Thavendiranathan, P.; Spears, D.; Hanneman, K. Cardiac MRI and Clinical Outcomes in TMEM43 Arrhythmogenic Cardiomyopathy. Radiol. Cardiothorac. Imaging 2023, 5, e230155. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.T.; Shenasa, M.; Boyle, N.G. Ventricular Arrhythmias and Sudden Cardiac Death. Card. Electrophysiol. Clin. 2017, 9, 693–708. [Google Scholar] [CrossRef] [PubMed]
- Huikuri, H.V.; Castellanos, A.; Myerburg, R.J. Sudden death due to cardiac arrhythmias. N. Engl. J. Med. 2001, 345, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Aune, D.; Schlesinger, S.; Norat, T.; Riboli, E. Body mass index, abdominal fatness, and the risk of sudden cardiac death: A systematic review and dose-response meta-analysis of prospective studies. Eur. J. Epidemiol. 2018, 33, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Tansey, D.K.; Aly, Z.; Sheppard, M.N. Fat in the right ventricle of the normal heart. Histopathology 2005, 46, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Sani, M.M.; Sung, E.; Engels, M.; Daimee, U.A.; Trayanova, N.; Wu, K.C.; Chrispin, J. Association of epicardial and intramyocardial fat with ventricular arrhythmias. Heart Rhythm 2023, 20, 1699–1705. [Google Scholar] [CrossRef] [PubMed]
- Zaleska-Kociecka, M.; Wojdynska, Z.; Kalisz, M.; Litwiniuk, A.; Maczewski, M.; Leszek, P.; Paterek, A. Epicardial fat and ventricular arrhythmias. Heart Rhythm 2024, 21, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Westaby, J.; Dalle-Carbonare, C.; Ster, I.C.; Sheppard, M.N. Obesity Cardiomyopathy in Sudden Cardiac Death: A Distinct Entity? A Comparative Study. JACC Adv. 2023, 2, 100414. [Google Scholar] [CrossRef] [PubMed]
- Iacobellis, G.; Ribaudo, M.C.; Assael, F.; Vecci, E.; Tiberti, C.; Zappaterreno, A.; Di Mario, U.; Leonetti, F. Echocardiographic epicardial adipose tissue is related to anthropometric and clinical parameters of metabolic syndrome: A new indicator of cardiovascular risk. J. Clin. Endocrinol. Metab. 2003, 88, 5163–5168. [Google Scholar] [CrossRef] [PubMed]
- Sommariva, E.; Stadiotti, I.; Perrucci, G.L.; Tondo, C.; Pompilio, G. Cell models of arrhythmogenic cardiomyopathy: Advances and opportunities. Dis. Models Mech. 2017, 10, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Bueno-Beti, C.; Asimaki, A. Histopathological Features and Protein Markers of Arrhythmogenic Cardiomyopathy. Front. Cardiovasc. Med. 2021, 8, 746321. [Google Scholar] [CrossRef] [PubMed]
- Burke, A.P.; Farb, A.; Tashko, G.; Virmani, R. Arrhythmogenic right ventricular cardiomyopathy and fatty replacement of the right ventricular myocardium: Are they different diseases? Circulation 1998, 97, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Zorn-Pauly, K.; Schaffer, P.; Pelzmann, B.; Bernhart, E.; Wei, G.; Lang, P.; Ledinski, G.; Greilberger, J.; Koidl, B.; Jurgens, G. Oxidized LDL induces ventricular myocyte damage and abnormal electrical activity—Role of lipid hydroperoxides. Cardiovasc. Res. 2005, 66, 74–83. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sommariva, E.; Stadiotti, I.; Casella, M.; Catto, V.; Dello Russo, A.; Carbucicchio, C.; Arnaboldi, L.; De Metrio, S.; Milano, G.; Scopece, A.; et al. Oxidized LDL-dependent pathway as new pathogenic trigger in arrhythmogenic cardiomyopathy. EMBO Mol. Med. 2021, 13, e14365. [Google Scholar] [CrossRef] [PubMed]
- Barriga, M.; Cal, R.; Cabello, N.; Llach, A.; Vallmitjana, A.; Benitez, R.; Badimon, L.; Cinca, J.; Llorente-Cortes, V.; Hove-Madsen, L. Low density lipoproteins promote unstable calcium handling accompanied by reduced SERCA2 and connexin-40 expression in cardiomyocytes. PLoS ONE 2013, 8, e58128. [Google Scholar] [CrossRef] [PubMed]
- DaTorre, S.D.; Creer, M.H.; Pogwizd, S.M.; Corr, P.B. Amphipathic lipid metabolites and their relation to arrhythmogenesis in the ischemic heart. J. Mol. Cell. Cardiol. 1991, 23 (Suppl. S1), 11–22. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, R.; Dong, J.; Rodriguez, G.; Bell, A.; Leung, T.K.; Schwartz, R.J.; Willerson, J.T.; Brugada, R.; Marian, A.J. Genetic fate mapping identifies second heart field progenitor cells as a source of adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ. Res. 2009, 104, 1076–1084. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Kohli, P.; Gulati, M. An update on exercise stress testing. Curr. Probl. Cardiol. 2012, 37, 177–202. [Google Scholar] [CrossRef] [PubMed]
- Dalal, D.; Nasir, K.; Bomma, C.; Prakasa, K.; Tandri, H.; Piccini, J.; Roguin, A.; Tichnell, C.; James, C.; Russell, S.D.; et al. Arrhythmogenic right ventricular dysplasia: A United States experience. Circulation 2005, 112, 3823–3832. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Ackerman, M.J.; Nishimura, R.A.; Pyeritz, R.E.; Towbin, J.A.; Udelson, J.E. Task Force 4: HCM and other cardiomyopathies, mitral valve prolapse, myocarditis, and Marfan syndrome. J. Am. Coll. Cardiol. 2005, 45, 1340–1345. [Google Scholar] [CrossRef] [PubMed]
- Fabritz, L.; Fortmueller, L.; Gehmlich, K.; Kant, S.; Kemper, M.; Kucerova, D.; Syeda, F.; Faber, C.; Leube, R.E.; Kirchhof, P.; et al. Endurance Training Provokes Arrhythmogenic Right Ventricular Cardiomyopathy Phenotype in Heterozygous Desmoglein-2 Mutants: Alleviation by Preload Reduction. Biomedicines 2024, 12, 985. [Google Scholar] [CrossRef] [PubMed]
- Kirchhof, P.; Fabritz, L.; Zwiener, M.; Witt, H.; Schafers, M.; Zellerhoff, S.; Paul, M.; Athai, T.; Hiller, K.H.; Baba, H.A.; et al. Age- and training-dependent development of arrhythmogenic right ventricular cardiomyopathy in heterozygous plakoglobin-deficient mice. Circulation 2006, 114, 1799–1806. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Basso, C.; Schiavon, M.; Thiene, G. Does sports activity enhance the risk of sudden cardiac death? J. Cardiovasc. Med. 2006, 7, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Estes, N.A., 3rd; Vetter, V.L.; Corrado, D. Clinical decisions. Cardiac screening before participation in sports. N. Engl. J. Med. 2013, 369, 2049–2053. [Google Scholar] [PubMed]
- Saberniak, J.; Hasselberg, N.E.; Borgquist, R.; Platonov, P.G.; Sarvari, S.I.; Smith, H.J.; Ribe, M.; Holst, A.G.; Edvardsen, T.; Haugaa, K.H. Vigorous physical activity impairs myocardial function in patients with arrhythmogenic right ventricular cardiomyopathy and in mutation positive family members. Eur. J. Heart Fail. 2014, 16, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Zorzi, A. Arrhythmogenic right ventricular cardiomyopathy and sports activity. Eur. Heart J. 2015, 36, 1708–1710. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Zorzi, A.; Sarto, P. Pre-participation screening for safe sports activity. Eur. Heart J. 2023, 44, 2258–2259. [Google Scholar] [CrossRef] [PubMed]
- Ruwald, A.C.; Marcus, F.; Estes, N.A., 3rd; Link, M.; McNitt, S.; Polonsky, B.; Calkins, H.; Towbin, J.A.; Moss, A.J.; Zareba, W. Association of competitive and recreational sport participation with cardiac events in patients with arrhythmogenic right ventricular cardiomyopathy: Results from the North American multidisciplinary study of arrhythmogenic right ventricular cardiomyopathy. Eur. Heart J. 2015, 36, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Basso, C.; Rizzoli, G.; Schiavon, M.; Thiene, G. Does sports activity enhance the risk of sudden death in adolescents and young adults? J. Am. Coll. Cardiol. 2003, 42, 1959–1963. [Google Scholar] [CrossRef] [PubMed]
- AbdelWahab, A.; Gardner, M.; Parkash, R.; Gray, C.; Sapp, J. Ventricular tachycardia ablation in arrhythmogenic right ventricular cardiomyopathy patients with TMEM43 gene mutations. J. Cardiovasc. Electrophysiol. 2018, 29, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Costantini, D.L.; Arruda, E.P.; Agarwal, P.; Kim, K.H.; Zhu, Y.; Zhu, W.; Lebel, M.; Cheng, C.W.; Park, C.Y.; Pierce, S.A.; et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell 2005, 123, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Choy, L.; Yeo, J.M.; Tse, V.; Chan, S.P.; Tse, G. Cardiac disease and arrhythmogenesis: Mechanistic insights from mouse models. Int. J. Cardiol. Heart Vasc. 2016, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Stroud, M.J.; Fang, X.; Zhang, J.; Guimaraes-Camboa, N.; Veevers, J.; Dalton, N.D.; Gu, Y.; Bradford, W.H.; Peterson, K.L.; Evans, S.M.; et al. Luma is not essential for murine cardiac development and function. Cardiovasc. Res. 2018, 114, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S. Catalpol reduces the production of inflammatory mediators via PPAR-gamma activation in human intestinal Caco-2 cells. J. Nat. Med. 2016, 70, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Xu, F.; Orgil, B.O.; Khuchua, Z.; Munkhsaikhan, U.; Johnson, J.N.; Alberson, N.R.; Pierre, J.F.; Black, D.D.; Dong, D.; et al. Systems Genetics Analysis Defines Importance Of TMEM43/LUMA For Cardiac And Metabolic Related Pathways. Physiol. Genom. 2021, 54, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Ashbrook, D.G.; Arends, D.; Prins, P.; Mulligan, M.K.; Roy, S.; Williams, E.G.; Lutz, C.M.; Valenzuela, A.; Bohl, C.J.; Ingels, J.F.; et al. A platform for experimental precision medicine: The extended BXD mouse family. Cell Syst. 2021, 12, 235–247.e9. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A. Nonalcoholic Fatty Liver Disease (NAFLD) and Risk of Cardiac Arrhythmias: A New Aspect of the Liver-heart Axis. J. Clin. Transl. Hepatol. 2017, 5, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.S.; Mathur, B.; Eblimit, Z.; Vasquez, H.; Taegtmeyer, H.; Karpen, S.J.; Penny, D.J.; Moore, D.D.; Anakk, S. Bile acid excess induces cardiomyopathy and metabolic dysfunctions in the heart. Hepatology 2017, 65, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Voiosu, A.; Wiese, S.; Voiosu, T.; Bendtsen, F.; Moller, S. Bile acids and cardiovascular function in cirrhosis. Liver Int. 2017, 37, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lim, G.; Kim, H.; Suh, D.; Choi, H.K.; Kim, H.P.; Yoon, H.G.; Park, S.W.; Kang, S.M.; Kwon, C.; et al. Generation of an induced pluripotent stem cell line from a patient with arrhythmogenic right ventricular cardiomyopathy harboring a TMEM43 splice-site variant. Stem Cell Res. 2024, 78, 103453. [Google Scholar] [CrossRef] [PubMed]
- Lalaguna, L.; Arevalo-Nunez de Arenas, M.; Lopez-Olaneta, M.; Villalba-Orero, M.; Jimenez-Rioboo, R.J.; Gomez-Gaviro, M.V.; Isern, J.; Munoz-Canoves, P.; Byrne, B.J.; Ochoa, J.P.; et al. Overexpression of Wild-Type TMEM43 Improves Cardiac Function in Arrhythmogenic Right Ventricular Cardiomyopathy Type 5. Circ. Res. 2025, 136, 830–844. [Google Scholar] [CrossRef] [PubMed]
- Naso, M.F.; Tomkowicz, B.; Perry, W.L., 3rd; Strohl, W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs 2017, 31, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, S.; Choi, E.; Shin, S.; Park, J. A novel SYNE2 mutation identified by whole exome sequencing in a Korean family with Emery-Dreifuss muscular dystrophy. Clin. Chim. Acta 2020, 506, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H. Novel candidate alleles associated with gene regulation for Emery-Dreifuss muscular dystrophy. eBioMedicine 2020, 52, 102620. [Google Scholar] [CrossRef] [PubMed]
- Schmit, K.; Michiels, C. TMEM Proteins in Cancer: A Review. Front. Pharmacol. 2018, 9, 1345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, W.; Yuan, Q.; Hong, W.; Yin, P.; Shen, T.; Fang, L.; Jiang, J.; Shi, F.; Chen, W. Illustrating the biological functions and diagnostic value of transmembrane protein family members in glioma. Front. Oncol. 2023, 13, 1145676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, J.; Mao, A.; Ning, G.; Cao, Y.; Zhang, W.; Wang, Q. Tmem88 confines ectodermal Wnt2bb signaling in pharyngeal arch artery progenitors for balancing cell cycle progression and cell fate decision. Nat. Cardiovasc. Res. 2023, 2, 234–250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yu, X.; Jiang, G.; Miao, Y.; Wang, L.; Zhang, Y.; Liu, Y.; Fan, C.; Lin, X.; Dong, Q.; et al. Cytosolic TMEM88 promotes invasion and metastasis in lung cancer cells by binding DVLS. Cancer Res. 2015, 75, 4527–4537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wan, J.X.; Ke, Z.P.; Wang, F.; Chai, H.X.; Liu, J.Q. TMEM88, CCL14 and CLEC3B as prognostic biomarkers for prognosis and palindromia of human hepatocellular carcinoma. Tumour Biol. 2017, 39, 1010428317708900. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Han, Y.; Jin, W.; Tian, M.; Chen, P.; Min, J.; Hu, H.; Xu, B.; Zhu, W.; Xiong, L.; et al. Overexpression and biological function of TMEM48 in non-small cell lung carcinoma. Tumour Biol. 2016, 37, 2575–2586. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Sun, W.; Zou, Y.; Cai, Z.; Wang, P.; Lin, X.; Huang, J.; Jiang, L.; Ding, X.; Hu, G. RNA interference against TMEM97 inhibits cell proliferation, migration, and invasion in glioma cells. Tumour Biol. 2015, 36, 8231–8238. [Google Scholar] [CrossRef] [PubMed]
- Hrasovec, S.; Hauptman, N.; Glavac, D.; Jelenc, F.; Ravnik-Glavac, M. TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Dis. Markers 2013, 34, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Cuajungco, M.P.; Podevin, W.; Valluri, V.K.; Bui, Q.; Nguyen, V.H.; Taylor, K. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 2012, 114, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Popescu, N.C.; Klein, G.; Imreh, S. The interferon-alpha responsive gene TMEM7 suppresses cell proliferation and is downregulated in human hepatocellular carcinoma. Cancer Genet. Cytogenet. 2007, 177, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Xin, S.; Ma, J.; Wang, H.; Zhang, H.; Liu, J. A three microRNA-based prognostic signature for small cell lung cancer overall survival. J. Cell. Biochem. 2019, 120, 8723–8730. [Google Scholar] [CrossRef] [PubMed]
- Toma, C.; Diaz-Gay, M.; Soares de Lima, Y.; Arnau-Collell, C.; Franch-Exposito, S.; Munoz, J.; Overs, B.; Bonjoch, L.; Carballal, S.; Ocana, T.; et al. Identification of a Novel Candidate Gene for Serrated Polyposis Syndrome Germline Predisposition by Performing Linkage Analysis Combined With Whole-Exome Sequencing. Clin. Transl. Gastroenterol. 2019, 10, e00100. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Fang, F.; Yang, C.; Chen, X.; Li, Q.; Shen, X. Lifting the veils on transmembrane proteins: Potential anticancer targets. Eur. J. Pharmacol. 2024, 963, 176225. [Google Scholar] [CrossRef] [PubMed]
- De Siati, R.D.; Rosenzweig, F.; Gersdorff, G.; Gregoire, A.; Rombaux, P.; Deggouj, N. Auditory Neuropathy Spectrum Disorders: From Diagnosis to Treatment: Literature Review and Case Reports. J. Clin. Med. 2020, 9, 1074. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Lee, C.J. Transmembrane proteins with unknown function (TMEMs) as ion channels: Electrophysiological properties, structure, and pathophysiological roles. Exp. Mol. Med. 2024, 56, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Corfas, G.; Stone, J.S. Inner ear supporting cells: Rethinking the silent majority. Semin. Cell Dev. Biol. 2013, 24, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, K.P.; Holden, R.G.; Escribano-Subias, P.M.; Cogolludo, A.; Veale, E.L.; Mathie, A. Characterization and regulation of wild-type and mutant TASK-1 two pore domain potassium channels indicated in pulmonary arterial hypertension. J. Physiol. 2019, 597, 1087–1101. [Google Scholar] [CrossRef] [PubMed]
- Wangemann, P. Supporting sensory transduction: Cochlear fluid homeostasis and the endocochlear potential. J. Physiol. 2006, 576 Pt 1, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Ratnavadivel, S.; Dammeier, J.; Gaertner, A.; de Toledo, M.A.S.; Zenke, M.; Gummert, J.; Bloch Rasmussen, T.; Klinke, N.; Jurgens, K.; Meyer, H.; et al. Generation of a TMEM43 knockout human induced pluripotent stem cell line (HDZi003-A-1) using CRISPR/Cas9. Stem Cell Res. 2024, 76, 103354. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orgil, B.-O.; Spaulding, M.S.; Smith, H.P.; Baba, Z.; Alberson, N.R.; Batsaikhan, E.; Towbin, J.A.; Purevjav, E. Transmembrane Protein 43: Molecular and Pathogenetic Implications in Arrhythmogenic Cardiomyopathy and Various Other Diseases. Int. J. Mol. Sci. 2025, 26, 6856. https://doi.org/10.3390/ijms26146856
Orgil B-O, Spaulding MS, Smith HP, Baba Z, Alberson NR, Batsaikhan E, Towbin JA, Purevjav E. Transmembrane Protein 43: Molecular and Pathogenetic Implications in Arrhythmogenic Cardiomyopathy and Various Other Diseases. International Journal of Molecular Sciences. 2025; 26(14):6856. https://doi.org/10.3390/ijms26146856
Chicago/Turabian StyleOrgil, Buyan-Ochir, Mekaea S. Spaulding, Harrison P. Smith, Zainab Baba, Neely R. Alberson, Enkhzul Batsaikhan, Jeffrey A. Towbin, and Enkhsaikhan Purevjav. 2025. "Transmembrane Protein 43: Molecular and Pathogenetic Implications in Arrhythmogenic Cardiomyopathy and Various Other Diseases" International Journal of Molecular Sciences 26, no. 14: 6856. https://doi.org/10.3390/ijms26146856
APA StyleOrgil, B.-O., Spaulding, M. S., Smith, H. P., Baba, Z., Alberson, N. R., Batsaikhan, E., Towbin, J. A., & Purevjav, E. (2025). Transmembrane Protein 43: Molecular and Pathogenetic Implications in Arrhythmogenic Cardiomyopathy and Various Other Diseases. International Journal of Molecular Sciences, 26(14), 6856. https://doi.org/10.3390/ijms26146856







