[1,3]Thiazolo[3,2-b][1,2,4]triazolium Salts as Effective Antimicrobial Agents: Synthesis, Biological Activity Evaluation, and Molecular Docking Studies
Abstract
1. Introduction
2. Results and Discussion
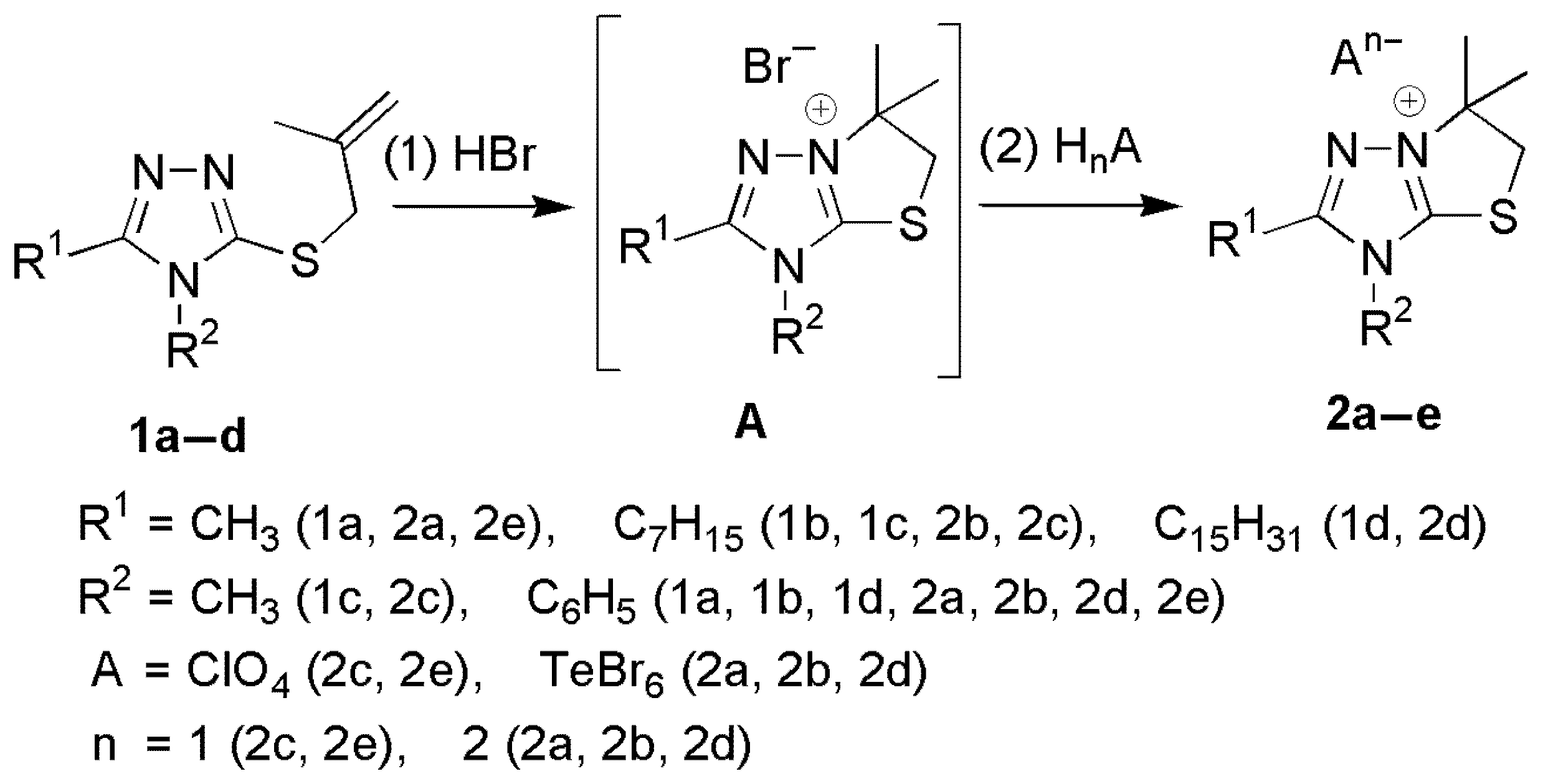
2.1. Synthesis of [1,3]Thiazolo[3,2-b][1,2,4]triazol-7-ium Salts 2a–e
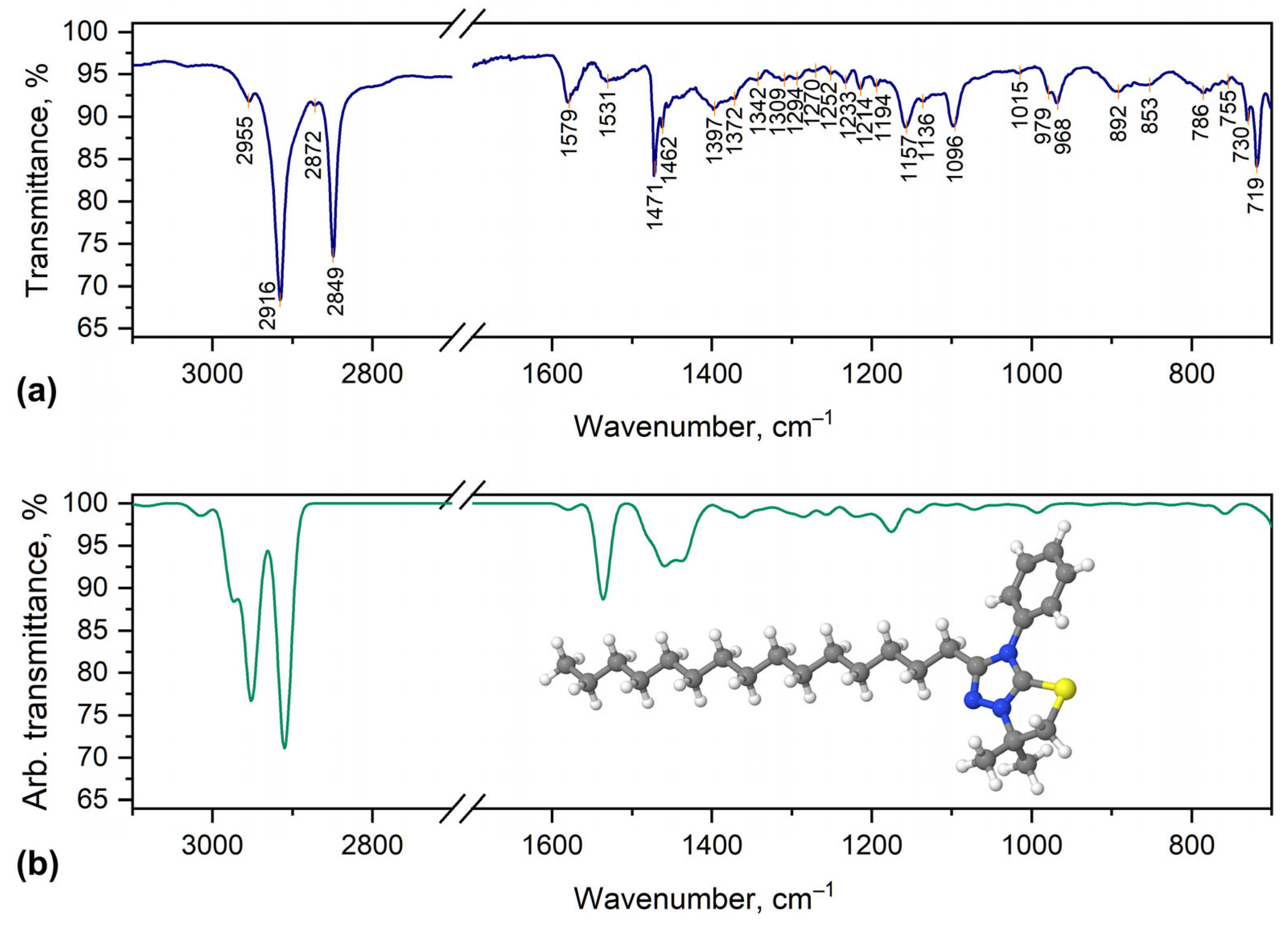
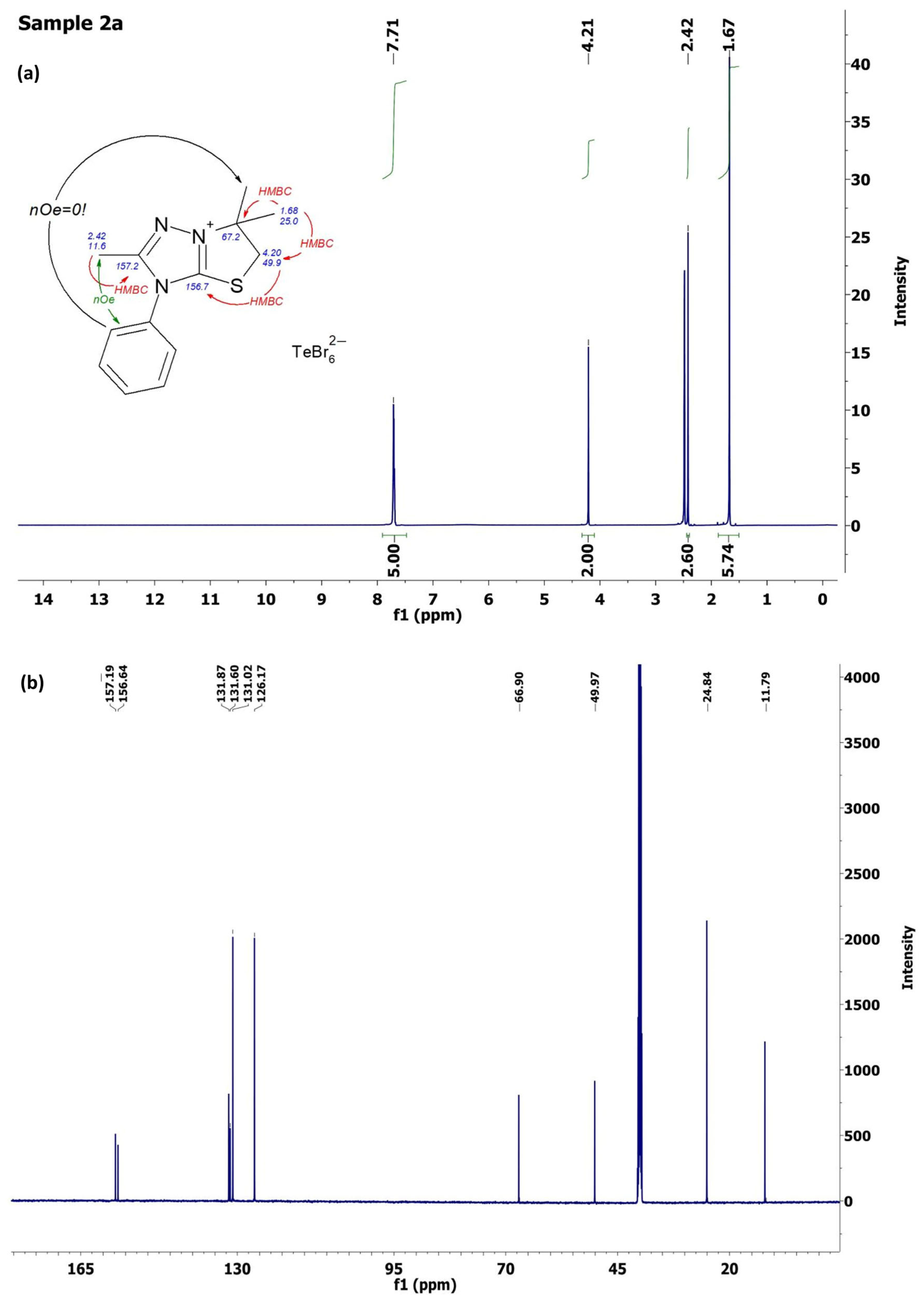
2.2. Characterization of [1,3]Thiazolo[3,2-b][1,2,4]triazol-7-ium Salts 2a–e
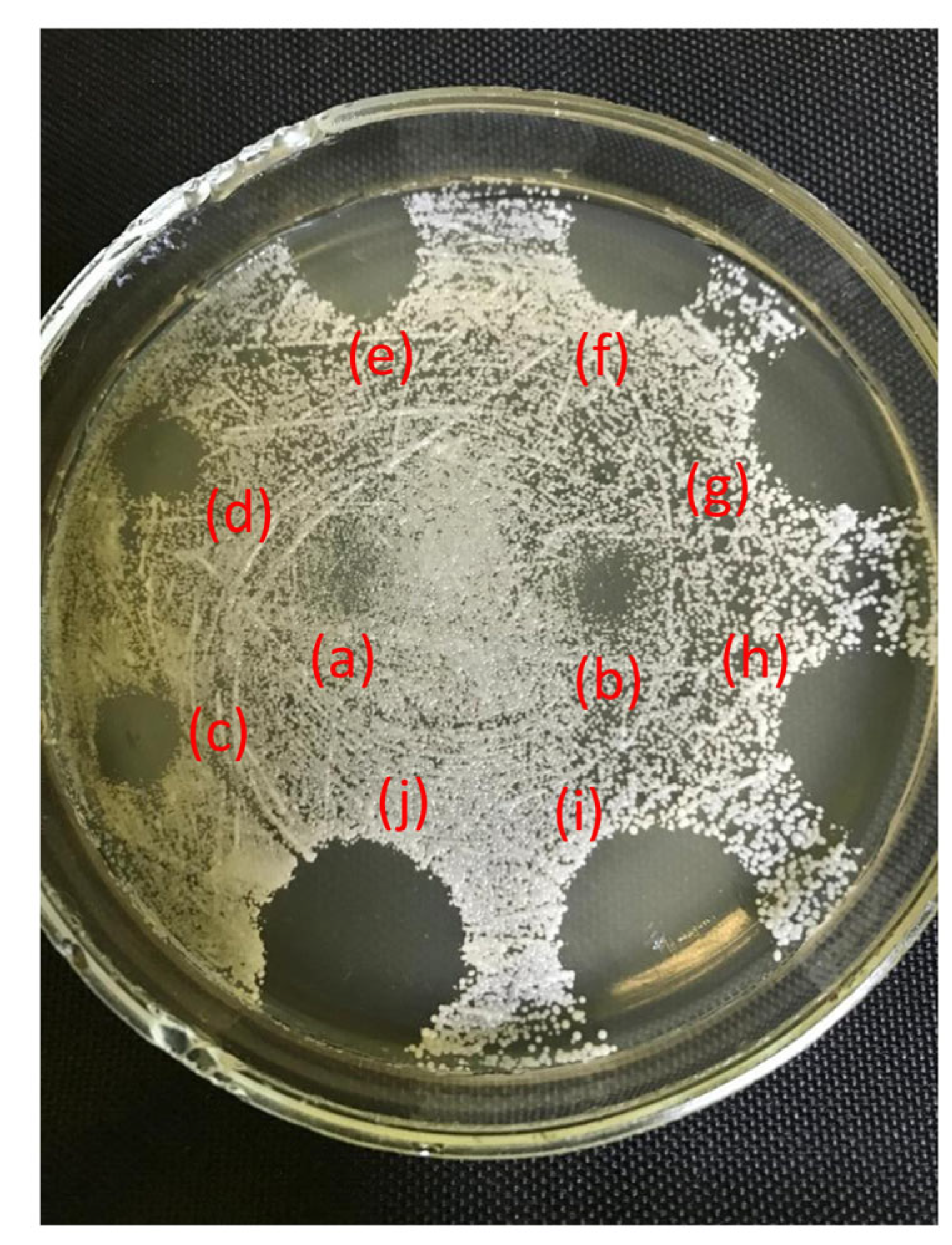
2.3. Antifungal Activity
2.4. Antibacterial Activity
2.5. Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Analysis
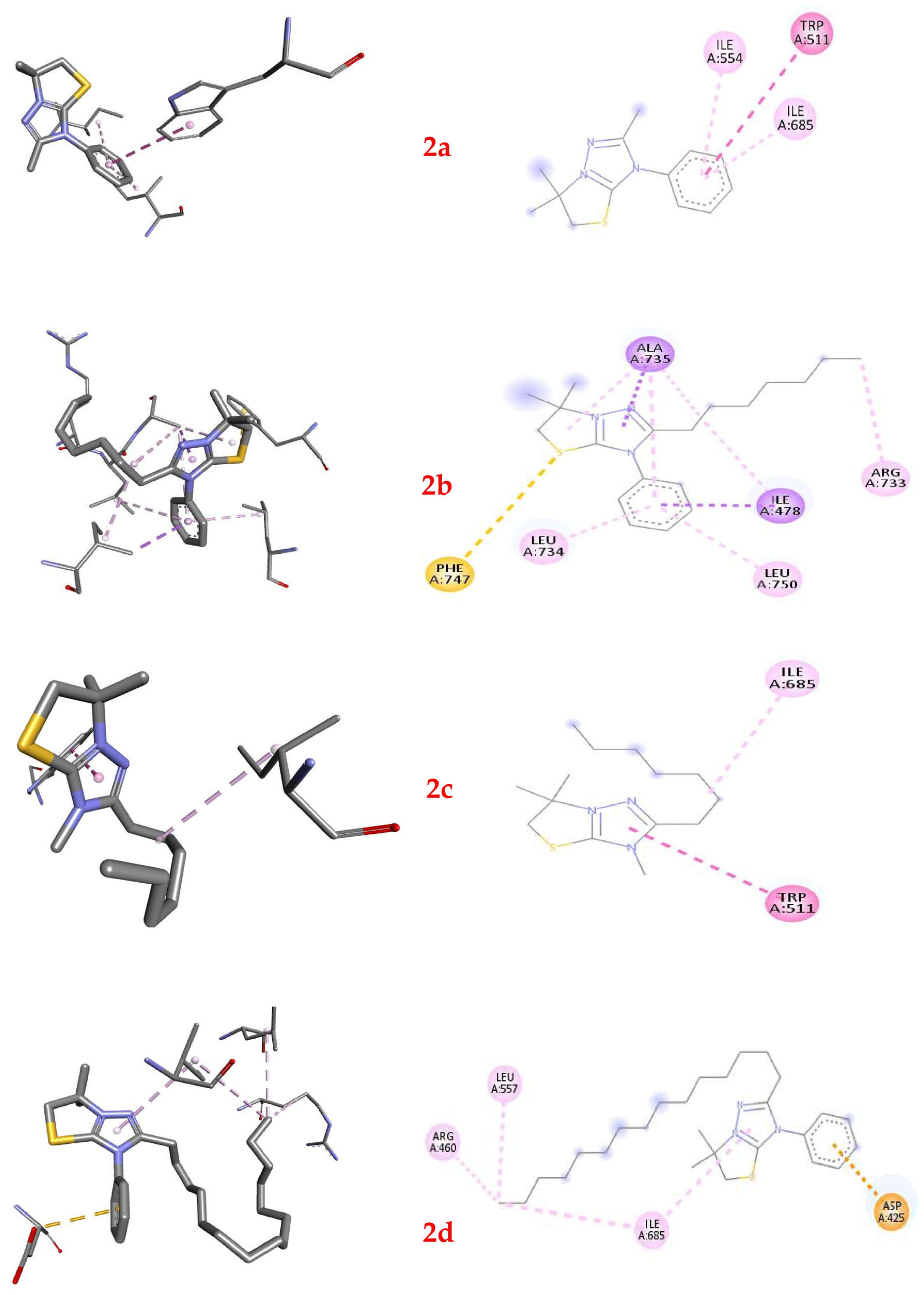
2.6. Molecular Docking of Thiazolo[3,2-b][1,2,4]triazolium Cations 2a–2d
3. Materials and Methods
3.1. Chemicals and Instruments
3.2. Synthesis of [1,3]Thiazolo[3,2-b][1,2,4]triazol-7-ium Salts 2a–e
3.3. Microbial Strains
3.4. Antimicrobial Assays
3.4.1. Antifungal Studies
3.4.2. Antibacterial Studies
3.5. Software
3.5.1. DFT Calculations
3.5.2. Molecular Docking Procedure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cella, E.; Giovanetti, M.; Benedetti, F.; Scarpa, F.; Johnston, C.; Borsetti, A.; Ceccarelli, G.; Azarian, T.; Zella, D.; Ciccozzi, M. Joining Forces against Antibiotic Resistance: The One Health Solution. Pathogens 2023, 12, 1074. [Google Scholar] [CrossRef]
- Gattu, R.; Ramesh, S.S.; Ramesh, S. Role of small molecules and nanoparticles in effective inhibition of microbial biofilms: A ray of hope in combating microbial resistance. Microb. Pathog. 2024, 188, 106543. [Google Scholar] [CrossRef] [PubMed]
- Loban’, G.; Faustova, M.; Dobrovolska, O.; Tkachenko, P. War in Ukraine: Incursion of antimicrobial resistance. Ir. J. Med. Sci. 2023, 192, 2905–2907. [Google Scholar] [CrossRef]
- Marinescu, M. Benzimidazole-Triazole Hybrids as Antimicrobial and Antiviral Agents: A Systematic Review. Antibiotics 2023, 12, 1220. [Google Scholar] [CrossRef]
- Ambade, S.S.; Gupta, V.K.; Bhole, R.P.; Khedekar, P.B.; Chikhale, R.V. A Review on Five and Six-Membered Heterocyclic Compounds Targeting the Penicillin-Binding Protein 2 (PBP2A) of Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules 2023, 28, 7008. [Google Scholar] [CrossRef]
- Hryhoriv, H.; Kovalenko, S.M.; Georgiyants, M.; Sidorenko, L.; Georgiyants, V. A Comprehensive Review on Chemical Synthesis and Chemotherapeutic Potential of 3-Heteroaryl Fluoroquinolone Hybrids. Antibiotics 2023, 12, 625. [Google Scholar] [CrossRef]
- Rusu, A.; Moga, I.-M.; Uncu, L.; Hancu, G. The Role of Five-Membered Heterocycles in the Molecular Structure of Antibacterial Drugs Used in Therapy. Pharmaceutics 2023, 15, 2554. [Google Scholar] [CrossRef]
- Tian, G.; Song, Q.; Liu, Z.; Guo, J.; Cao, S.; Long, S. Recent advances in 1,2,3- and 1,2,4-triazole hybrids as antimicrobials and their SAR: A critical review. Eur. J. Med. Chem. 2023, 259, 115603. [Google Scholar] [CrossRef]
- Ungureanu, D.; Tiperciuc, B.; Nastasă, C.; Ionuț, I.; Marc, G.; Oniga, I.; Oniga, O. An Overview of the Structure–Activity Relationship in Novel Antimicrobial Thiazoles Clubbed with Various Heterocycles (2017–2023). Pharmaceutics 2024, 16, 89. [Google Scholar] [CrossRef]
- Slivka, M.V.; Korol, N.I.; Fizer, M.M. Fused bicyclic 1,2,4-triazoles with one extra sulfur atom: Synthesis, properties, and biological activity. J. Heterocycl. Chem. 2020, 57, 3236–3254. [Google Scholar] [CrossRef]
- Korol, N.I.; Slivka, M.V. Recent progress in the synthesis of thiazolo[3,2-b][1,2,4]triazoles (microreview). Chem. Heterocycl. Compd. 2017, 53, 852–854. [Google Scholar] [CrossRef]
- Fotopoulou, E.; Anastasiou, P.K.; Tomza, C.; Neochoritis, C.G. The Ugi reaction as the green alternative towards active pharmaceutical ingredients. Tetrahedron Green Chem 2024, 3, 100044. [Google Scholar] [CrossRef]
- Larghi, E.L.; Bracca, A.B.J.; Simonetti, S.O.; Kaufman, T.S. Relevant Developments in the Use of Three-Component Reactions for the Total Synthesis of Natural Products. The last 15 Years. ChemistryOpen 2024, 13, e202300306. [Google Scholar] [CrossRef]
- Chebanov, V.A.; Desenko, S.M.; Lipson, V.V. Heterocycles on the crest of microwaves and ultrasonics in the Institute for Single Crystals of National Academy of Sciences of Ukraine: Chemistry and history. Chem. Heterocycl. Compd. 2023, 59, 386–405. [Google Scholar] [CrossRef]
- Slivka, M.; Onysko, M. The use of electrophilic cyclization for the preparation of condensed heterocycles. Synthesis 2021, 53, 3497–3512. [Google Scholar] [CrossRef]
- Neuhaus, J.D.; Willis, M.C. Homogeneous rhodium(i)-catalysis in: De novo heterocycle syntheses. Org. Biomol. Chem. 2016, 14, 4986–5000. [Google Scholar] [CrossRef] [PubMed]
- Frecentese, F.; Sodano, F.; Corvino, A.; Schiano, M.E.; Magli, E.; Albrizio, S.; Sparaco, R.; Andreozzi, G.; Nieddu, M.; Rimoli, M.G. The Application of Microwaves, Ultrasounds, and Their Combination in the Synthesis of Nitrogen-Containing Bicyclic Heterocycles. Int. J. Mol. Sci. 2023, 24, 10722. [Google Scholar] [CrossRef]
- Kamanna, K.; Amaregouda, Y. Water Mediated Green Method Synthesis of Bioactive Heterocyclic Reported Between 2012–2021 Accelerated by Microwave Irradiation: A Decennary Update. Curr. Organocatalysis 2023, 10, 160–179. [Google Scholar] [CrossRef]
- Mantovani, A.C.; Hernandez, J.G.; Bolm, C. Synthesis of 3-iodobenzofurans by electrophilic cyclization under solventless conditions in a ball mill. Eur. J. Org. Chem. 2018, 20–21, 2458–2461. [Google Scholar] [CrossRef]
- Slivka, M.; Korol, N.; Pantyo, V.; Baumer, V.; Lendel, V. Regio- and stereoselective synthesis of [1,3]thiazolo[3,2-b][1,2,4]triazol-7-ium salts via electrophilic heterocyclization of 3-S-propargylthio-4H-1,2,4-triazoles and their antimicrobial activity. Heterocycl. Commun. 2017, 23, 109–113. [Google Scholar] [CrossRef]
- Slivka, M.; Fizer, M.; Mariychuk, R.; Ostafin, M.; Moyzesh, O.; Koval, G.; Holovko-Kamoshenkova, O.; Rusyn, I.; Lendel, V. Synthesis and Antimicrobial Activity of Functional Derivatives of thiazolo[2,3-c][1,2,4]triazoles. Lett. Drug Des. Discov. 2022, 19, 791–799. [Google Scholar] [CrossRef]
- Slivka, M.; Korol, N.; Fizer, M.; Baumer, V.; Lendel, V. Thiazolo[3,2-b][1,2,4]triazol-7-ium salts: Synthesis, properties and structural studies. Heterocycl. Commun. 2018, 24, 197–203. [Google Scholar] [CrossRef]
- Kut, D.; Kut, M.; Komarovska-Porokhnyavets, O.; Kurka, M.; Onysko, M.; Lubenets, V. Antimicrobial Activity of Halogen- and Chalcogen-Functionalized Thiazoloquinazolines. Lett. Drug Des. Discov. 2024, 21, 2490–2496. [Google Scholar] [CrossRef]
- Khattab, S.M.; Altowyan, M.S.; El-Faham, A.; Barakat, A.; Haukkaorsy, M.; Abu-Youssef, M.A.M.; Soliman, S.M. A New Bromo-Mn(II) Complex with 1,3,5-Triazine Derivative: Synthesis, Crystal Structure, DFT and Biological Studies. Inorganics 2024, 12, 284. [Google Scholar] [CrossRef]
- Mtar, S.; Sellami, S.; Chniba-Boudjada, N.; Boujelbene, M. Structural study, optical properties, thermal stability, and biological activity of a non-centrosymmetric hybrid compound 3,4-Diaminopyridinium perchlorate (3,4-DAPP). Results Chem. 2025, 14, 102078. [Google Scholar] [CrossRef]
- Bardiya, N.; Bae, J.-H. Dissimilatory perchlorate reduction: A review. Microbiol. Res. 2011, 166, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Vávrová, S.; Struhárňanská, E.; Turňa, J.; Stuchlík, S. Tellurium: A Rare Element with Influence on Prokaryotic and Eukaryotic Biological Systems. Int. J. Mol. Sci. 2021, 22, 5924. [Google Scholar] [CrossRef]
- Fizer, M.; Slivka, M.; Korol, N.; Fizer, O. Identifying and explaining the regioselectivity of alkylation of 1,2,4-triazole-3-thiones using NMR, GIAO and DFT methods. J. Mol. Struct. 2021, 1223, 128973. [Google Scholar] [CrossRef]
- Fizer, M.; Slivka, M.; Fizer, O. Synthetic strategy and structure characterization of a new [1,3]thiazolo[3,2-b][1,2,4]triazolium cationic surfactant. Biointerface Res. Appl. Chem. 2021, 11, 13885–13892. [Google Scholar] [CrossRef]
- Fizer, M.; Slivka, M.; Baumer, V. Efficient synthesis of substituted [1,3]thiazolo[3,2-b][1,2,4]triazol-7-ium hexabromotellurates. J. Organomet. Chem. 2021, 952, 122044. [Google Scholar] [CrossRef]
- Fizer, O.; Dujnič, V.; Sidey, V.; Baumer, V.; Slivka, M.; Fizer, M. New 2-amino-[1,3]thiazolo[3,2-b][1,2,4]triazol-7-ium hexabromotellurates: Experimental and computational structural studies. J. Mol. Struct. 2024, 1295, 136632. [Google Scholar] [CrossRef]
- Fizer, O.; Fizer, M.; Filep, M.; Sidey, V.; Mariychuk, R. On the structure of cetylpyridinium perchlorate: A combined XRD, NMR, IR and DFT study. J. Mol. Liq. 2022, 368, 120659. [Google Scholar] [CrossRef]
- CLSI. Method for Antifungal Disk Diffusion Susceptibility Testing of Yeasts, 3rd ed.; CLSI guideline M44; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; Available online: https://clsi.org/media/2634/m44ed3_sample.pdf (accessed on 28 August 2021).
- Krauß, J.; Müller, C.; Klimt, M.; Valero, L.J.; Martínez, J.F.; Müller, M.; Bartel, K.; Binder, U.; Bracher, F. Synthesis, Biological Evaluation, and Structure–Activity Relationships of 4-Aminopiperidines as Novel Antifungal Agents Targeting Ergosterol Biosynthesis. Molecules 2021, 26, 7208. [Google Scholar] [CrossRef] [PubMed]
- Doğan, I.S.; Saraç, S.; Sari, S.; Kart, D.; Eşsiz Gökhan, Ş.; Vural, İ.; Dalkara, S. New Azole Derivatives Showing Antimicrobial Effects and Their Mechanism of Antifungal Activity by Molecular Modeling Studies. Eur. J. Med. Chem. 2017, 130, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Kontoyiannis, D.P.; Lewis, R.E. Antifungal drug resistance of pathogenic fungi. Lancet 2002, 359, 1135–1144. [Google Scholar] [CrossRef]
- Krogh-Madsen, M.; Arendrup, M.C.; Heslet, L.; Knudsen, J.D. Amphotericin B and caspofungin resistance in Candida glabrata isolates recovered from a critically ill patient. Clin. Infect. Dis. 2006, 42, 938–944. [Google Scholar] [CrossRef]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef]
- Enache-Angoulvant, A.; Hennequin, C. Invasive Saccharomyces Infection: A Comprehensive Review. Clin. Infect. Dis. 2005, 41, 1559–1568. [Google Scholar] [CrossRef]
- Steigenberger, J.; Verleysen, Y.; Geudens, N.; Martins, J.C.; Heerklotz, H. The Optimal Lipid Chain Length of a Membrane-Permeabilizing Lipopeptide Results From the Balance of Membrane Partitioning and Local Damage. Front. Microbiol. 2021, 12, 669709. [Google Scholar] [CrossRef]
- Wang, S.Q.; Wang, Y.-F.; Xu, Z. Tetrazole Hybrids and Their Antifungal Activities. Eur. J. Med. Chem. 2019, 170, 225–234. [Google Scholar] [CrossRef]
- Kim, S.-H.; Hind, C.K.; Fernandes, G.F.S.; Wu, J.; Semenya, D.; Clifford, M.; Marsh, C.; Anselmi, S.; Mason, A.J.; Bruce, K.D.; et al. Development of Novel Membrane-Disrupting Lipoguanidine Compounds Sensitizing Gram-Negative Bacteria to Antibiotics. ACS Med. Chem. Lett. 2024, 15, 239–249. [Google Scholar] [CrossRef]
- Saxena, D.; Maitra, R.; Bormon, R.; Czekanska, M.; Meiers, J.; Titz, A.; Verma, S.; Chopra, S. Tackling the outer membrane: Facilitating compound entry into Gram-negative bacterial pathogens. npj Antimicrob Resist. 2023, 1, 17. [Google Scholar] [CrossRef]
- Manrique, P.D.; Leus, I.V.; López, C.A.; Mehla, J.; Malloci, G.; Gervasoni, S.; Vargiu, A.V.; Kinthada, R.K.; Herndon, L.; Hengartner, N.W.; et al. Predicting Permeation of Compounds across the Outer Membrane of Pseudomonas aeruginosa Using Molecular Descriptors. Commun. Chem. 2024, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- ADMETlab 3.0. Available online: https://admetlab3.scbdd.com/server/evaluation (accessed on 2 December 2024).
- Arendrup, M.C.; Meletiadis, J.; Mouton, J.W.; Lagrou, K.; Hamal, P.; Guinea, J.; Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID, European Committee for Antimicrobial Susceptibility Testing (EUCAST). Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts. EUCAST E. Def 7.4 October 2023. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E.Def_7.4_Yeast_definitive_revised_2023.pdf (accessed on 16 February 2024).
- Hamon, Y.; Péron, Y. Description de quelques épreuves simples permettant de distinguer les bactériocines sensu-stricto des autres agents antibactériens produits par les bactéries. C.R. Acad. Sci. Paris Ser. D 1977, 285, 1215–1217. [Google Scholar]
- Sasoni, N.; Caracciolo, B.; Cabeza, M.S.; Gamarra, S.; Carnovale, S.; Garcia-Effron, G. Antifungal susceptibility testing following the CLSI M27 document, along with the measurement of MFC/MIC ratio, could be the optimal approach to detect amphotericin B resistance in Clavispora (Candida) lusitaniae. Susceptibility patterns of contemporary isolates of this species. Antimicrob. Agents Chemother. 2024, 68, e0096823. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard, 11th ed.; CLSI document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; Available online: https://clsi.org/media/1928/m07ed11_sample.pdf (accessed on 14 March 2021).
- Sader, H.S.; Fritsche, T.R.; Jones, R.N. Daptomycin bactericidal activity and correlation between disk and broth microdilution method results in testing of Staphylococcus aureus strains with decreased susceptibility to vancomycin. Antimicrob. Agents Chemother. 2006, 50, 2330–2336. [Google Scholar] [CrossRef]
- French, G.L. Bactericidal agents in the treatment of MRSA infections—The potential role of daptomycin. J. Antimicrob. Chemother. 2006, 58, 1107–1117. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced Basis Sets of Split Valence, Triple Zeta Valence and Quadruple Zeta Valence Quality for H to Rn: Design and Assessment of Accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297. [Google Scholar] [CrossRef]
- Zheng, J.; Xu, X.; Truhlar, D.G. Minimally augmented Karlsruhe basis sets. Theor. Chem. Acc. 2011, 128, 295. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Frank, N. Software update: The ORCA program system—Version 5.0. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2022, 12, e1606. [Google Scholar] [CrossRef]
- Teixeira, F.; Cordeiro, M.N.D.S. Improving Vibrational Mode Interpretation Using Bayesian Regression. Chem. Theory Comput. 2019, 15, 456. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- The Scripps Research Institute. AutoDock Vina (version 1.2.0); San Diego, CA, USA. Available online: https://vina.scripps.edu/ (accessed on 20 July 2021).
- Dassault Systèmes BIOVIA. Discovery Studio Visualizer, Release 2021, Dassault Systèmes, San Diego, CA, USA. 2021. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 21 December 2021).
- Hyperchem, Inc. Gainesville, FL, USA. Available online: http://www.hypercubeusa.com/ (accessed on 5 May 2022).
- BIOVIA Discovery Studio, Dassault Systèmes, France. Available online: https://www.3ds.com/products/biovia/discovery-studio (accessed on 5 May 2023).



| Antifungal Compounds | 6 mm Disk Content µg | Zone of Inhibition, mm * | ||
|---|---|---|---|---|
| Candida albicans ATCC 885–653 | Candida albicans BS3 | Saccharomyces cerevisiae BS3 | ||
| Compound 2a | 20 | 20.3 ± 0.1 | 20.4 ± 0.2 | 20 ± 0.2 |
| Amphotericin B | 100 | 12.0 ± 0.2 | 11.5 ± 0.1 | 22 ± 0.3 |
| Nystatin | 80 | 13.2 ± 0.1 | 12.1 ± 0.2 | 21 ± 0.1 |
| Clotrimazole | 10 | 26.1 ± 0.2 | 24.5 ± 0.2 | 19.2 ± 0.2 |
| Fluconazole | 20 | 29.4 ± 0.1 | 29.5 ± 0.3 | 14.0 ± 0.3 |
| Itraconazole | 10 | 26.4 ± 0.3 | 25.5 ± 0.1 | 14.1 ± 0.2 |
| Ketoconazole | 20 | 29.1 ± 0.1 | 29.0 ± 0.1 | 13.7± 0.2 |
| Fungi | Thiazolotriazole 2a, µg/mL | Fluconazole, µg/mL | ||
|---|---|---|---|---|
| MFC | MIC | MFC | MIC | |
| Candida albicans ATCC 885–653 | 125 | 31.25 | - | 31.25 |
| Candida albicans BS3 | 125 | 31.25 | - | 31.25 |
| Saccharomyces cerevisiae BS3 | 125 | 31.25 | - | 500 |
| Microorganism |  2a |  2b |  2c |  2d |  2e | Str | Amp | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | |
| Gram positive | ||||||||||||||
| E. faecalis ATCC 29212 | 31.25 | 15.63 | - | - | 31.25 | 15.63 | - | - | 500 | 250 | 31.25 | 15.63 | 15.63 | 7.81 |
| S. aureus ATCC 12600 | 62.5 | 31.25 | - | - | 125 | 62.5 | - | - | - | - | 31.25 | 15.63 | 3.9 | 1.95 |
| S. aureus ATCC 29213 | 62.5 | 31.25 | - | - | 125 | 62.5 | - | - | - | - | 31.25 | 15.63 | 3.9 | 1.95 |
| S. sapro-phyticus ATCC 15305 | 125 | 62.5 | - | - | 31.25 | 15.63 | 250 | 125 | 125 | 62.5 | 1.95 | 0.97 | 62.5 | 31.25 |
| S. epidermidis ATCC 14990 | 62.5 | 31.25 | - | - | 15.63 | 7.81 | - | - | 62.5 | 31.25 | 31.25 | 15.63 | 31.25 | 1.95 |
| Gram negative | ||||||||||||||
| P. aeruginosa ATCC 27853 | - | - | 500 | 250 | - | - | 125 | 62.5 | 500 | 250 | 62.5 | 31.25 | >500 | >500 |
| E. coli ATCC 25922 | 3.9 | 1.95 | 125 | 62.5 | 125 | 62.5 | 31.25 | 15.63 | 62.5 | 7.81 | 31.25 | 15.63 | 15.63 | 7.81 |
| K. pneumoniae CSES 23/821 | 15.63 | 7.81 | 15.63 | 1.95 | - | - | 62.5 | 31.25 | 62.5 | 31.25 | 31.25 | 15.63 | 125 | 62.5 |
| Sh. flexneri NCTC 9725 | 15.63 | 7.81 | 31.25 | 15.6 | 500 | 250 | 62.5 | 31.25 | 3.9 | 1.95 | 15.63 | 7.81 | 31.25 | 7.81 |
| Parameter | Cation | |||
|---|---|---|---|---|
| 2a | 2b | 2c | 2d | |
| Molecular Weight, g/mol | 246.11 | 330.2 | 268.18 | 442.33 |
| Topological Polar Surface Area (TPSA), Å2 | 21.7 | 21.7 | 21.7 | 21.7 |
| Aqueous Solubility (LogS) | −3.299 | −4.772 | 3.525 | 6.399 |
| Lipophilicity (LogP) | 2.393 | 4.256 | 3.409 | 7.59 |
| Distribution (LogD) | 2.584 | 3.704 | 3.079 | 4.844 |
| Caco-2 Permeability | 5.089 | −5.032 | 5.069 | −5.131 |
| Intestinal Absorption (HIA) | HIA- (Low) | HIA- (Very Low) | HIA+ (Moderate) | HIA- (Very Low) |
| Plasma Protein Binding, % | 80.217 | 98.808 | 94.149 | 100.551 |
| BBB Penetration | High (0.885) | Low (0.247) | Low (0.102) | Very Low (0.029) |
| P-gp Inhibition | Strong Inhibitor (0.931) | Strong Inhibitor (0.99) | Weak Inhibitor (0.227) | Strong Inhibitor (0.867) |
| Human Liver Microsomal (HLM) Stability | Unstable (0.991) | Unstable (0.979) | Moderate Stability (0.402) | Unstable (0.914) |
| Half-life (T1/2), hours | 0.411 | 0.231 | 0.274 | 0.717 |
| Plasma Clearance, mL/min/kg | 7.558 | 5.778 | 5.981 | 5.078 |
| Bioavailability (F50%) | Low (0.868) | Low (0.976) | Low (0.995) | Very Low (0.997) |
| Drug-Induced Liver Injury (DILI) | High (0.959) | High (0.844) | High (0.771) | High (0.811) |
| Skin Sensitization | High (0.996) | High (0.994) | High (0.997) | High (0.999) |
| Respiratory Toxicity | Moderate (0.833) | High (0.895) | High (0.896) | High (0.971) |
| Carcinogenicity | Moderate (0.771) | Moderate (0.457) | Moderate (0.538) | Moderate (0.369) |
| Genotoxicity | Moderate (0.979) | Low (0.274) | Moderate (0.403) | Low (0.002) |
| Cardiotoxicity (hERG Inhibition) | Low (0.095) | Moderate (0.431) | Moderate (0.253) | High (0.697) |
| Compound | Affinity, Kcal/Mol | Interacted Residue | Distance, Å |
|---|---|---|---|
| 2a | −6.0 | TRP511 | 5.24641 |
| ILE554 | 5.05906 | ||
| ILE685 | 5.36218 | ||
| 2b | −6.8 | ILE478 | 3.73107 |
| ALA735 | 3.82162 | ||
| PHE747 | 5.46826 | ||
| ILE478 | 4.47169 | ||
| ALA735 | 5.09851 | ||
| ALA735 | 3.66844 | ||
| ARG733 | 4.64739 | ||
| LEU734 | 5.41669 | ||
| ALA735 | 5.47675 | ||
| LEU750 | 4.76871 | ||
| 2c | −5.3 | TRP511 | 5.04091 |
| ILE685 | 4.7875 | ||
| 2d | −5.1 | ASP425 | 3.45701 |
| ARG460 | 4.28296 | ||
| LEU557 | 4.83272 | ||
| ILE685 | 5.27912 | ||
| ILE685 | 4.88259 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slivka, M.; Sharga, B.; Pylypiv, D.; Aleksyk, H.; Korol, N.; Fizer, M.; Fedurcya, O.I.; Pshenychnyi, O.G.; Mariychuk, R. [1,3]Thiazolo[3,2-b][1,2,4]triazolium Salts as Effective Antimicrobial Agents: Synthesis, Biological Activity Evaluation, and Molecular Docking Studies. Int. J. Mol. Sci. 2025, 26, 6845. https://doi.org/10.3390/ijms26146845
Slivka M, Sharga B, Pylypiv D, Aleksyk H, Korol N, Fizer M, Fedurcya OI, Pshenychnyi OG, Mariychuk R. [1,3]Thiazolo[3,2-b][1,2,4]triazolium Salts as Effective Antimicrobial Agents: Synthesis, Biological Activity Evaluation, and Molecular Docking Studies. International Journal of Molecular Sciences. 2025; 26(14):6845. https://doi.org/10.3390/ijms26146845
Chicago/Turabian StyleSlivka, Mykhailo, Boris Sharga, Daryna Pylypiv, Hanna Aleksyk, Nataliya Korol, Maksym Fizer, Olena I. Fedurcya, Oleksandr G. Pshenychnyi, and Ruslan Mariychuk. 2025. "[1,3]Thiazolo[3,2-b][1,2,4]triazolium Salts as Effective Antimicrobial Agents: Synthesis, Biological Activity Evaluation, and Molecular Docking Studies" International Journal of Molecular Sciences 26, no. 14: 6845. https://doi.org/10.3390/ijms26146845
APA StyleSlivka, M., Sharga, B., Pylypiv, D., Aleksyk, H., Korol, N., Fizer, M., Fedurcya, O. I., Pshenychnyi, O. G., & Mariychuk, R. (2025). [1,3]Thiazolo[3,2-b][1,2,4]triazolium Salts as Effective Antimicrobial Agents: Synthesis, Biological Activity Evaluation, and Molecular Docking Studies. International Journal of Molecular Sciences, 26(14), 6845. https://doi.org/10.3390/ijms26146845







