Unraveling ADAR-Mediated Protein Recoding: A Proteogenomic Exploration in Model Organisms and Human Pathology
Abstract
1. Introduction. RNA Editing by ADAR Adenosine Deaminases
2. Protein Recoding via A-to-I RNA Editing
3. Functional Significance of Protein Recoding
4. Searching for Recoded Proteins Using the Proteogenomic Approach
4.1. Methodological Aspects
4.2. Results of Proteogenomic Searches for ADAR-Mediated Protein Recoding
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADAR | Adenosine deaminase acting on RNA |
| MRM | Multiple reaction monitoring |
| PRM | Parallel reaction monitoring |
| sORF | short open-reading frame |
| LINE | Long interspersed nuclear elements |
| IFN I | Interferon I |
| AMPA | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| AMPARs | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors |
| NMDARs | N-methyl-D-aspartate receptors |
| CCNI | Cyclin-I |
| AZIN1 | Antizyme Inhibitor 1 |
| GRIA | Glutamate Ionotropic Receptor AMPA |
| GluA | Glutamate transport ATP-binding protein GluA |
| KAR | Kainate receptor |
| FLNA | Filamin A |
| FLNB | Filamin B |
| CYFIP2 | Cytoplasmic FMR1 Interacting Protein 2 |
| CADPS | Calcium Dependent Secretion Activator |
| RIMS2B | regulating synaptic membrane exocytosis 2b |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| Syx1A | Syntaxin 1 A |
| Syt1 | Synaptotagmin-1 |
| EndoA | endophilin A |
| cpx | complexin |
| FUS protein | Fused in Sarcoma protein |
| Neat1 | Nuclear Paraspeckle Assembly Transcript 1 |
| HSPA1L | Heat shock 70 kDa protein 1L |
| CNS | central nervous system |
| CACNG8 | Calcium Voltage-Gated Channel Auxiliary Subunit Gamma 8 |
| TARP γ8 | transmembrane AMPA receptor regulatory protein subunit γ8 |
| IGFBP7 | insulin-like growth factor-binding protein 7 |
References
- Smith, L.M.; Kelleher, N.L. Consortium for Top Down Proteomics Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [PubMed]
- Lisitsa, A.; Moshkovskii, S.; Chernobrovkin, A.; Ponomarenko, E.; Archakov, A. Profiling proteoforms: Promising follow-up of proteomics for biomarker discovery. Expert Rev. Proteom. 2014, 11, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L.; Weintraub, H. A developmentally regulated activity that unwinds RNA duplexes. Cell 1987, 48, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L.; Weintraub, H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 1988, 55, 1089–1098. [Google Scholar] [CrossRef]
- Zinshteyn, B.; Nishikura, K. Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 202–209. [Google Scholar] [CrossRef]
- Keegan, L.P.; Hajji, K.; O’Connell, M.A. Adenosine Deaminase Acting on RNA (ADAR) Enzymes: A Journey from Weird to Wondrous. Acc. Chem. Res. 2023, 56, 3165–3174. [Google Scholar] [CrossRef]
- Morse, D.P.; Aruscavage, P.J.; Bass, B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. USA 2002, 99, 7906–7911. [Google Scholar] [CrossRef]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004, 2, e391. [Google Scholar] [CrossRef]
- Liu, H.; Golji, J.; Brodeur, L.K.; Chung, F.S.; Chen, J.T.; deBeaumont, R.S.; Bullock, C.P.; Jones, M.D.; Kerr, G.; Li, L.; et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 2019, 25, 95–102. [Google Scholar] [CrossRef]
- Ishizuka, J.J.; Manguso, R.T.; Cheruiyot, C.K.; Bi, K.; Panda, A.; Iracheta-Vellve, A.; Miller, B.C.; Du, P.P.; Yates, K.B.; Dubrot, J.; et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 2019, 565, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Goncharov, A.O.; Shender, V.O.; Kuznetsova, K.G.; Kliuchnikova, A.A.; Moshkovskii, S.A. Interplay between A-to-I Editing and Splicing of RNA: A Potential Point of Application for Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 5240. [Google Scholar] [CrossRef] [PubMed]
- Liddicoat, B.J.; Piskol, R.; Chalk, A.M.; Ramaswami, G.; Higuchi, M.; Hartner, J.C.; Li, J.B.; Seeburg, P.H.; Walkley, C.R. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015, 349, 1115–1120. [Google Scholar] [CrossRef]
- Rice, G.I.; Kasher, P.R.; Forte, G.M.A.; Mannion, N.M.; Greenwood, S.M.; Szynkiewicz, M.; Dickerson, J.E.; Bhaskar, S.S.; Zampini, M.; Briggs, T.A.; et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 2012, 44, 1243–1248. [Google Scholar] [CrossRef]
- Hood, J.L.; Emeson, R.B. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr. Top. Microbiol. Immunol. 2012, 353, 61–90. [Google Scholar] [CrossRef]
- Jepson, J.E.C.; Reenan, R.A. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta 2008, 1779, 459–470. [Google Scholar] [CrossRef]
- Yang, L.; Yi, L.; Yang, J.; Zhang, R.; Xie, Z.; Wang, H. Temporal landscape and translational regulation of A-to-I RNA editing in mouse retina development. BMC Biol. 2024, 22, 106. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q.; Zhao, F. Synonymous but not silent: The codon usage code for gene expression and protein folding. Annu. Rev. Biochem. 2021, 90, 375–401. [Google Scholar] [CrossRef]
- Walsh, I.M.; Bowman, M.A.; Soto Santarriaga, I.F.; Rodriguez, A.; Clark, P.L. Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc. Natl. Acad. Sci. USA 2020, 117, 3528–3534. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, H.G.; Beal, P.A. Structural and functional effects of inosine modification in mRNA. RNA 2024, 30, 512–520. [Google Scholar] [CrossRef]
- Zhang, M.; Fritsche, J.; Roszik, J.; Williams, L.J.; Peng, X.; Chiu, Y.; Tsou, C.-C.; Hoffgaard, F.; Goldfinger, V.; Schoor, O.; et al. RNA editing derived epitopes function as cancer antigens to elicit immune responses. Nat. Commun. 2018, 9, 3919. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Nakahama, T.; Wu, Y.; Inoue, M.; Kim, J.I.; Todo, H.; Shibuya, T.; Kato, Y.; Kawahara, Y. RNA editing of AZIN1 coding sites is catalyzed by ADAR1 p150 after splicing. J. Biol. Chem. 2023, 299, 104840. [Google Scholar] [CrossRef] [PubMed]
- Zambrano-Mila, M.S.; Witzenberger, M.; Rosenwasser, Z.; Uzonyi, A.; Nir, R.; Ben-Aroya, S.; Levanon, E.Y.; Schwartz, S. Dissecting the basis for differential substrate specificity of ADAR1 and ADAR2. Nat. Commun. 2023, 14, 8212. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K.; Sato, S.; Lazinski, D.W. Substrate recognition by ADAR1 and ADAR2. RNA 2001, 7, 846–858. [Google Scholar] [CrossRef]
- Källman, A.M.; Sahlin, M.; Ohman, M. ADAR2 A-->I editing: Site selectivity and editing efficiency are separate events. Nucleic Acids Res. 2003, 31, 4874–4881. [Google Scholar] [CrossRef]
- Moldovan, M.A.; Chervontseva, Z.S.; Nogina, D.S.; Gelfand, M.S. A hierarchy in clusters of cephalopod mRNA editing sites. Sci. Rep. 2022, 12, 3447. [Google Scholar] [CrossRef]
- Lehmann, K.A.; Bass, B.L. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 2000, 39, 12875–12884. [Google Scholar] [CrossRef]
- Eggington, J.M.; Greene, T.; Bass, B.L. Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2011, 2, 319. [Google Scholar] [CrossRef]
- Stefl, R.; Xu, M.; Skrisovska, L.; Emeson, R.B.; Allain, F.H.-T. Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure 2006, 14, 345–355. [Google Scholar] [CrossRef]
- Tan, M.H.; Li, Q.; Shanmugam, R.; Piskol, R.; Kohler, J.; Young, A.N.; Liu, K.I.; Zhang, R.; Ramaswami, G.; Ariyoshi, K.; et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 2017, 550, 249–254. [Google Scholar] [CrossRef]
- Shanmugam, R.; Zhang, F.; Srinivasan, H.; Richard, J.L.C.; Liu, K.I.; Zhang, X.; Woo, C.W.A.; Chua, Z.H.M.; Buschdorf, J.P.; Meaney, M.J.; et al. SRSF9 selectively represses ADAR2-mediated editing of brain-specific sites in primates. Nucleic Acids Res. 2018, 46, 7379–7395. [Google Scholar] [CrossRef]
- Eisenberg, E. Proteome diversification by RNA editing. Methods Mol. Biol. 2021, 2181, 229–251. [Google Scholar] [CrossRef]
- Sommer, B.; Köhler, M.; Sprengel, R.; Seeburg, P.H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 1991, 67, 11–19. [Google Scholar] [CrossRef]
- Lomeli, H.; Mosbacher, J.; Melcher, T.; Höger, T.; Geiger, J.R.; Kuner, T.; Monyer, H.; Higuchi, M.; Bach, A.; Seeburg, P.H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 1994, 266, 1709–1713. [Google Scholar] [CrossRef]
- Burns, C.M.; Chu, H.; Rueter, S.M.; Hutchinson, L.K.; Canton, H.; Sanders-Bush, E.; Emeson, R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 1997, 387, 303–308. [Google Scholar] [CrossRef]
- Higuchi, M.; Single, F.N.; Köhler, M.; Sommer, B.; Sprengel, R.; Seeburg, P.H. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell 1993, 75, 1361–1370. [Google Scholar] [CrossRef]
- Seeburg, P.H.; Hartner, J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol. 2003, 13, 279–283. [Google Scholar] [CrossRef]
- Kawahara, Y.; Ito, K.; Sun, H.; Aizawa, H.; Kanazawa, I.; Kwak, S. Glutamate receptors: RNA editing and death of motor neurons. Nature 2004, 427, 801. [Google Scholar] [CrossRef]
- Kwak, S.; Kawahara, Y. Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. J. Mol. Med. 2005, 83, 110–120. [Google Scholar] [CrossRef]
- Yang, Q.; Li, X. Pan-cancer analysis of ADAR1 with its prognostic relevance in low-grade glioma. Immunobiology 2024, 229, 152855. [Google Scholar] [CrossRef]
- Higuchi, M.; Maas, S.; Single, F.N.; Hartner, J.; Rozov, A.; Burnashev, N.; Feldmeyer, D.; Sprengel, R.; Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000, 406, 78–81. [Google Scholar] [CrossRef]
- Horsch, M.; Seeburg, P.H.; Adler, T.; Aguilar-Pimentel, J.A.; Becker, L.; Calzada-Wack, J.; Garrett, L.; Götz, A.; Hans, W.; Higuchi, M.; et al. Requirement of the RNA-editing enzyme ADAR2 for normal physiology in mice. J. Biol. Chem. 2011, 286, 18614–18622. [Google Scholar] [CrossRef]
- Marion, S.; Weiner, D.M.; Caron, M.G. RNA editing induces variation in desensitization and trafficking of 5-hydroxytryptamine 2c receptor isoforms. J. Biol. Chem. 2004, 279, 2945–2954. [Google Scholar] [CrossRef]
- Niswender, C.M.; Copeland, S.C.; Herrick-Davis, K.; Emeson, R.B.; Sanders-Bush, E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem. 1999, 274, 9472–9478. [Google Scholar] [CrossRef] [PubMed]
- Price, R.D.; Weiner, D.M.; Chang, M.S.; Sanders-Bush, E. RNA editing of the human serotonin 5-HT2C receptor alters receptor-mediated activation of G13 protein. J. Biol. Chem. 2001, 276, 44663–44668. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Grimberg, A.; Teegarden, S.; Mombereau, C.; Liu, S.; Bale, T.L.; Blendy, J.A.; Nishikura, K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J. Neurosci. 2008, 28, 12834–12844. [Google Scholar] [CrossRef] [PubMed]
- Englander, M.T.; Dulawa, S.C.; Bhansali, P.; Schmauss, C. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J. Neurosci. 2005, 25, 648–651. [Google Scholar] [CrossRef]
- Iwamoto, K.; Bundo, M.; Kato, T. Serotonin receptor 2C and mental disorders: Genetic, expression and RNA editing studies. RNA Biol. 2009, 6, 248–253. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, J. Human coding RNA editing is generally nonadaptive. Proc. Natl. Acad. Sci. USA 2014, 111, 3769–3774. [Google Scholar] [CrossRef]
- Paz-Yaacov, N.; Bazak, L.; Buchumenski, I.; Porath, H.T.; Danan-Gotthold, M.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015, 13, 267–276. [Google Scholar] [CrossRef]
- Chan, T.H.M.; Qamra, A.; Tan, K.T.; Guo, J.; Yang, H.; Qi, L.; Lin, J.S.; Ng, V.H.E.; Song, Y.; Hong, H.; et al. ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer. Gastroenterology 2016, 151, 637–650.e10. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Diao, L.; Yu, S.; Xu, X.; Li, J.; Zhang, R.; Yang, Y.; Werner, H.M.J.; Eterovic, A.K.; Yuan, Y.; et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 2015, 28, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Moya, J.; Baker, A.R.; Slack, F.J.; Santisteban, P. ADAR1-mediated RNA editing is a novel oncogenic process in thyroid cancer and regulates miR-200 activity. Oncogene 2020, 39, 3738–3753. [Google Scholar] [CrossRef]
- Cenci, C.; Barzotti, R.; Galeano, F.; Corbelli, S.; Rota, R.; Massimi, L.; Di Rocco, C.; O’Connell, M.A.; Gallo, A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 2008, 283, 7251–7260. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.; Patt, S.; Schrey, M.; Rich, A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. USA 2001, 98, 14687–14692. [Google Scholar] [CrossRef]
- Chan, T.H.M.; Lin, C.H.; Qi, L.; Fei, J.; Li, Y.; Yong, K.J.; Liu, M.; Song, Y.; Chow, R.K.K.; Ng, V.H.E.; et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut 2014, 63, 832–843. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, H.; Xu, J.; Gao, W. ADAR, the carcinogenesis mechanisms of ADAR and related clinical applications. Ann. Transl. Med. 2019, 7, 686. [Google Scholar] [CrossRef]
- Jiang, D.; Zhang, J. The preponderance of nonsynonymous A-to-I RNA editing in coleoids is nonadaptive. Nat. Commun. 2019, 10, 5411. [Google Scholar] [CrossRef]
- Popitsch, N.; Huber, C.D.; Buchumenski, I.; Eisenberg, E.; Jantsch, M.; von Haeseler, A.; Gallach, M. A-to-I RNA Editing Uncovers Hidden Signals of Adaptive Genome Evolution in Animals. Genome Biol. Evol. 2020, 12, 345–357. [Google Scholar] [CrossRef]
- Moldovan, M.; Chervontseva, Z.; Bazykin, G.; Gelfand, M.S. Adaptive evolution at mRNA editing sites in soft-bodied cephalopods. PeerJ 2020, 8, e10456. [Google Scholar] [CrossRef]
- Picimbon, J.-F. A new view of genetic mutations. Australas. Med. J. 2017, 10, 701–715. [Google Scholar] [CrossRef]
- Shoshan, Y.; Liscovitch-Brauer, N.; Rosenthal, J.J.C.; Eisenberg, E. Adaptive Proteome Diversification by Nonsynonymous A-to-I RNA Editing in Coleoid Cephalopods. Mol. Biol. Evol. 2021, 38, 3775–3788. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Leutert, M.; Entwisle, S.W.; Villén, J. Decoding Post-Translational Modification Crosstalk With Proteomics. Mol. Cell. Proteom. 2021, 20, 100129. [Google Scholar] [CrossRef]
- Mun, D.-G.; Bhat, F.A.; Joshi, N.; Sandoval, L.; Ding, H.; Jain, A.; Peterson, J.A.; Kang, T.; Pujari, G.P.; Tomlinson, J.L.; et al. Diversity of post-translational modifications and cell signaling revealed by single cell and single organelle mass spectrometry. Commun. Biol. 2024, 7, 884. [Google Scholar] [CrossRef] [PubMed]
- Liscovitch-Brauer, N.; Alon, S.; Porath, H.T.; Elstein, B.; Unger, R.; Ziv, T.; Admon, A.; Levanon, E.Y.; Rosenthal, J.J.C.; Eisenberg, E. Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods. Cell 2017, 169, 191–202.e11. [Google Scholar] [CrossRef]
- Xuan, N.; Bu, X.; Liu, Y.Y.; Yang, X.; Liu, G.X.; Fan, Z.X.; Bi, Y.P.; Yang, L.Q.; Lou, Q.N.; Rajashekar, B.; et al. Molecular evidence of RNA editing in Bombyx chemosensory protein family. PLoS ONE 2014, 9, e86932. [Google Scholar] [CrossRef]
- Duan, Y.; Tang, X.; Lu, J. Evolutionary driving forces of A-to-I editing in metazoans. Wiley Interdiscip. Rev. RNA 2022, 13, e1666. [Google Scholar] [CrossRef]
- Gommans, W.M.; Mullen, S.P.; Maas, S. RNA editing: A driving force for adaptive evolution? Bioessays 2009, 31, 1137–1145. [Google Scholar] [CrossRef]
- Terajima, H.; Yoshitane, H.; Ozaki, H.; Suzuki, Y.; Shimba, S.; Kuroda, S.; Iwasaki, W.; Fukada, Y. ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat. Genet. 2017, 49, 146–151. [Google Scholar] [CrossRef]
- Robinson, J.E.; Paluch, J.; Dickman, D.K.; Joiner, W.J. ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat. Commun. 2016, 7, 10512. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Vukic, D.; Michalík, D.; O’Connell, M.A.; Keegan, L.P. ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 2017, 136, 1265–1278. [Google Scholar] [CrossRef]
- Garrett, S.; Rosenthal, J.J.C. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science 2012, 335, 848–851. [Google Scholar] [CrossRef]
- Alon, S.; Garrett, S.C.; Levanon, E.Y.; Olson, S.; Graveley, B.R.; Rosenthal, J.J.C.; Eisenberg, E. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. eLife 2015, 4, e05198. [Google Scholar] [CrossRef] [PubMed]
- Birk, M.A.; Liscovitch-Brauer, N.; Dominguez, M.J.; McNeme, S.; Yue, Y.; Hoff, J.D.; Twersky, I.; Verhey, K.J.; Sutton, R.B.; Eisenberg, E.; et al. Temperature-dependent RNA editing in octopus extensively recodes the neural proteome. Cell 2023, 186, 2544–2555.e13. [Google Scholar] [CrossRef]
- Forni, D.; Mozzi, A.; Pontremoli, C.; Vertemara, J.; Pozzoli, U.; Biasin, M.; Bresolin, N.; Clerici, M.; Cagliani, R.; Sironi, M. Diverse selective regimes shape genetic diversity at ADAR genes and at their coding targets. RNA Biol. 2015, 12, 149–161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Porath, H.T.; Schaffer, A.A.; Kaniewska, P.; Alon, S.; Eisenberg, E.; Rosenthal, J.; Levanon, E.Y.; Levy, O. A-to-I RNA Editing in the Earliest-Diverging Eumetazoan Phyla. Mol. Biol. Evol. 2017, 34, 1890–1901. [Google Scholar] [CrossRef]
- Perivolidi, V.-I.; Violitzi, F.; Ioannidou, E.; Rinotas, V.; Stamatakis, G.; Samiotaki, M.; Panayotou, G.; Douni, E. Proteomic Identification of the SLC25A46 Interactome in Transgenic Mice Expressing SLC25A46-FLAG. J. Proteome Res. 2022, 21, 375–394. [Google Scholar] [CrossRef]
- Song, Y.-C.; Das, D.; Zhang, Y.; Chen, M.-X.; Fernie, A.R.; Zhu, F.-Y.; Han, J. Proteogenomics-based functional genome research: Approaches, applications, and perspectives in plants. Trends Biotechnol. 2023, 41, 1532–1548. [Google Scholar] [CrossRef]
- Liu, H.; Li, Y.; Chen, D.; Qi, Z.; Wang, Q.; Wang, J.; Jiang, C.; Xu, J.-R. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2017, 114, E7756–E7765. [Google Scholar] [CrossRef]
- Teichert, I.; Dahlmann, T.A.; Kück, U.; Nowrousian, M. RNA editing during sexual development occurs in distantly related filamentous ascomycetes. Genome Biol. Evol. 2017, 9, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Q.; He, Y.; Chen, L.; Hao, C.; Jiang, C.; Li, Y.; Dai, Y.; Kang, Z.; Xu, J.-R. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 2016, 26, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, J.-R.; Liu, H. A-to-I RNA editing independent of ADARs in filamentous fungi. RNA Biol. 2016, 13, 940–945. [Google Scholar] [CrossRef]
- Kuznetsova, K.G.; Kliuchnikova, A.A.; Ilina, I.U.; Chernobrovkin, A.L.; Novikova, S.E.; Farafonova, T.E.; Karpov, D.S.; Ivanov, M.V.; Goncharov, A.O.; Ilgisonis, E.V.; et al. Proteogenomics of Adenosine-to-Inosine RNA Editing in the Fruit Fly. J. Proteome Res. 2018, 17, 3889–3903. [Google Scholar] [CrossRef]
- Kliuchnikova, A.A.; Goncharov, A.O.; Levitsky, L.I.; Pyatnitskiy, M.A.; Novikova, S.E.; Kuznetsova, K.G.; Ivanov, M.V.; Ilina, I.Y.; Farafonova, T.E.; Zgoda, V.G.; et al. Proteome-Wide Analysis of ADAR-Mediated Messenger RNA Editing during Fruit Fly Ontogeny. J. Proteome Res. 2020, 19, 4046–4060. [Google Scholar] [CrossRef]
- Nasaev, S.S.; Kopeykina, A.S.; Kuznetsova, K.G.; Levitsky, L.I.; Moshkovskii, S.A. Proteomic Analysis of Zebrafish Protein Recoding via mRNA Editing by ADAR Enzymes. Biochemistry 2022, 87, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Levitsky, L.I.; Kliuchnikova, A.A.; Kuznetsova, K.G.; Karpov, D.S.; Ivanov, M.V.; Pyatnitskiy, M.A.; Kalinina, O.V.; Gorshkov, M.V.; Moshkovskii, S.A. Adenosine-to-Inosine RNA Editing in Mouse and Human Brain Proteomes. Proteomics 2019, 19, e1900195. [Google Scholar] [CrossRef]
- Levitsky, L.I.; Ivanov, M.V.; Goncharov, A.O.; Kliuchnikova, A.A.; Bubis, J.A.; Lobas, A.A.; Solovyeva, E.M.; Pyatnitskiy, M.A.; Ovchinnikov, R.K.; Kukharsky, M.S.; et al. Massive Proteogenomic Reanalysis of Publicly Available Proteomic Datasets of Human Tissues in Search for Protein Recoding via Adenosine-to-Inosine RNA Editing. J. Proteome Res. 2023, 22, 1695–1711. [Google Scholar] [CrossRef]
- Gabay, O.; Shoshan, Y.; Kopel, E.; Ben-Zvi, U.; Mann, T.D.; Bressler, N.; Cohen-Fultheim, R.; Schaffer, A.A.; Roth, S.H.; Tzur, Z.; et al. Landscape of adenosine-to-inosine RNA recoding across human tissues. Nat. Commun. 2022, 13, 1184. [Google Scholar] [CrossRef]
- Paro, S.; Li, X.; O’Connell, M.A.; Keegan, L.P. Regulation and functions of ADAR in drosophila. Curr. Top. Microbiol. Immunol. 2012, 353, 221–236. [Google Scholar] [CrossRef]
- Buchumenski, I.; Holler, K.; Appelbaum, L.; Eisenberg, E.; Junker, J.P.; Levanon, E.Y. Systematic identification of A-to-I RNA editing in zebrafish development and adult organs. Nucleic Acids Res. 2021, 49, 4325–4337. [Google Scholar] [CrossRef] [PubMed]
- Licht, K.; Kapoor, U.; Mayrhofer, E.; Jantsch, M.F. Adenosine to Inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 2016, 44, 6398–6408. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, E.; Shaikh, S.A.; Berka, V.; Durham, R.J.; Litwin, D.B.; Lee, G.; MacLean, D.M.; Nowak, L.M.; Jayaraman, V. Mechanism of modulation of AMPA receptors by TARP-γ8. J. Gen. Physiol. 2020, 152, jgp.201912451. [Google Scholar] [CrossRef]
- Zhang, Y.; Kang, H.; Lee, Y.; Kim, Y.; Lee, B.; Kim, J.Y.; Jin, C.; Kim, S.; Kim, H.; Han, K. Smaller Body Size, Early Postnatal Lethality, and Cortical Extracellular Matrix-Related Gene Expression Changes of Cyfip2-Null Embryonic Mice. Front. Mol. Neurosci. 2018, 11, 482. [Google Scholar] [CrossRef] [PubMed]
- Levanon, E.Y.; Hallegger, M.; Kinar, Y.; Shemesh, R.; Djinovic-Carugo, K.; Rechavi, G.; Jantsch, M.F.; Eisenberg, E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005, 33, 1162–1168. [Google Scholar] [CrossRef]
- Sie, C.G.; Hesler, S.; Maas, S.; Kuchka, M. IGFBP7’s susceptibility to proteolysis is altered by A-to-I RNA editing of its transcript. FEBS Lett. 2012, 586, 2313–2317. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S.; et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef]
- Jain, M.; Mann, T.D.; Stulić, M.; Rao, S.P.; Kirsch, A.; Pullirsch, D.; Strobl, X.; Rath, C.; Reissig, L.; Moreth, K.; et al. RNA editing of Filamin A pre-mRNA regulates vascular contraction and diastolic blood pressure. EMBO J. 2018, 37, e94813. [Google Scholar] [CrossRef]
- Peng, X.; Xu, X.; Wang, Y.; Hawke, D.H.; Yu, S.; Han, L.; Zhou, Z.; Mojumdar, K.; Jeong, K.J.; Labrie, M.; et al. A-to-I RNA Editing Contributes to Proteomic Diversity in Cancer. Cancer Cell 2018, 33, 817–828.e7. [Google Scholar] [CrossRef]
- Wang, S.-Y.; Zhang, L.-J.; Chen, G.-J.; Ni, Q.-Q.; Huang, Y.; Zhang, D.; Han, F.-Y.; He, W.-F.; He, L.-L.; Ding, Y.-Q.; et al. COPA A-to-I RNA editing hijacks endoplasmic reticulum stress to promote metastasis in colorectal cancer. Cancer Lett. 2023, 553, 215995. [Google Scholar] [CrossRef]
- Song, Y.; An, O.; Ren, X.; Chan, T.H.M.; Tay, D.J.T.; Tang, S.J.; Han, J.; Hong, H.; Ng, V.H.E.; Ke, X.; et al. RNA editing mediates the functional switch of COPA in a novel mechanism of hepatocarcinogenesis. J. Hepatol. 2021, 74, 135–147. [Google Scholar] [CrossRef]
- Fu, Y.-F.; Liu, X.; Gao, M.; Zhang, Y.-N.; Liu, J. Endoplasmic reticulum stress induces autophagy and apoptosis while inhibiting proliferation and drug resistance in multiple myeloma through the PI3K/Akt/mTOR signaling pathway. Oncotarget 2017, 8, 61093–61106. [Google Scholar] [CrossRef]
- Rosenthal, J.J.C.; Bezanilla, F. Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron 2002, 34, 743–757. [Google Scholar] [CrossRef]
- Rieder, L.E.; Savva, Y.A.; Reyna, M.A.; Chang, Y.-J.; Dorsky, J.S.; Rezaei, A.; Reenan, R.A. Dynamic response of RNA editing to temperature in Drosophila. BMC Biol. 2015, 13, 1. [Google Scholar] [CrossRef]
- Fierro-Monti, I.; Fröhlich, K.; Schori, C.; Schmidt, A. Assessment of Data-Independent Acquisition Mass Spectrometry (DIA-MS) for the Identification of Single Amino Acid Variants. Proteomes 2024, 12, 33. [Google Scholar] [CrossRef]
- Wright, B.W.; Yi, Z.; Weissman, J.S.; Chen, J. The dark proteome: Translation from noncanonical open reading frames. Trends Cell Biol. 2022, 32, 243–258. [Google Scholar] [CrossRef]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.-L.; et al. The translational landscape of the human heart. Cell 2019, 178, 242–260.e29. [Google Scholar] [CrossRef]
- Chen, J.; Brunner, A.-D.; Cogan, J.Z.; Nuñez, J.K.; Fields, A.P.; Adamson, B.; Itzhak, D.N.; Li, J.Y.; Mann, M.; Leonetti, M.D.; et al. Pervasive functional translation of noncanonical human open reading frames. Science 2020, 367, 1140–1146. [Google Scholar] [CrossRef]
- Piazzi, M.; Bavelloni, A.; Salucci, S.; Faenza, I.; Blalock, W.L. Alternative splicing, RNA editing, and the current limits of next generation sequencing. Genes 2023, 14, 1386. [Google Scholar] [CrossRef]

| Codon | Amino Acid | Edited Codons | Amino Acid After Recoding | Possible Amino Acid Substitutions |
|---|---|---|---|---|
| UAU UAC | Tyrosine | UGU UGC | Cysteine | Y>C |
| AAU AAC | Asparagine | AGU AGC | Serine | N>S |
| AAU AAC | Asparagine | GAU GAC | Aspartic acid | N>D * |
| CAA CAG | Glutamine | CGA, CGG CGG | Arginine | Q>R |
| AAU AAC | Asparagine | GGU GGC | Glycine | N>G |
| AGU AGC | Serine | GGU GGC | Glycine | S>G |
| ACC ACA ACG ACU | Threonine | GCC GCA, GCG GCG GCU | Alanine | T>A |
| CAU CAC | Histidine | CGU CGC | Arginine | H>R |
| AAA AAG | Lysine | GAA, GAG GAG | Glutamic acid | K>E ** |
| AAA AAG | Lysine | AGA, AGG AGG | Arginine | K>R |
| AAA AAG | Lysine | GGA, GGG GGG | Glycine | K>G |
| AGA AGG | Arginine | GGA, GGG GGG | Glycine | R>G |
| GAU GAC | Aspartic acid | GGU GGC | Glycine | D>G |
| GAA GAG | Glutamic acid | GGA, GGG GGG | Glycine | E>G |
| AUU AUC AUA | Isoleucine | GUU GUC GUA, GUG | Valine | I>V |
| AUA | Isoleucine | AUG | Methionine | I>M |
| AUG | Methionine | GUG | Valine | M>V |
| UAA UAG UGA | Stop codon | UAG, UGG, UGA UGG UGG | Tryptophan | STOP>W |
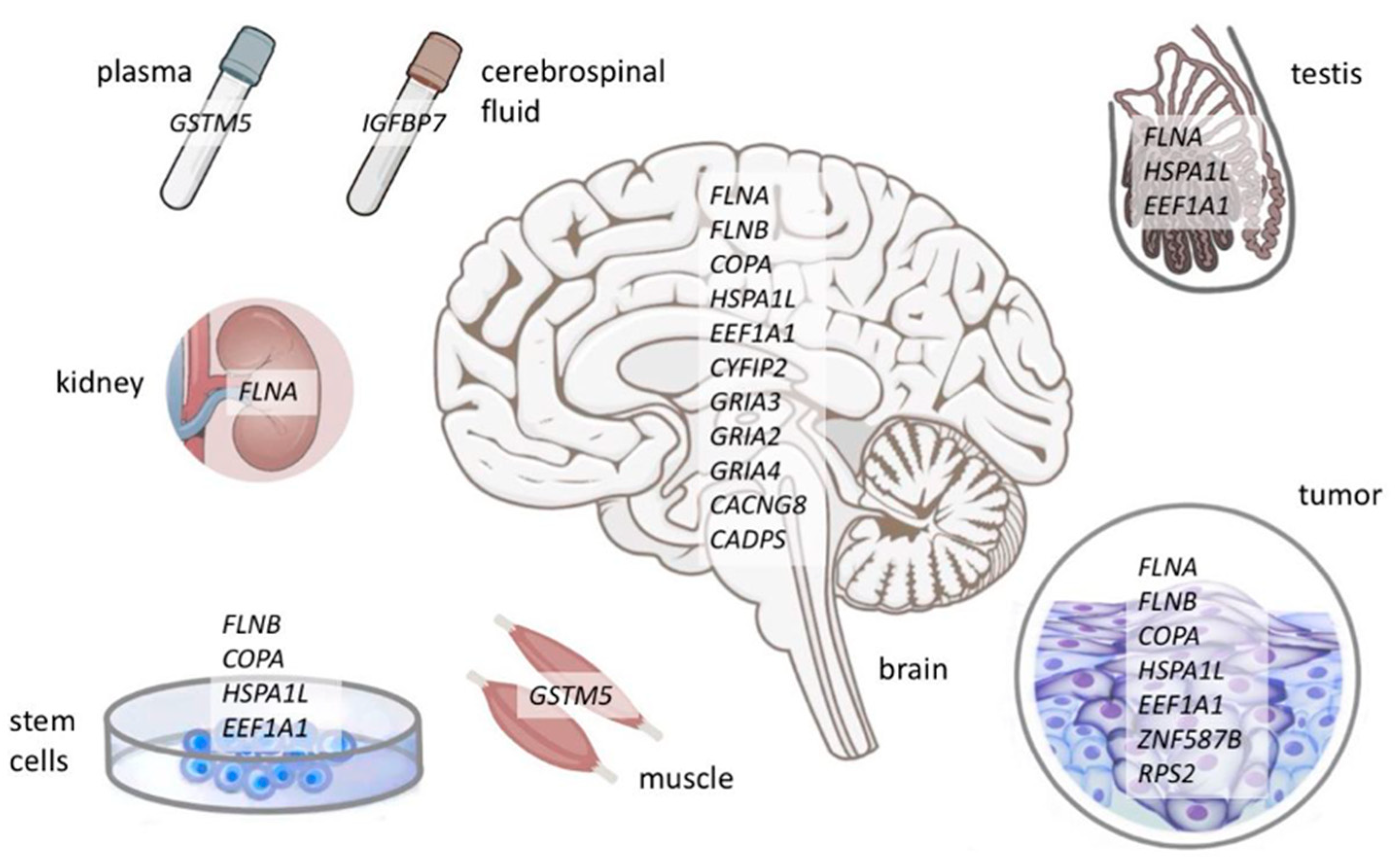
| Recoded Tryptic Peptide | Protein | Gene | Tissues | Found in Mouse Brain Samples | Identified in [89]: Transcriptome/Proteome |
|---|---|---|---|---|---|
| tissue-nonspecific recoding events | |||||
| LTVSSL[Q>R] | filamin A | FLNA | brain, testis, kidney, pancreas ductal adenocarcinoma cell line, triple-negative breast cancer, B-cell lymphoma | ✓ | ✓/✓ |
| LTV[M>V]SLQESGLK | filamin B | FLNB | brain, ovarian cancer cells, meningioma, mesenchymal stem cells | ✗ | ✓/✓ |
| VWD[I>V]SGLR | coatomer subunit alpha | COPA | brain, glioma-derived stem cells, ovarian cancer cells, triple-negative breast cancer, mesenchymal stem cells, meningioma | ✓ | ✓/✗ |
| QTQIF[T>A]TYSDNQPGVLIQVYEGER | heat shock 70 kDa protein 1-like | HSPA1L | brain, B-cell lymphoma, testis, glioma-derived stem cells, ovarian cancer cells, | ✗ | ✗/✗ |
| QTQIFT[T>A]YSDNQPGVLIQVYEGER | brain, B-cell lymphoma, mesenchymal stem cells, testis | ✗ | ✗/✗ | ||
| Y[Y>C]VTIIDAPGHR | eukaryotic translation elongation factor 1 alpha 1 | EEF1A1 | brain, glioma-derived stem cells, pancreas ductal adenocarcinoma cell line, ovarian cancer cells | ✗ | ✗/✗ |
| NMI[T>A]GTSQADCAVLIVAAGVGEFEAGISK | glioma-derived stem cells, mesenchymal stem cells, testis | ✗ | ✗/✗ | ||
| TSLGN[I>M]VK | zinc finger protein 587B | ZNF587B | glioma-derived stem cells, brain, ovarian cancer cells | ✗ | ✗/✗ |
| TYS[Y>C]LTPDLWK | 40S ribosomal protein S2 | RPS2 | ovarian cancer cells, meningioma | ✗ | ✗/✗ |
| HNLCGETEEE[K>R] | glutathione S-transferase Mu 5 | GSTM5 | muscle, plasma | ✗ | ✓/✗ |
| neural-specific recoding events | |||||
| YI[K>E]TSAHYEENK | cytoplasmic FMR1-interacting protein 2 | CYFIP2 | brain | another peptide of this protein was found | ✓/✓ |
| GSAL[R>G]NAVNLAVLK | glutamate receptor 3 (GluR-3), flop isoform | GRIA3 | brain | ✓ | ✓/✓ |
| GSAL[R>G]TPVNLAVLK | glutamate receptor 3 (GluR-3), flip isoform | brain | ✓ | ✓/✓ | |
| GSSL[R>G]NAVNLAVLK | glutamate receptors 2 and 4 (GluR2, GluR4), flop isoforms | GRIA2, GRIA4 | brain | ✗ | ✓/✗ |
| GSSL[R>G]TPVNLAVLK | glutamate receptors 2 and 4 (GluR2, GluR4), flop isoforms | brain | ✓ | ✓/✗ | |
| AGGGAGG[S>G]GGSGPSAILR | voltage-dependent calcium channel gamma-8 subunit | CACNG8 | brain | ✗ | ✓/✗ |
| AGGGRPS[S>G]PSPSVVSEK | calcium-dependent secretion activator 1 | CADPS | brain | another peptide of this protein was found | ✓/✗ |
| GEGEPCGGGGAG[R>G]GYCAPGMECVK | insulin-like growth factor-binding protein 7 | IGFBP7 | cerebrospinal fluid | ✓ | ✗/✗ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudriavskii, V.V.; Kliuchnikova, A.A.; Goncharov, A.O.; Ilgisonis, E.V.; Moshkovskii, S.A. Unraveling ADAR-Mediated Protein Recoding: A Proteogenomic Exploration in Model Organisms and Human Pathology. Int. J. Mol. Sci. 2025, 26, 6837. https://doi.org/10.3390/ijms26146837
Kudriavskii VV, Kliuchnikova AA, Goncharov AO, Ilgisonis EV, Moshkovskii SA. Unraveling ADAR-Mediated Protein Recoding: A Proteogenomic Exploration in Model Organisms and Human Pathology. International Journal of Molecular Sciences. 2025; 26(14):6837. https://doi.org/10.3390/ijms26146837
Chicago/Turabian StyleKudriavskii, Viacheslav V., Anna A. Kliuchnikova, Anton O. Goncharov, Ekaterina V. Ilgisonis, and Sergei A. Moshkovskii. 2025. "Unraveling ADAR-Mediated Protein Recoding: A Proteogenomic Exploration in Model Organisms and Human Pathology" International Journal of Molecular Sciences 26, no. 14: 6837. https://doi.org/10.3390/ijms26146837
APA StyleKudriavskii, V. V., Kliuchnikova, A. A., Goncharov, A. O., Ilgisonis, E. V., & Moshkovskii, S. A. (2025). Unraveling ADAR-Mediated Protein Recoding: A Proteogenomic Exploration in Model Organisms and Human Pathology. International Journal of Molecular Sciences, 26(14), 6837. https://doi.org/10.3390/ijms26146837






