In Silico Evaluation of Quinolone–Triazole and Conazole–Triazole Hybrids as Promising Antimicrobial and Anticancer Agents
Abstract
1. Introduction
2. Results and Discussion
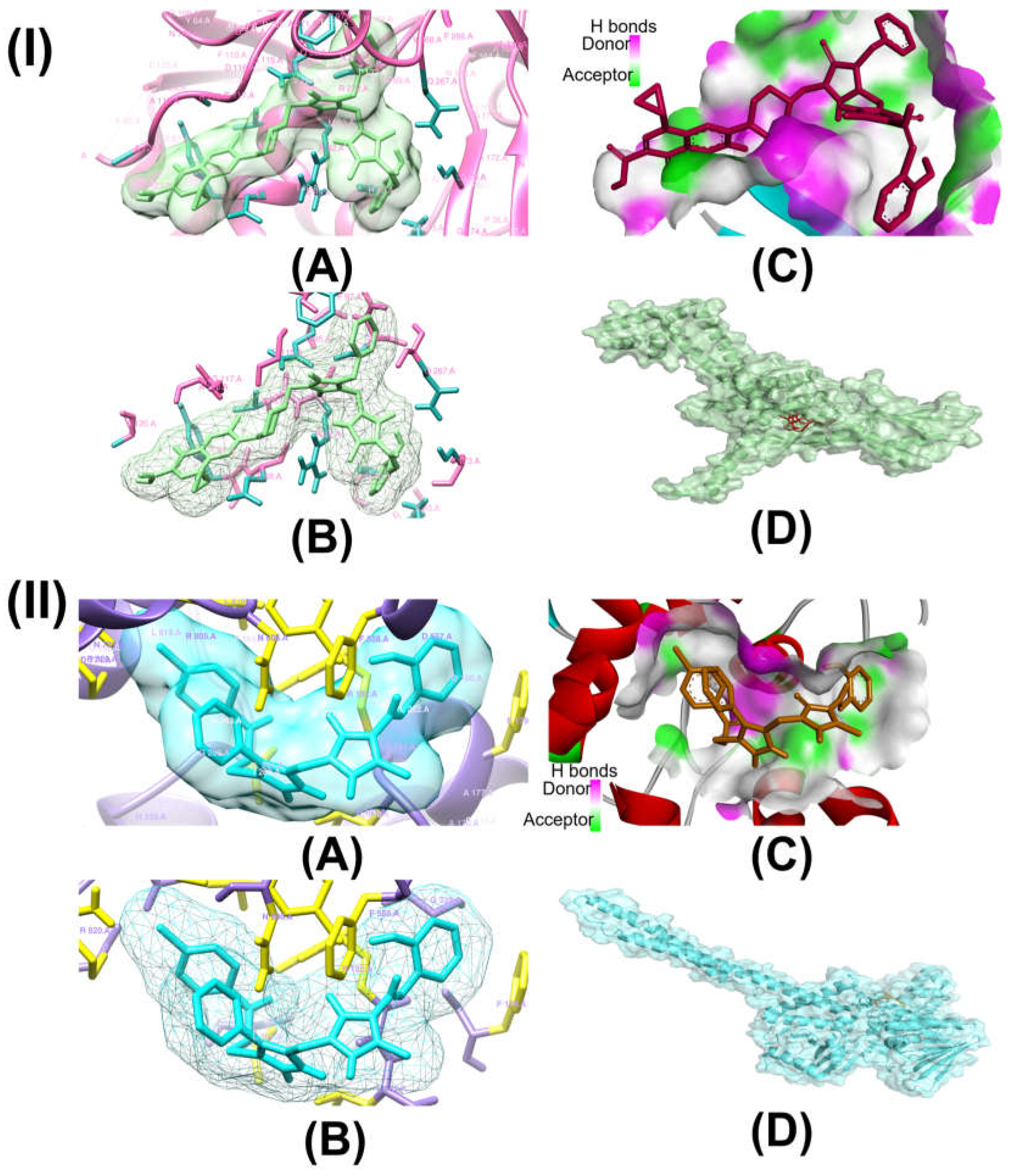
2.1. In Silico Molecular Docking
2.2. Physicochemical and Pharmacokinetic Characteristics Analysis
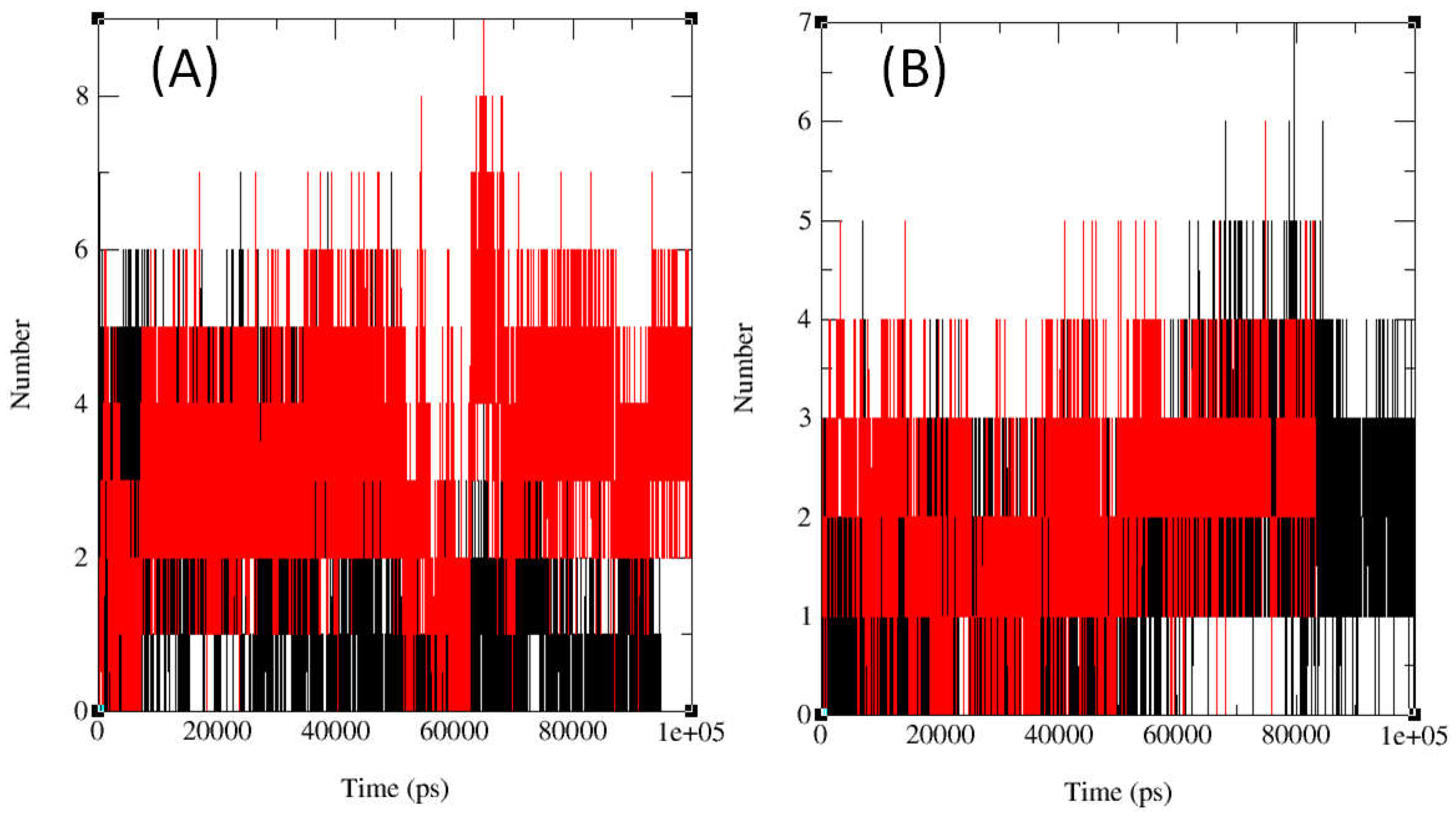
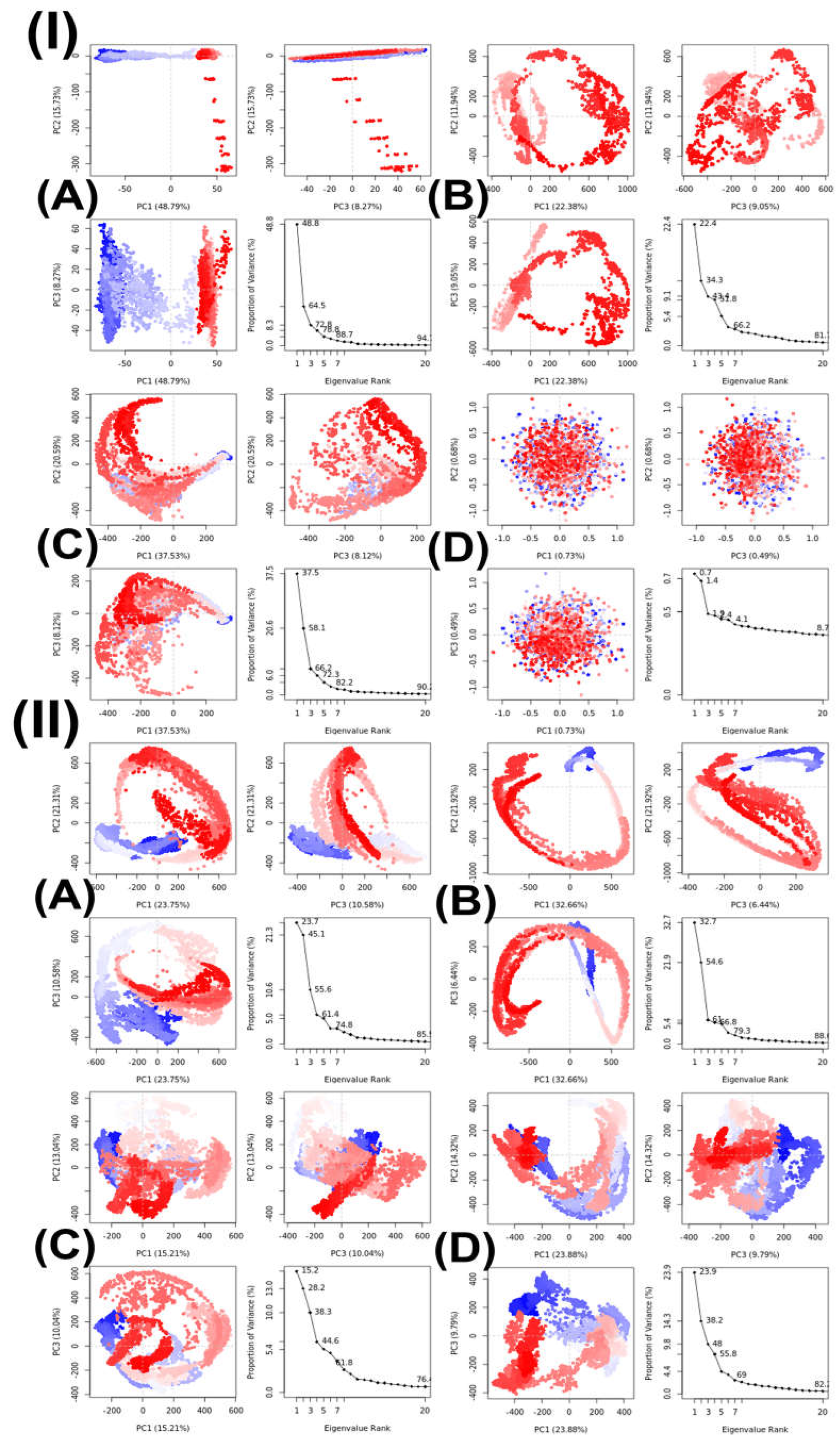
2.3. In Silico Molecular Dynamics (MD)
3. Materials and Methods
3.1. Computational Analysis
3.2. Evaluation of Physicochemical and Pharmacokinetic Properties
3.3. Pharmacological Attributes
3.4. Molecular Docking
3.4.1. Compound Preparation
3.4.2. Target Protein Preparation
3.4.3. Protein and Compound Docking
3.5. Molecular Dynamics Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Kakkar, S.; Kumar, S.; Lim, S.M.; Ramasamy, K.; Mani, V.; Shah, S.A.A.; Narasimhan, B. Design, synthesis and biological evaluation of 3-(2-aminooxazol-5-yl)-2H-chromen-2-one derivatives. BMC Chem. 2018, 12, 130. [Google Scholar] [CrossRef]
- Ma, X.; Yu, H. Cancer issue: Global burden of cancer. Yale J. Biol. Med. 2006, 79, 85–94. [Google Scholar]
- Nesaragi, A.R.; Kamble, R.R.; Bayannavar, P.K.; Shaikh, S.K.J.; Hoolageri, S.R.; Kodasi, B.; Joshi, S.D.; Kumbar, V.M. Microwave assisted regioselective synthesis of quinoline appended triazoles as potent anti-tubercular and antifungal agents via copper catalyzed cycloaddition. Bioorgan. Med. Chem. Lett. 2021, 41, 127984. [Google Scholar] [CrossRef] [PubMed]
- Ueda, S.; Nagasawa, H. Facile synthesis of 1, 2, 4-triazoles via a copper-catalyzed tandem addition oxidative cyclization. J. Am. Chem. Soc. 2009, 131, 15080–15081. [Google Scholar] [CrossRef] [PubMed]
- Kaplancikli, Z.A.; Turan-Zitouni, G.; Ozdemir, A.; Revial, G. New triazole and triazolothiadiazine derivatives as possible antimicrobial agents. Eur. J. Med. Chem. 2008, 43, 155–159. [Google Scholar] [CrossRef]
- Nesaragi, A.R.; Kamble, R.R.; Bayannavar, P.K.; Metre, T.V.; Kariduraganavar, M.Y.; Margankop, S.B.; Joshi, S.D.; Kumbar, V.M. Microwave facilitated one-pot three component triazoles: Antimicrobial evaluation, molecular docking and in silico ADME studie. Synth. Commun. 2021, 51, 3460–3472. [Google Scholar] [CrossRef]
- Kavaklı, C.; Kavaklı, P.A.; Güven, O. Preparation and characterization of glycidyl methacrylate grafted 4-amino-1, 2, 4-triazole modified nonwoven fiber adsorbent for environmental application. Radiat. Phys. Chem. 2014, 94, 111–114. [Google Scholar] [CrossRef]
- Zhou, C.H.; Wang, Y. Recent researches in triazole compounds as medicinal drugs. Curr. Med. Chem. 2012, 19, 239–280. [Google Scholar] [CrossRef]
- Fang, B.; Zhou, C.H.; Rao, X.C. Synthesis and biological activities of novel amine-derived bis-azoles as potential antibacterial and antifungal agents. Eur. J. Med. Chem. 2010, 45, 4388–4398. [Google Scholar] [CrossRef]
- Nesaragi, A.R.; Algethami, J.S.; Alsaiari, M.; Alsareii, S.A.; Mathada, B.S.; Ningaiah, S.; Sasidhar, B.S.; Harraz, F.A.; Patil, S.A. A comprehensive overview of coumarinyl-triazole hybrids as anticancer agents. J. Mol. Struc. 2024, 1302, 137478. [Google Scholar] [CrossRef]
- Saeed, A.; Shaheen, U.; Hameed, A.; Kazmi, F. Synthesis and antimicrobial activity of some novel 2-(substituted fluorobenzoylimino)-3-(substituted fluorophenyl)-4- methyl-1,3-thiazolines. J. Fluor. Chem. 2010, 131, 333–339. [Google Scholar] [CrossRef]
- Lemilemu, F.; Bitew, M.; Demissie, T.B.; Eswaramoorthy, R.; Endale, M. Synthesis, antibacterial and antioxidant activities of Thiazole-based Schiff base derivatives: A combined experimental and computational study. BMC Chem. 2021, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Yurttaş, L.; Özkay, Y.; Karaca, H.; Çevik, U.A. Synthesis of some new thiazole derivatives and their biological activity evaluation. J. Chem. 2015. [Google Scholar] [CrossRef]
- Morsy, M.A.; Ali, E.M.; Kandeel, M.; Venugopala, K.N.; Nair, A.B.; Greish, K.; El-Daly, M. Screening and molecular docking of novel benzothiazole derivatives as potential antimicrobial agents. Antibiotics 2020, 9, 221. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Gahlyan, P.; Verma, A.; Jain, R.; Das, S.; Konwar, R.; Prasad, A.K. Design and synthesis of fluorescent symmetric bis-triazolylated-1,4-dihydropyridines as potent antibreast cancer agents. Synth. Commun. 2018, 48, 778–785. [Google Scholar] [CrossRef]
- Kumbhare, R.M.; Kosurkar, U.B.; Bagul, P.K.; Kanwal, A.; Appalanaidu, K.; Dadmal, T.L.; Banerjee, S.K. Synthesis and evaluation of novel triazoles and mannich bases functionalized 1,4-dihydropyridine as angiotensin converting enzyme (ACE) inhibitors. Bioorgan. Med. Chem. 2014, 22, 5824–5830. [Google Scholar] [CrossRef]
- Singh, H.; Sindhu, J.; Khurana, J.M.; Sharma, C.; Aneja, K.R. A facile eco-friendly one-pot five-component synthesis of novel 1,2,3-triazole-linked pentasubstituted 1,4- dihydropyridines and their biological and photophysical studies. Aust. J. Chem. 2013, 66, 1088–1096. [Google Scholar] [CrossRef]
- Vijesh, A.M.; Isloor, A.M.; Peethambar, S.K.; Shivananda, K.N.; Arulmoli, T.; Isloor, N.A. Hantzsch reaction: Synthesis and characterization of some new 1,4- dihydropyridine derivatives as potent antimicrobial and antioxidant agents. Eur. J. Med. Chem. 2011, 46, 5591–5597. [Google Scholar] [CrossRef]
- Khan, M.A.; Kola, V.B.; Noor, B.; Acco, J. The antioxidant activity of dihydropyridine derivatives. Curr. Res. Bioorgan. Org. Chem. 2020, 3, 124. [Google Scholar]
- da Costa Cabrera, D.; Santa-Helena, E.; Leal, H.P.; de Moura, R.R.; Nery, L.E.M.; Goncalves, C.A.N.; Russowsky, D.; D’Oca, M.G.M. Synthesis and antioxidant activity of new lipophilic dihydropyridines. Bioorgan. Chem. 2019, 84, 1–16. [Google Scholar] [CrossRef]
- Raju, R.; Rajasekar, S.; Raghunathan, R.; Arumugam, N.; Almansour, A.I.; Kumar, R.S. Regioselective synthesis and antioxidant activity of a novel class of mono and C2- symmetric bis-1,2,3-triazole and acridinedione grafted macromolecules. J. Saudi Chem. Soc. 2020, 24, 934–941. [Google Scholar] [CrossRef]
- Hadjipavlou-Litina, D.; Głowacka, I.E.; Marco-Contelles, J.; Piotrowska, D.G. Synthesis and antioxidant properties of novel 1,2,3-triazole-containing nitrones. Antioxidants 2023, 12, 36. [Google Scholar] [CrossRef]
- Khare, S.P.; Deshmukh, T.R.; Sangshetti, J.N.; Krishna, V.S.; Sriram, D.; Khedkar, V.M.; Shingate, B.B. Design, synthesis and molecular docking studies of novel triazole-chromene conjugates as antitubercular, antioxidant and antifungal agents. ChemistrySelect 2018, 3, 13113–13122. [Google Scholar] [CrossRef]
- Khare, S.P.; Deshmukh, T.R.; Akolkar, S.V.; Sangshetti, J.N.; Khedkar, V.M.; Shingate, B.B. New 1,2,3-triazole-linked tetrahydrobenzo[b]pyran derivatives: Facile synthesis, biological evaluation and molecular docking study. Res. Chem. Intermed. 2019, 45, 5159–5182. [Google Scholar] [CrossRef]
- Khare, S.P.; Deshmukh, T.R.; Sangshetti, J.N.; Khedkar, V.M.; Shingate, B.B. Ultrasound assisted rapid synthesis, biological evaluation, and molecular docking study of new 1,2,3-triazolyl pyrano[2,3-c]pyrazoles as antifungal and antioxidant agent. Synth. Commun. 2019, 49, 2521–2537. [Google Scholar] [CrossRef]
- Danne, A.B.; Lathi, K.V.; Sangshetti, J.N.; Khedkar, V.M.; Khalse, L.D.; Shingate, B.B. New 1,2,3-Triazole Tethered-1,4-Dihydropyridines as Potential Antioxidant Agents: Synthesis and Molecular Docking Study. J. Mol. Struct. 2023, 1299, 137129. [Google Scholar] [CrossRef]
- Chen, G.L.; Guo, L.; Yang, S.; Ji, D.M. Cancer risk in tuberculosis patients in a high endemic area. BMC Cancer 2021, 21, 679. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.J.; Yang, H.Y.; Chung, C.H.; Chang, W.C.; Yang, S.S.; Sun, C.A.; Chien, W.C.; Su, R.Y.; Subbiah, S.K. Increased risk of secondary lung cancer in patients with tuberculosis: A nationwide, population-based cohort study. PLoS ONE 2021, 16, e0250531. [Google Scholar] [CrossRef]
- Luczynski, P.; Poulin, P.; Romanowski, K.; Johnston, J.C.; Duell, E.J. Tuberculosis and risk of cancer: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0278661. [Google Scholar] [CrossRef]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef]
- Saghazadeh, A.; Rezaei, N.; Rottenberg, M.E. Vascular endothelial growth factor levels in tuberculosis: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0268543. [Google Scholar] [CrossRef]
- Hoolageri, S.R.; Kamble, R.R.; Nesaragi, A.R.; Bheemayya, L.; Nadoni, V.B.; Dixit, S.; Vootla, S.; Joshi, S.D. Cu (I) catalyzed A3 cascade coupling via C-H functionalization followed by cyclization: Synthesis, in silico, in vitro, and toxicity studies of imidazo[2,1-b]thiazoles. Appl. Organomet. Chem. 2022, 36, e6801. [Google Scholar] [CrossRef]
- Sashidhara, K.V.; Kumar, A.; Chatterjee, M.; Rao, K.B.; Singh, S.; Verma, A.K.; Palit, G. Discovery and synthesis of novel 3-phenylcoumarin derivatives as antidepressant agents. Bioorgan. Med. Chem. Lett. 2011, 21, 1937–1941. [Google Scholar] [CrossRef]
- Nargotra, A.; Sharma, S.; Alam, M.I.; Ahmed, Z.; Bhagat, A.; Taneja, S.C.; Qazi, G.N.; Koul, S. In silico identification of viper phospholipaseA2 inhibitors: Validation by in vitro, in vivo studies. J. Mol. Model. 2011, 17, 3063–3073. [Google Scholar] [CrossRef] [PubMed]
- Alipour, M.; Khoobi, M.; Moradi, A.; Nadri, H.; Moghadam, F.H.; Emami, S.; Hasanpour, Z.; Foroumadi, A.; Shafiee, A. Synthesis and anti-cholinesterase activity of new 7-hydroxycoumarin derivatives. Eur. J. Med. Chem. 2014, 82, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.K.; Singh, J.; Ara, T.; Koul, S.; Farooq, S.; Kaul, A. Synthesis and biological evaluation of novel isoxazoles and triazoles linked 6-hydroxycoumarin as potent cytotoxic agents. Bioorgan. Med. Chem. Lett. 2014, 24, 4243–4246. [Google Scholar]
- Thigulla, Y.; Kumar, T.U.; Trivedi, P.; Ghosh, B.; Bhattacharya, A. One-step synthesis of fused chromeno[4,3-b]pyrrolo[3,2-h]quinolin-7(1H)-one compounds and their anticancer activity evaluation. ChemistrySelect 2017, 2, 2718–2721. [Google Scholar] [CrossRef]
- Peyressatre, M.; Prével, C.; Pellerano, M.; Morris, M. Targeting cyclin-dependent kinases in human cancers: From small molecules to Peptide inhibitors. Cancers 2015, 7, 179–237. [Google Scholar] [CrossRef]
- Smalley, K.S.M.; Contractor, R.; Nguyen, T.K.; Xiao, M.; Edwards, R.; Muthusamy, V.; King, A.J.; Flaherty, K.T.; Bosenberg, M.; Herlyn, M.; et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res. 2008, 68, 5743–5752. [Google Scholar] [CrossRef]
- Dobashi, Y.; Goto, A.; Fukayama, M.; Abe, A.; Ooi, A. Overexpression of cdk4/cyclin D1, a possible mediator of apoptosis and an indicator of prognosis in human primary lung carcinoma. Int. J. Cancer 2004, 110, 532–541. [Google Scholar] [CrossRef]
- Graf, F.; Mosch, B.; Koehler, L.; Bergmann, R.; Wuest, F.; Pietzsch, J. Cyclin-dependent kinase 4/6 (cdk4/6) inhibitors: Perspectives in cancer therapy and imaging. Mini Rev. Med. Chem. 2010, 10, 527–539. [Google Scholar] [CrossRef]
- Ribnikar, D.; Volovat, S.R.; Cardoso, F. Targeting CDK4/6 pathways and beyond in breast cancer. Breast 2019, 43, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Farghaly, T.A.; Pashameah, R.A.; Bayazeed, A.; Al-Soliemy, A.M.; Alsaedi, A.M.R.; Harras, M.F. Design and Synthesis of New bis-oxindole and Spiro(triazole-oxindole) as CDK4 Inhibitors with Potent Anti-breast Cancer Activity. Med. Chem. 2024, 20, 63–77. [Google Scholar] [CrossRef]
- Chamduang, C.; Pingaew, R.; Prachayasittikul, V.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Novel triazole-tetrahydroisoquinoline hybrids as human aromatase inhibitors. Bioorgan. Chem. 2019, 93, 103327. [Google Scholar] [CrossRef]
- Doiron, J.; Richard, R.; Touré, M.M.; Picot, N.; Richard, R.; Čuperlović-Culf, M.; Robichaud, G.A.; Touaibia, M. Synthesis and structure–activity relationship of 1-and 2-substituted-1, 2, 3-triazole letrozole-based analogues as aromatase inhibitors. Eur. J. Med. Chem. 2011, 46, 4010–4024. [Google Scholar] [CrossRef] [PubMed]
- Henneberta, O.; Montes, M.; Favre-Reguillon, A.; Chermetted, H.; Ferroudc, C.; Mortin, R. Epimerase activity of the human 11β-hydroxysteroid dehydrogenase type 1 on 7-hydroxylated C19-steroids. J. Steroid Biochem. Mol. Biol. 2009, 114, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Leechaisit, R.; Pingaew, R.; Prachayasittikul, V.; Worachartcheewan, A.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Synthesis, molecular docking, and QSAR study of bis-sulfonamide derivatives as potential aromatase inhibitors. Bioorgan. Med. Chem. 2019, 27, 115040. [Google Scholar] [CrossRef]
- Brueggemeier, R.W.; Hackett, J.C.; Diaz-Cruz, E.S. Aromatase inhibitors in the treatment of breast cancer. Endocr. Rev. 2005, 26, 331–345. [Google Scholar] [CrossRef]
- Cepa, M.M.; da Silva, E.J.T.; Correia-da-Silva, G.; Roleira, F.M.; Teixeira, N.A. Synthesis and biochemical studies of 17-substituted androst-3-enes and 3,4-epoxyandrostanes as aromatase inhibitors. Steroids 2008, 73, 1409–1415. [Google Scholar] [CrossRef]
- Çevik, U.A.; Sağlık, B.N.; Osmaniye, D.; Levent, S.; Çavuşoğlu, B.K.; Karaduman, A.B.; Ozkay, Y.; Kaplancıklı, Z.A. Synthesis and docking study of benzimidazole–triazolothiadiazine hybrids as aromatase inhibitors. Arch. Pharm. 2020, 353, e2000008. [Google Scholar] [CrossRef]
- Çevik, U.A.; Çavuşoğlu, B.K.; Sağlık, B.N.; Osmaniye, D.; Levent, S.; Ilgın, S.; Özkay, Y.; Kaplancıklı, Z.A. Synthesis, Docking Studies and Biological Activity of New Benzimidazole- Triazolothiadiazine Derivatives as Aromatase Inhibitor. Molecules 2020, 25, 1642. [Google Scholar] [CrossRef]
- Sahoo, R.; Babu, V.C.; Okaly, G.V.P.; Rao, S.; Nargund, A.; Venkataswamy, E.; Rao, R.; Kumar, B.S. Screening for EGFR mutations in lung cancer, a report from India. Lung Cancer 2011, 73, 316–319. [Google Scholar] [CrossRef]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Gariganti, N.; Loke, S.K.; Pagadala, E.; Chinta, P.; Poola, B.; Chetti, P.; Bansal, A.; Ramachandran, B.; Srinivasadesikan, V.; Kottalanka, R.K. Design, synthesis, anticancer activity of new amide derivatives derived from 1,2,3-triazole-benzofuran hybrids: An insights from molecular docking, molecular dynamics simulation and DFT studies. J. Mol. Struct. 2023, 1273, 134250. [Google Scholar] [CrossRef]
- Liang, T.; Sun, X.; Li, W.; Hou, G.; Gao, F. 1,2,3-triazole-containing compounds as anti-lung cancer agents: Current developments, mechanisms of action, and structure-activity relationship. Front. Pharmacol. 2021, 12, 1374. [Google Scholar] [CrossRef]
- Othman, E.M.; Fayed, E.A.; Husseiny, E.M.; Abulkhair, H.S. Rationale design, synthesis, cytotoxicity evaluation, and in silico mechanistic studies of novel 1,2,3-triazoles with potential anticancer activity. New J. Chem. 2022, 46, 12206–12216. [Google Scholar] [CrossRef]
- Özdemir, A.; Sever, B.; Altıntop, M.D.; Temel, H.E.; Atlı, Ö.; Baysal, M.; Demirci, F. Synthesis and Evaluation of New Oxadiazole, Thiadiazole, and Triazole Derivatives as Potential Anticancer Agents Targeting MMP-9. Molecules 2017, 22, 1109. [Google Scholar] [CrossRef] [PubMed]
- Hamdy, R.; Jones, A.T.; El-Sadek, M.; Hamoda, A.M.; Shakartalla, S.B.; ALShareef, Z.M.; Soliman, S.S.M.; Westwell, A.D. New Bioactive Fused Triazolothiadiazoles as Bcl-2-Targeted Anticancer Agents. Int. J. Mol. Sci. 2021, 22, 12272. [Google Scholar] [CrossRef]
- Zheng, Y.C.; Duan, Y.C.; Ma, J.L.; Xu, R.M.; Zi, X.; Lv, W.L.; Wang, M.M.; Ye, X.W.; Zhu, S.; Mobley, D.; et al. Triazole-dithiocarbamate based selective lysine specific demethylase 1 (LSD1) inactivators inhibit gastric cancer cell growth, invasion, and migration. J. Med. Chem. 2013, 56, 8543–8560. [Google Scholar] [CrossRef]
- Mou, Z.; Gao, J.; Miao, H.; Zhang, L.; Su, L.; Wang, B.; Luan, Y. Design and synthesis of novel histone deacetylase 6 inhibitors with benzyl-triazole as the core skeleton. Biosci. Trends 2019, 13, 267–272. [Google Scholar] [CrossRef]
- Esha, N.J.I.; Quayum, S.T.; Saif, M.Z.; Almatarneh, M.H.; Rahman, S.; Alodhayb, A.; Poirier, R.A.; Uddin, K.M. Exploring the potential of fluoro-flavonoid derivatives as anti-lung cancer agents: DFT, molecular docking, and molecular dynamics techniques. Int. J. Quantum Chem. 2024, 124, E27274. [Google Scholar] [CrossRef]
- Cebeci, Y.U.; Ceylan, S.; Karaoglu, S.A.; Altun, M. An efficient microwave-assisted synthesis of novel quinolone–triazole and conazole–triazole hybrid derivatives as antimicrobial and anticancer agents. J. Heterocycl. Chem. 2023, 60, 47. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Khan, T.; Dixit, S.; Ahmad, R.; Raza, S.; Azad, I.; Joshi, S.; Khan, A.R. Molecular docking, PASS analysis, bioactivity score prediction, synthesis, characterization and biological activity evaluation of a functionalized 2-butanone thiosemicarbazone ligand and its complexes. J. Chem. Biol. 2017, 10, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Fukunishi, Y.; Nakamura, H. Definition of Drug-Likeness for Compound Affinity. J. Chem. Inf. Model. 2011, 51, 1012–1016. [Google Scholar] [CrossRef]
- Gurung, A.B.; Bhattacharjee, A.; Ali, M.A. Exploring the Physicochemical Profile and the Binding Patterns of Selected Novel Anticancer Himalayan Plant Derived Active Compounds with Macromolecular Targets. Inform. Med. Unlocked 2016, 5, 1–14. [Google Scholar] [CrossRef]
- Shadrack, D.M.; Ndesendo, V.M.K. Molecular Docking and ADMET Study of Emodin Derivatives as Anticancer Inhibitors of NAT2, COX2 and TOP1 Enzymes. Comput. Mol. Biosci. 2017, 7, 1–18. [Google Scholar] [CrossRef]
- Ozkurt, T.E.; Akgul, T.; Baykut, S. Principal Component Analysis of the Fractional Brownian Motion For 0 < H < 0.5. In Proceedings of the 2006 IEEE International Conference on Acoustics Speech and Signal Processing Proceedings, Toulouse, France, 14–19 May 2006; p. III. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Uddin, K.M.; Alrawashdeh, A.I.; Henry, D.J.; Warburton, P.L.; Poirier, R.A. Hydrolytic Deamination Reactions of Amidine and Nucleobase Derivatives. Int. J. Quantum Chem. 2019, 120, e26059. [Google Scholar] [CrossRef]
- Uddin, K.M.; Henry, D.J.; Alrawashdeh, A.I.; Warburton, P.L.; Poirier, R.A. Mechanism for the deamination of ammeline, guanine, and their analogues. Struct. Chem. 2017, 28, 146. [Google Scholar] [CrossRef]
- Uddin, K.M.; Almatarneh, M.H.; Shaw, D.M.; Poirier, R.A. Mechanistic study of the deamination reaction of guanine: A computational study. J. Phys. Chem. A 2011, 115, 2065–2076. [Google Scholar] [CrossRef] [PubMed]
- Uddin, K.M.; Poirier, R.A. Computational study of the deamination of 8-oxoguanine. J. Phys. Chem. B 2011, 115, 9151–9159. [Google Scholar] [CrossRef] [PubMed]
- Uddin, K.M.; Flinn, C.G.; Poirier, R.A.; Warburton, P.L. Comparative computational investigation of the reaction mechanism for the hydrolytic deamination of cytosine, cytosine butane dimer and 5, 6-saturated cytosine analogues. Comput. Theor. Chem. 2014, 1027, 91–102. [Google Scholar] [CrossRef]
- Alrawashdeh, A.I.; Almatarneh, M.H.; Poirier, R.A. Computational study on the deamination reaction of adenine with OH−/nH2O (n = 0, 1, 2, 3) and 3H2O. Can. J. Chem. 2013, 91, 518–526. [Google Scholar] [CrossRef]
- Almatarneh, M.H.; Flinn, C.G.; Poirier, R.A.; Sokalski, W.A. Computational Study of the Deamination Reaction of Cytosine with H2O and OH−. J. Phys. Chem. A 2006, 110, 8227–8234. [Google Scholar] [CrossRef]
- Uddin, K.M.; Hosen, M.A.; Khan, M.F.; Ozeki, Y.; Kawsar, S.M.A. Investigation of Structural, Physicochemical, Pharmacokinetics, PASS Prediction, and Molecular Docking Analysis of Methyl 6-O-Myristoyl-α-D-Glucopyranoside Derivatives against SARS-CoV-2. Philipp. J. Sci. 2022, 151, 2215–2231. [Google Scholar] [CrossRef]
- Chamizo, J.A.; Morgado, J.; Sosa, P. Organometallic Aromaticity. Organometallics 1993, 12, 5005–5007. [Google Scholar] [CrossRef]
- Glasstone, S.; Laidler, K.J.; Eyring, H. The Theory of Rate Processes: The Kinetics of Chemical Reactions, Viscosity, Diffusion and Electrochemical Phenomena; McGraw-Hill Book Company, Incorporated: New York, NY, USA, 1941. [Google Scholar]
- Alberty, R.A. The Foundations of Chemical Kinetics. J. Chem. Educ. 1960, 37, 660. [Google Scholar] [CrossRef]
- Parr, R.G.; von Szentpály, L.; Liu, S. Electrophilicity Index. J. Am. Chem. Soc. 1999, 121, 1922–1924. [Google Scholar] [CrossRef]
- Pal, R.; Chattaraj, P.K. Electrophilicity Index Revisited. J. Comput. Chem. 2023, 44, 278–297. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. AdmetSAR 2.0: Web-Service for Prediction and Optimization of Chemical Admet Properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Druzhilovskiy, D.S.; Rudik, A.V.; Filimonov, D.A.; Gloriozova, T.A.; Lagunin, A.A.; Dmitriev, A.V.; Pogodin, P.V.; Dubovskaya, V.I.; Ivanov, S.M.; Tarasova, O.A.; et al. Computational Platform Way2Drug: From the Prediction of Biological Activity to Drug Repurposing. Russ. Chem. Bull. 2017, 66, 1832–1841. [Google Scholar] [CrossRef]
- Zardecki, C.; Dutta, S.; Goodsell, D.S.; Voigt, M.; Burley, S.K. RCSB Protein Data Bank: A Resource for Chemical, Biochemical, and Structural Explorations of Large and Small Biomolecules. J. Chem. Educ. 2016, 93, 569–575. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. Methods Mol. Biol. 2014, 1263, 243–250. [Google Scholar]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Adobe Systems Inc. Postscript Language Reference Manual; Addison-Wesley: Boston, MA, USA, 1985. [Google Scholar]
- Allen, F.H.; Bellard, S.; Brice, M.D.; Cartwright, B.A.; Doubleday, A.; Higgs, H.; Hummelink, T.; Hummelink-Peters, B.G.; Kennard, O.; Motherwell, W.D.S.; et al. The Cambridge Crystallographic Data Centre: Computer-Based Search, Retrieval, Analysis and Display of Information. Acta Cryst. 1979, 35, 2331–2339. [Google Scholar] [CrossRef]
- Bernstein, F.C.; Koetzle, T.F.; Williams, G.J.B.; Meyer, E.F., Jr.; Brice, M.D.; Rogers, J.R.; Kennard, O.; Shimanouchi, T.; Tasumi, M. The Protein Data Bank: A Computer-Based Archival File for Macromolecular Structures. J. Mol. Biol. 1977, 112, 535–542. [Google Scholar] [CrossRef]
- Engh, R.A.; Huber, R. Accurate Bond and Angle Parameters for X-Ray Protein Structure Refinement. Acta Cryst. 1991, 47, 392–400. [Google Scholar] [CrossRef]
- IUPAC-IUB Commission on Biochemical Nomenclature Abbreviations and Symbols for the Description of the Conformation of Polypeptide Chains. J. Mol. Biol. 1970, 52, 1–17. [CrossRef]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Macarthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Morris, A.L.; Macarthur, M.W.; Hutchinson, E.G.; Thornton, J.M. Stereochemical Quality of Protein Structure Coordinates. Proteins 1992, 12, 345–364. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, K.; Ooi, T. Radial Locations of Amino-Acid Residues in A Globular Protein: Correlation with the Sequence. J. Biochem. 1986, 100, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A Method to Identify Protein Sequences That Fold into A Known Three-Dimensional Structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of Protein Models with Three-Dimensional Profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Touw, W.G.; Baakman, C.; Black, J.; te Beek, T.A.; Krieger, E.; Joosten, R.P.; Vriend, G. A Series of Pdb Related Databanks for Everyday Needs. Nucleic Acids Res. 2015, 43, D364–D368. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Wikipedia Contributors. DSSP (Algorithm); Wikipedia: San Francisco, CA, USA, 2024. [Google Scholar]
- Sippl, M.J. Recognition of Errors in Three-Dimensional Structures of Proteins. Proteins 1993, 17, 355–362. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 28, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Mering, C.V.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Ferrin, T.E. Visualizing Density Maps with UCSF Chimera. J. Struct. Biol. 2007, 157, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Using PyMOL as a Platform for Computational Drug Design. WIREs Comput. Mol. Sci. 2017, 7, E1298. [Google Scholar] [CrossRef]
- Baroroh, U.; Biotek, M.; Muscifa, Z.S.; Destiarani, W.; Rohmatullah, F.G.; Yusuf, M. Molecular Interaction Analysis and Visualization of Protein-Compound Docking Using Biovia Discovery Studio Visualizer. Indones. J. Comput. Biol. (IJCB) 2023, 2, 22–30. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. Gromacs: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Showalter, S.A.; Brüschweiler, R. Validation of Molecular Dynamics Simulations of Biomolecules Using NMR Spin Relaxation as Benchmarks: Application to the AMBER99SB Force Field. J. Chem. Theory Comput. 2007, 3, 961–975. [Google Scholar] [CrossRef]
- Bray, S.A.; Lucas, X.; Kumar, A.; Grüning, B.A. The Chemicaltoolbox: Reproducible, User-Friendly Cheminformatics Analysis on the Galaxy Platform. J. Cheminf. 2020, 12, 40. [Google Scholar] [CrossRef]
- Cuendet, M.A.; van Gunsteren, W.F. On the Calculation of Velocity-Dependent Properties in Molecular Dynamics Simulations Using the Leapfrog Integration Algorithm. J. Chem. Phys. 2007, 127, 184102. [Google Scholar] [CrossRef]
- Uddin, K.M.; Sakib, M.; Siraji, S.; Uddin, R.; Rahman, S.; Alodhayb, A.; Alibrahim, K.A.; Kumer, A.; Matin, M.M.; Bhuiyan, M.M. Synthesis of New Derivatives of Benzylidinemalononitrile and Ethyl 2-Cyano-3-phenylacrylate: In Silico Anticancer Evaluation. ACS Omega 2023, 8, 25817–25831. [Google Scholar] [CrossRef]
- Quayum, S.T.; Esha, N.J.I.; Siraji, S.; Abbad, S.S.A.; Alsunaidi, Z.H.A.; Almatarneh, M.H.; Rahman, S.; Alodhayb, A.N.; Alibrahim, K.A.; Kawsar, S.M.A.; et al. Exploring the effectiveness of flavone derivatives for treating liver diseases: Utilizing DFT, molecular docking, and molecular dynamics techniques. MethodsX 2024, 12, 102537. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3D: An R Package for the Comparative Analysis of Protein Structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Awasthi, A.; Kumari, A.; Sood, D.; Jain, P.; Singh, T.; Sharma, N.; Grover, A.; Chandra, R. Antitussive Noscapine and Antiviral Drug Conjugates as Arsenal Against COVID-19: A Comprehensive Chemoinformatics Analysis. J. Biomol. Struct. Dyn. 2022, 40, 101–116. [Google Scholar] [CrossRef]
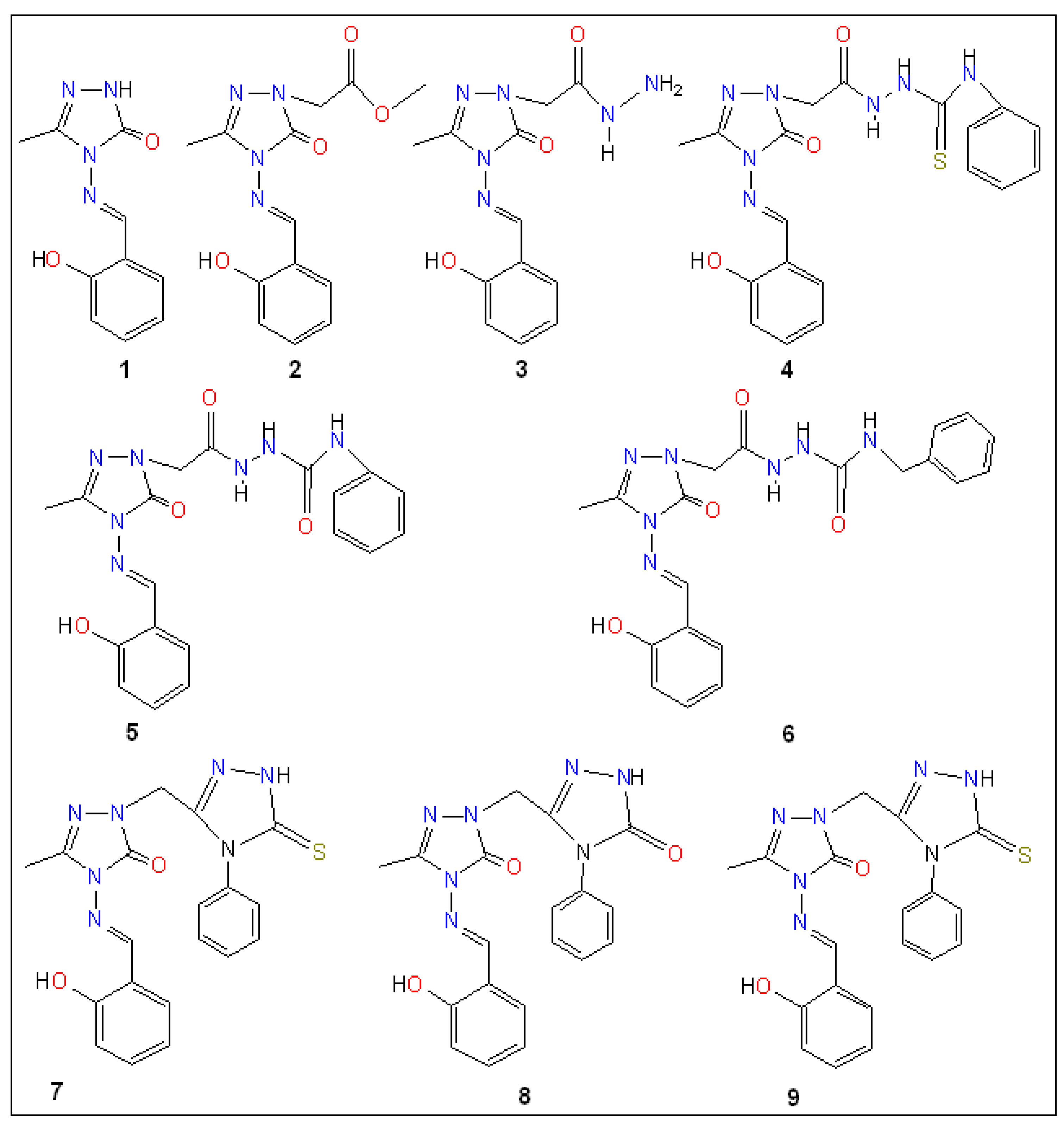
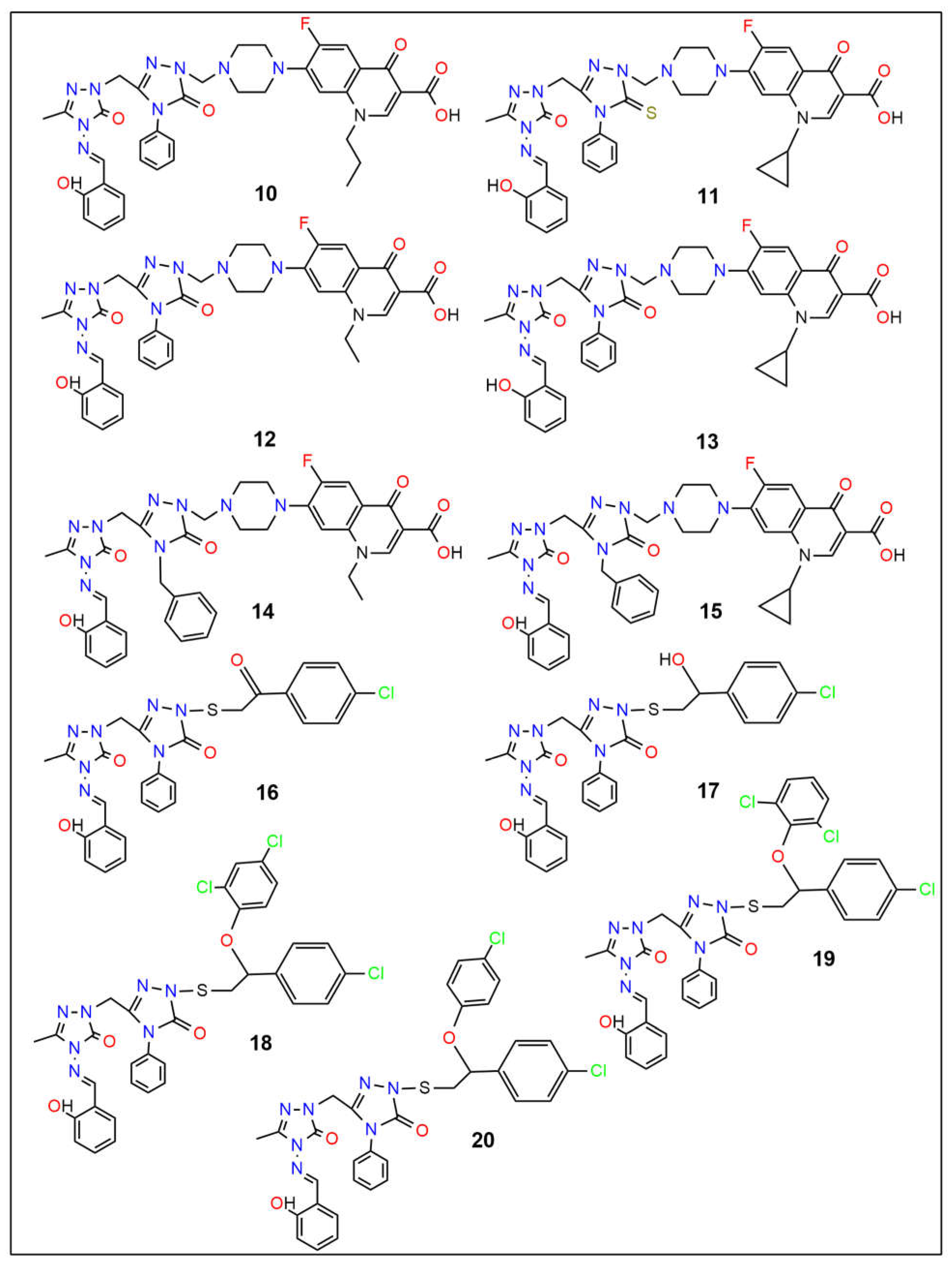
| Binding Affinity (kcal mol−1) | ||||||
|---|---|---|---|---|---|---|
| Ligand | 4KSG | 5BS8 | 5CDQ | 6RKU | 6ZT3 | 8BN6 |
| 1 | −8.1 | −6.8 | −6.3 | −6.6 | −6.4 | −7.9 |
| 2 | −7.8 | −6.5 | −6.6 | −6.3 | −8.1 | −7.5 |
| 3 | −7.7 | −6.5 | −9.2 | −6.9 | −8.9 | −6.6 |
| 4 | −7.3 | −7.0 | −7.1 | −7.2 | −7.4 | −8.0 |
| 5 | −8.2 | −7.8 | −7.1 | −7.9 | −8.5 | −8.7 |
| 6 | −8 | −6.5 | −8.6 | −7.9 | −7.6 | −8.1 |
| 7 | −7.2 | −8.8 | −7.5 | −7.2 | −7.4 | −6.9 |
| 8 | −7.9 | −8.6 | −7.6 | −7.7 | −8.8 | −7.8 |
| 9 | −7.7 | −9.4 | −8.2 | −7.5 | −6.9 | −6.7 |
| 10 | −8.2 | −10.2 | −9.1 | −7.9 | −9.2 | −7.8 |
| 11 | −7.5 | −8.6 | −9.1 | −8.5 | −8.6 | −8.4 |
| 12 | −8.7 | −9.3 | −9.8 | −9.0 | −8.1 | −9.3 |
| 13 | −8.8 | −9.4 | −9.3 | −8.5 | −7.8 | −9.4 |
| 14 | −8.9 | −9.2 | −10.1 | −7.8 | −7.7 | −8.0 |
| 15 | −9.2 | −9.1 | −10.6 | −8.9 | −8.1 | −8.3 |
| 16 | −8.5 | −7.6 | −9.1 | −8.2 | −8.4 | −9.6 |
| 17 | −9.1 | −6.4 | −7.9 | −8.8 | −8.7 | −8.9 |
| 18 | −9.0 | −8.7 | −8.8 | −8.5 | −7.1 | −9.8 |
| 19 | −7.7 | −6.7 | −5.8 | −6.4 | −7.6 | −7.7 |
| 20 | −8.0 | −8.6 | −9.2 | −8.6 | −7.7 | −9.0 |
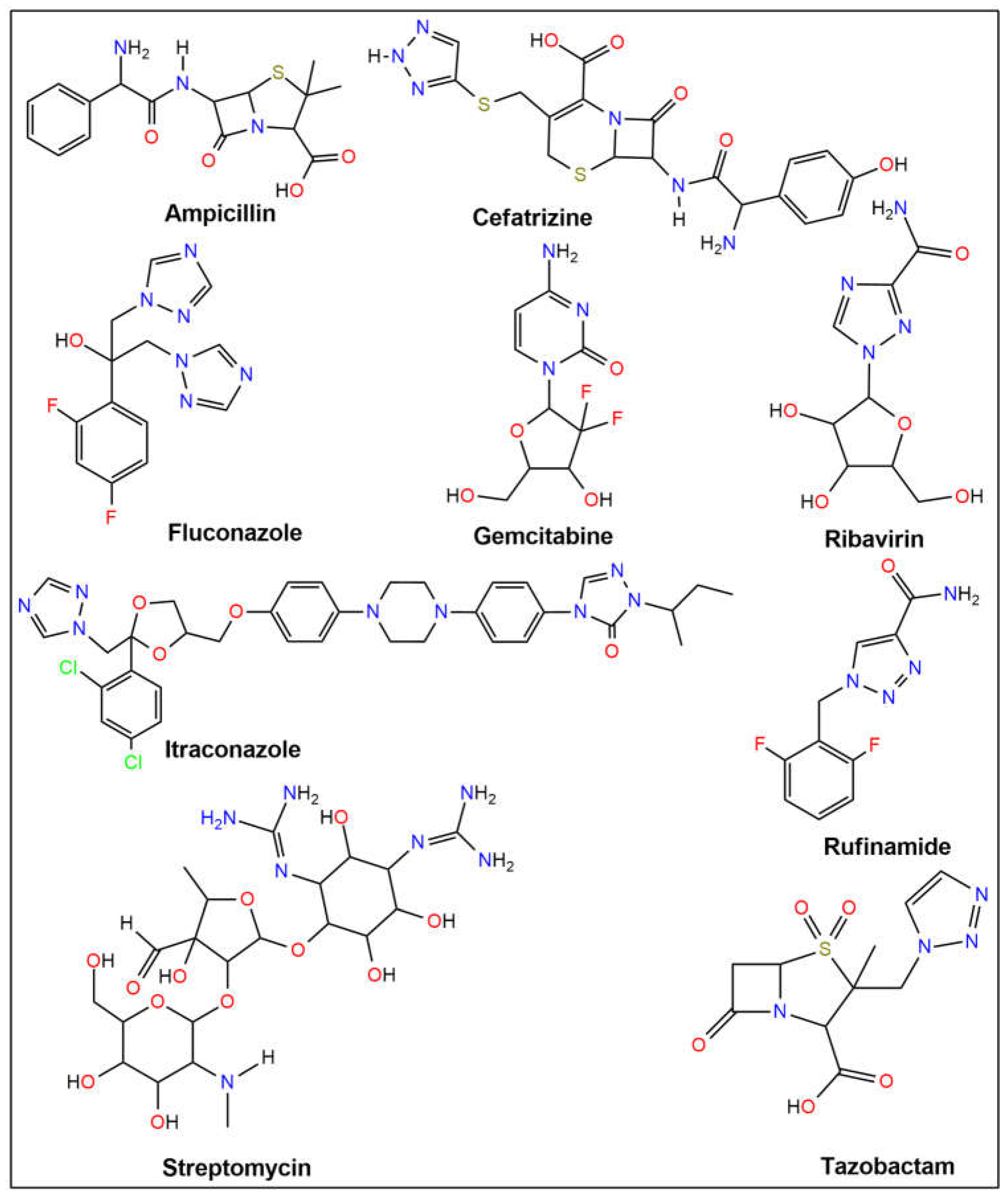
| Ampicillin | −6.2 | −6.9 | −6.5 | −6.6 | −6.9 | −6.9 |
| Cefatrizine | −7.7 | −7.1 | −8.2 | −7.3 | −7.1 | −8.0 |
| Fluconazole | −7.0 | −6.8 | −7.2 | −6.3 | −5.8 | −6.9 |
| Gemcitabine | −6.2 | −6.1 | −6.6 | −5.9 | −5.3 | −6.3 |
| Itraconazole | −8.3 | −8.1 | −9.9 | −9.0 | −8.1 | −8.2 |
| Ribavirin | −6.5 | −6.4 | −6.1 | −6.0 | −6.5 | −6.6 |
| Rufinamide | −7.4 | −6.8 | −7.0 | −6.5 | −6.0 | −6.6 |
| Streptomycin | −6.8 | −8.2 | −7.4 | −6.9 | −6.3 | −7.2 |
| Tazobactam | −6.1 | −6.7 | −6.1 | −6.1 | −6.0 | −5.7 |
| Binding Affinity (kcal mol−1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | 1M17 | 1MHL | 1Y6A | 2EUF | 2J0T | 2O21 | 2Z3Y | 3PHD | 3S79 | 4NRE |
| 1 | −7.0 | −6.1 | −6.8 | −7.4 | −7.1 | −6.2 | −7.5 | −6.8 | −7.1 | −7.7 |
| 2 | −6.7 | −6.1 | −6.5 | −7.8 | −6.8 | −6.3 | −6.3 | −6.3 | −7 | −7.3 |
| 3 | −7.1 | −6.4 | −6.9 | −8.0 | −7.5 | −6.3 | −6.4 | −6.1 | −7.9 | −8.8 |
| 4 | −7.8 | −6.4 | −6.9 | −8.9 | −7.4 | −6.9 | −7.5 | −6.8 | −8 | −7.1 |
| 5 | −8.1 | −7.7 | −8.3 | −9.3 | −8.2 | −8.2 | −9.6 | −7.7 | −9.1 | −9.8 |
| 6 | −7.9 | −7.3 | −8.0 | −9.9 | −7.9 | −6.7 | −7.3 | −7.0 | −8.7 | −9.0 |
| 7 | −7.8 | −7.2 | −7.5 | −7.1 | −8.6 | −7.8 | −9.0 | −6.4 | −8.4 | −8.1 |
| 8 | −8.4 | −7.6 | −7.8 | −8.5 | −8.7 | −8.5 | −9.4 | −6.5 | −8.9 | −8.1 |
| 9 | −7.5 | −7.4 | −7.9 | −9.4 | −7.9 | −7.8 | −7.8 | −6.5 | −8.2 | −7.6 |
| 10 | −9.7 | −8.9 | −8.8 | −9.9 | −9.8 | −9.3 | −9.5 | −7.2 | −9.1 | −9.7 |
| 11 | −9.9 | −8.8 | −8.8 | −9.6 | −9.3 | −9.6 | −10.9 | −7.5 | −9.5 | −8.3 |
| 12 | −9.9 | −9.2 | −8.7 | −9.9 | −10.1 | −9.6 | −10.6 | −7.4 | −9.3 | −10.7 |
| 13 | −10.1 | −9.5 | −8.9 | −10.1 | −9.4 | −9.7 | −11.4 | −7.4 | −9.8 | −10.9 |
| 14 | −9.8 | −8.7 | −8.7 | −9.6 | −9.3 | −7.9 | −9.9 | −7.9 | −8.9 | −9.8 |
| 15 | −9.7 | −8.8 | −7.9 | −9.9 | −8.8 | −9.2 | −10.9 | −7.6 | −8.4 | −9.8 |
| 16 | −9.2 | −8.2 | −8.3 | −10.0 | −10.2 | −8.8 | −12.0 | −7.1 | −10.4 | −10.9 |
| 17 | −8.9 | −7.9 | −8.1 | −10.1 | −10.0 | −8.7 | −11.2 | −6.4 | −8.7 | −9.8 |
| 18 | −10.0 | −8.6 | −9.1 | −10.7 | −8.9 | −8.5 | −11.8 | −6.9 | −8.3 | −9.3 |
| 19 | −6.9 | −6.2 | −6.5 | −7.7 | −7.1 | −6.4 | −6.5 | −6.3 | −6.6 | −7.6 |
| 20 | −9.5 | −8.4 | −7.2 | −8.3 | −8.8 | −8.8 | −8.4 | −6.8 | −9.6 | −7.5 |
| Ampicillin | −7.9 | −6.5 | −6.7 | −8.2 | −6.8 | −6.9 | −7.0 | −6.6 | −8 | −7.7 |
| Cefatrizine | −8.2 | −7.1 | −7.4 | −8.3 | −7.4 | −6.9 | −7.1 | −6.6 | −7.8 | −8.1 |
| Fluconazole | −7.3 | −6.5 | −6.6 | −8.3 | −6.8 | −6.8 | −7.6 | −5.1 | −7.1 | −8.2 |
| Gemcitabine | −6.6 | −6.0 | −5.7 | −6.7 | −5.9 | −6.0 | −6.5 | −5.1 | −7.6 | −6.2 |
| Itraconazole | −8.7 | −7.4 | −8.3 | −8.8 | −8.4 | −9.0 | −10.7 | −7.4 | −8.7 | −8.6 |
| Ribavirin | −6.3 | −5.8 | −5.8 | −6.6 | −6.9 | −5.8 | −6.9 | −5.1 | −6.4 | −6.8 |
| Rufinamide | −7.0 | −5.8 | −6.2 | −7.9 | −6.7 | −6.6 | −6.9 | −7.0 | −7.2 | −7.5 |
| Streptomycin | −6.4 | −7.2 | −6.1 | −6.2 | −7.3 | −6.9 | −7.7 | −5.7 | −7.2 | −7.3 |
| Tazobactam | −6.0 | −5.8 | −6.0 | −5.7 | −5.8 | −5.6 | −6.4 | −5.3 | −6.5 | −6.5 |
| Complex | Interacting Residues | Distance (Å) | Type of Interaction | 2D Diagram of Interaction |
|---|---|---|---|---|
| Compound 15-Topo II | ALA A:118 | 4.88 | Pi-Alkyl | 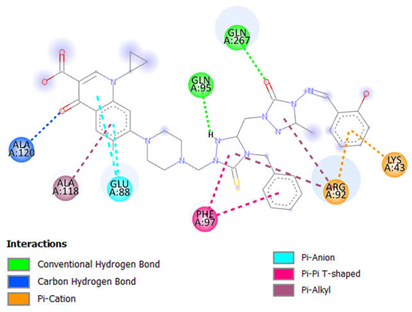 |
| ALA A:120 | 2.62 | Carbon-Hydrogen Bond | ||
| ARG A:92 | 4.64 | Pi-Cation | ||
| 4.99, 5.37 | Pi-Alkyl | |||
| GLN A:95 | 2.48 | Conventional Hydrogen Bond | ||
| GLN A:267 | 2.31 | Conventional Hydrogen Bond | ||
| GLU A:88 | 3.79, 3.88 | Pi-Anion | ||
| LYS A:43 | 2.78 | Pi-Cation; Pi-Donor Hydrogen Bond | ||
| PHE A:97 | 5.08, 5.78 | Pi-Pi T-shaped | ||
| Compound 16-LSD1 | ARG A:182 | 1.89, 2.58 | Conventional Hydrogen Bond |  |
| 4.19 | Pi-Alkyl | |||
| ARG A:820 | 2.51 | Carbon-Hydrogen Bond | ||
| 3.73 | Alkyl | |||
| 5.19 | Pi-Alkyl | |||
| ILE A:804 | 4.24 | Alkyl | ||
| LEU A:816 | 4.36 | Pi-Alkyl | ||
| 4.99 | Alkyl | |||
| PHE A:179 | 5.09 | Pi-Pi T-shaped | ||
| PHE A:558 | 2.97 | Carbon-Hydrogen Bond | ||
| 4.95 | Pi-Pi T-shaped | |||
| TYR A:807 | 2.62 | Conventional Hydrogen Bond |
| Ligand | MW | mLogP | nHBD | nHBA | Lipinski’s Violations | Veber’s Violations | TPSA Å2 | nRotb | PAINS #Alerts | Brenk #Alerts |
|---|---|---|---|---|---|---|---|---|---|---|
| Lipinski | ≤500 | ≤5 | ≤5 | ≤10 | - | - | - | - | - | - |
| Veber | - | - | - | - | - | - | ≤140 | ≤10 | - | - |
| 1 | 218.21 | 1.2 | 2 | 4 | 0 | 0 | 83.27 | 2 | 1 | 1 |
| 2 | 304.3 | 1.46 | 1 | 6 | 0 | 0 | 98.71 | 6 | 1 | 1 |
| 3 | 290.28 | 0.55 | 3 | 6 | 0 | 0 | 127.53 | 5 | 1 | 3 |
| 4 | 425.46 | 1.92 | 4 | 5 | 0 | 1 | 157.66 | 9 | 1 | 2 |
| 5 | 409.4 | 1.97 | 4 | 6 | 1 | 1 | 142.64 | 9 | 1 | 1 |
| 6 | 423.43 | 1.93 | 4 | 6 | 1 | 1 | 142.64 | 10 | 1 | 1 |
| 7 | 407.45 | 1.77 | 2 | 5 | 0 | 0 | 138.11 | 5 | 1 | 2 |
| 8 | 391.38 | 1.82 | 2 | 6 | 0 | 0 | 123.09 | 5 | 1 | 1 |
| 9 | 421.48 | 1.73 | 2 | 5 | 0 | 0 | 138.11 | 6 | 1 | 2 |
| 10 | 738.79 | 2.28 | 2 | 10 | 2 | 1 | 193.03 | 10 | 1 | 2 |
| 11 | 750.8 | 2.46 | 2 | 10 | 2 | 1 | 193.03 | 10 | 1 | 2 |
| 12 | 722.72 | 1.95 | 2 | 11 | 2 | 1 | 178.01 | 10 | 1 | 1 |
| 13 | 734.74 | 2.13 | 2 | 11 | 2 | 1 | 178.01 | 10 | 1 | 1 |
| 14 | 752.82 | 2.19 | 2 | 10 | 2 | 2 | 193.03 | 11 | 1 | 2 |
| 15 | 764.83 | 2.36 | 2 | 10 | 2 | 2 | 193.03 | 11 | 1 | 2 |
| 16 | 560.03 | 3.65 | 1 | 7 | 1 | 1 | 145.49 | 9 | 1 | 1 |
| 17 | 562.04 | 3.72 | 2 | 7 | 1 | 1 | 148.65 | 9 | 1 | 1 |
| 18 | 721.06 | 5.73 | 1 | 7 | 2 | 1 | 137.65 | 12 | 1 | 1 |
| 19 | 721.06 | 5.73 | 1 | 7 | 2 | 1 | 137.65 | 12 | 1 | 1 |
| 20 | 686.61 | 5.29 | 1 | 7 | 2 | 1 | 137.65 | 12 | 1 | 1 |
| Ampicillin | 349.4 | 0.75 | 3 | 5 | 0 | 0 | 138.03 | 5 | 0 | 0 |
| Cefatrizine | 462.5 | 0.02 | 5 | 8 | 1 | 1 | 225.13 | 8 | 0 | 0 |
| Fluconazole | 306.27 | 1.47 | 1 | 7 | 0 | 0 | 81.65 | 5 | 0 | 0 |
| Gemcitabine | 263.2 | −1.2 | 3 | 7 | 0 | 0 | 110.6 | 2 | 0 | 0 |
| Itraconazole | 705.63 | 4.21 | 0 | 7 | 3 | 1 | 104.7 | 11 | 2 | 0 |
| Ribavirin | 244.2 | −2.94 | 4 | 7 | 0 | 1 | 143.72 | 3 | 0 | 0 |
| Rufinamide | 238.19 | 1.44 | 1 | 5 | 0 | 0 | 73.8 | 3 | 0 | 0 |
| Streptomycin | 581.57 | −5.96 | 12 | 15 | 3 | 1 | 336.43 | 9 | 0 | 3 |
| Tazobactam | 300.29 | −0.44 | 1 | 7 | 0 | 0 | 130.84 | 3 | 0 | 0 |
| Ligand | GPCR | ICM | KI | NRL | EI | PI |
|---|---|---|---|---|---|---|
| 1 | −1.28 | −1.20 | −1.07 | −1.56 | −1.54 | −1.05 |
| 2 | −0.79 | −1.14 | −0.98 | −0.92 | −0.85 | −0.73 |
| 3 | −1.02 | −1.54 | −1.08 | −1.45 | −0.98 | −0.83 |
| 4 | −0.95 | −1.17 | −1.03 | −1.26 | −0.86 | −0.87 |
| 5 | −0.60 | −0.97 | −0.66 | −1.11 | −0.62 | −0.68 |
| 6 | −0.51 | −0.94 | −0.73 | −1.02 | −0.48 | −0.61 |
| 7 | −0.76 | −1.14 | −1.10 | −0.98 | −0.96 | −0.87 |
| 8 | −0.52 | −0.82 | −0.69 | −0.76 | −0.75 | −0.69 |
| 9 | −0.72 | −1.11 | −1.04 | −0.90 | −0.88 | −0.75 |
| 10 | −1.49 | −2.80 | −2.24 | −2.32 | −1.51 | −1.89 |
| 11 | −1.68 | −2.98 | −2.47 | −2.59 | −1.60 | −2.01 |
| 12 | −1.35 | −2.66 | −2.08 | −2.20 | −1.38 | −1.78 |
| 13 | −1.53 | −2.85 | −2.30 | −2.48 | −1.47 | −1.91 |
| 14 | −1.63 | −2.99 | −2.41 | −2.48 | −1.59 | −2.01 |
| 15 | −1.82 | −3.19 | −2.65 | −2.76 | −1.69 | −2.15 |
| 16 | −0.71 | −1.27 | −0.94 | −1.04 | −0.79 | −0.74 |
| 17 | −0.59 | −1.09 | −0.86 | −0.82 | −0.73 | −0.71 |
| 18 | −0.89 | −1.89 | −1.44 | −1.41 | −0.81 | −1.23 |
| 19 | −0.86 | −1.88 | −1.44 | −1.44 | −0.81 | −1.22 |
| 20 | −0.80 | −1.73 | −1.31 | −1.27 | −0.72 | −1.09 |
| Ampicillin | 0.04 | −0.47 | −0.71 | −0.61 | 0.87 | 0.25 |
| Cefatrizine | −0.23 | −0.72 | −0.91 | −0.98 | 0.34 | 0.06 |
| Fluconazole | 0.04 | 0.01 | −0.09 | 0.23 | −0.09 | 0.03 |
| Gemcitabine | 0.58 | 0.11 | 0.33 | −1.00 | 0.15 | 1.06 |
| Itraconazole | −0.40 | −1.50 | −1.30 | −1.31 | −0.66 | −0.97 |
| Ribavirin | 0.31 | 0.21 | −0.21 | −1.46 | −0.20 | 0.71 |
| Rufinamide | −0.19 | −0.27 | −0.16 | −1.01 | −0.48 | −0.03 |
| Streptomycin | 0.09 | −0.16 | −0.17 | −0.18 | 0.65 | 0.38 |
| Tazobactam | 0.18 | −0.27 | −0.29 | −0.45 | 1.35 | 1.04 |
| Ligand | aHIA | aBBB | ahERG_ pIC50 | aCYP2C19 Inhibition | aCYP3A4 Inhibition | aSubcellular Localization | bSA Score |
|---|---|---|---|---|---|---|---|
| 1 | +1.0000 | +0.9391 | 0.7734 | 0.8711 | 0.9465 | M—0.7809 | 2.59 |
| 2 | +0.9483 | +0.8026 | 0.7528 | 0.6314 | 0.9628 | M—0.7452 | 3.31 |
| 3 | +0.9964 | +0.9002 | 0.8610 | 0.8958 | 0.9421 | M—0.8127 | 3.19 |
| 4 | +0.9625 | +0.6926 | 0.8919 | 0.7727 | 0.8210 | M—0.7928 | 3.71 |
| 5 | +0.9493 | +0.6651 | 0.8382 | 0.7965 | 0.9006 | M—0.8182 | 3.66 |
| 6 | +0.9126 | +0.6327 | 0.7789 | 0.7407 | 0.8156 | M—0.7846 | 3.72 |
| 7 | +0.9875 | +0.8430 | 0.8850 | 0.6117 | 0.6447 | M—0.7725 | 3.64 |
| 8 | +0.9968 | +0.8669 | 0.7863 | 0.7570 | 0.8270 | M—0.9005 | 3.65 |
| 9 | +0.9475 | +0.7982 | 0.7144 | 0.6524 | 0.5131 | M—0.7270 | 3.75 |
| 10 | +0.9638 | −0.8199 | 0.6779 | 0.7168 | 0.6288 | M—0.4652 | 5.29 |
| 11 | +0.9335 | −0.7924 | 0.6345 | 0.7440 | 0.7540 | M—0.5160 | 5.33 |
| 12 | +0.9947 | −0.7511 | 0.5613 | 0.7627 | 0.5000 | M—0.5847 | 5.30 |
| 13 | +0.9900 | −0.7126 | 0.6078 | 0.8121 | 0.6447 | M—0.6287 | 5.34 |
| 14 | +0.9638 | −0.8199 | 0.6779 | 0.7168 | 0.6288 | M—0.4652 | 5.39 |
| 15 | +0.9335 | −0.7924 | 0.6345 | 0.7440 | 0.7540 | M—0.5160 | 5.43 |
| 16 | +0.9895 | +0.7942 | 0.8315 | 0.5315 | 0.6792 | M—0.7179 | 4.25 |
| 17 | +0.9925 | +0.6399 | 0.8949 | 0.5000 | 0.5979 | M—0.7157 | 4.83 |
| 18 | +0.9904 | +0.8728 | 0.8437 | 0.6195 | 0.6509 | M—0.7188 | 5.41 |
| 19 | +0.9904 | +0.8728 | 0.8437 | 0.6195 | 0.6509 | M—0.7188 | 5.41 |
| 20 | +0.9904 | +0.8728 | 0.8437 | 0.6195 | 0.6509 | M—0.7188 | 5.39 |
| Ampicillin | −0.9270 | −0.9961 | 0.9998 | 0.9399 | 0.8309 | L—0.5707 | 4.16 |
| Cefatrizine | −0.9522 | −0.9833 | 0.9613 | 0.7493 | 0.7150 | M—0.6074 | 4.87 |
| Fluconazole | +0.9894 | +0.9382 | 0.8229 | 0.5320 | 0.8196 | M—0.8498 | 2.45 |
| Gemcitabine | +0.9814 | +0.9693 | 0.9948 | 0.8478 | 0.9032 | N—0.4165 | 3.71 |
| Itraconazole | +0.9973 | −0.6151 | 0.5782 | 0.5703 | 0.5279 | M—0.4776 | 5.77 |
| Ribavirin | +0.9852 | +0.9381 | 0.9948 | 0.9095 | 0.9535 | M—0.4619 | 3.89 |
| Rufinamide | +1.0000 | +0.9777 | 0.9484 | 0.5561 | 0.7995 | M—0.8591 | 2.22 |
| Streptomycin | −0.8824 | −0.9712 | 0.9934 | 0.9026 | 0.8867 | M—0.4518 | 6.92 |
| Tazobactam | −0.6161 | −0.9659 | 0.9939 | 0.7143 | 0.8596 | M—0.4356 | 4.23 |
| Principal Components | |||||
|---|---|---|---|---|---|
| Complex | Temperature | PC1 (%) | PC2 (%) | PC3 (%) | Cosine Value |
| Topo II-compound 15 | 300 | 48.79 | 15.73 | 8.27 | 0.09 |
| 305 | 22.38 | 11.94 | 9.05 | 0.48 | |
| 310 | 37.53 | 20.59 | 8.12 | 0.70 | |
| 320 | 0.73 | 0.68 | 0.49 | 0.01 | |
| LSD1-compound 16 | 300 | 23.75 | 21.31 | 10.58 | 0.83 |
| 305 | 32.66 | 21.92 | 6.44 | 0.79 | |
| 310 | 15.21 | 13.04 | 10.04 | 0.78 | |
| 320 | 23.88 | 14.32 | 9.79 | 0.32 | |
| Minimal Inhibition Concentration Values (μg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Compound | Ec | Yp | Pa | Sa | Ef | Bc | Ms | Ca | Sc |
| 10 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | - | - |
| 11 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | - | - |
| 12 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | - | - |
| 13 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | - | - |
| 14 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | - | - |
| 15 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | <0.24 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suha, H.N.; Almatarneh, M.H.; Poirier, R.A.; Uddin, K.M. In Silico Evaluation of Quinolone–Triazole and Conazole–Triazole Hybrids as Promising Antimicrobial and Anticancer Agents. Int. J. Mol. Sci. 2025, 26, 6752. https://doi.org/10.3390/ijms26146752
Suha HN, Almatarneh MH, Poirier RA, Uddin KM. In Silico Evaluation of Quinolone–Triazole and Conazole–Triazole Hybrids as Promising Antimicrobial and Anticancer Agents. International Journal of Molecular Sciences. 2025; 26(14):6752. https://doi.org/10.3390/ijms26146752
Chicago/Turabian StyleSuha, Humaera Noor, Mansour H. Almatarneh, Raymond A. Poirier, and Kabir M. Uddin. 2025. "In Silico Evaluation of Quinolone–Triazole and Conazole–Triazole Hybrids as Promising Antimicrobial and Anticancer Agents" International Journal of Molecular Sciences 26, no. 14: 6752. https://doi.org/10.3390/ijms26146752
APA StyleSuha, H. N., Almatarneh, M. H., Poirier, R. A., & Uddin, K. M. (2025). In Silico Evaluation of Quinolone–Triazole and Conazole–Triazole Hybrids as Promising Antimicrobial and Anticancer Agents. International Journal of Molecular Sciences, 26(14), 6752. https://doi.org/10.3390/ijms26146752






