Global DNA Methylation in Poorly Controlled Type 2 Diabetes Mellitus: Association with Redox and Inflammatory Biomarkers
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. Laboratory Analyses
4.3. Assessment of Global DNA Methylation
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 5mC | 5-methylcytosine |
| AGE | Advanced Glycation End Products |
| AOPP | Advanced Oxidation Protein Products |
| AU | Arbitrary Units |
| BMI | Body Mass Index |
| CBC | Complete Blood Count |
| CG | Control Group |
| CVD | Cardiovascular Disease |
| dC | Deoxycytidine |
| DNMTs | DNA Methyltransferases |
| DPP-4 | Dipeptidyl Peptidase-4 |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| F-AGEs | Fluorescent Advanced Glycation End Products |
| HbA1c | Glycated Hemoglobin |
| HDL-C | High-Density Lipoprotein Cholesterol |
| HPLC/UHPLC-DAD | (Ultra) High-Performance Liquid Chromatography with Diode Array Detection |
| hsCRP | High-Sensitivity C-Reactive Protein |
| IMA | Ischemia-Modified Albumin |
| KMO | Kaiser-Meyer-Olkin |
| LDL-C | Low-Density Lipoprotein Cholesterol |
| LUMA | Luminometric Methylation Assay |
| LWR | Lymphocyte-to-White Blood Cell Ratio |
| MAPK | Mitogen-Activated Protein Kinase |
| MLR | Monocyte-to-Lymphocyte Ratio |
| MWR | Monocyte-to-White Blood Cell Ratio |
| NF-κB | Nuclear Factor kappa B |
| NLR | Neutrophil-to-Lymphocyte Ratio |
| NNMT | Nicotinamide N-methyltransferase |
| NWR | Neutrophil-to-White Blood Cell Ratio |
| OHAs | Oral Hypoglycemic Agents |
| PAB | Prooxidant-Antioxidant Balance |
| PBMC | Peripheral Blood Mononuclear Cells |
| PBS | Phosphate-Buffered Saline |
| PCA | Principal Component Analysis |
| PLR | Platelet-to-Lymphocyte Ratio |
| RAGE | Receptor for Advanced Glycation End Products |
| ROS | Reactive Oxygen Species |
| SAM | S-adenosylmethionine |
| SH-groups | Sulfhydryl Groups |
| SOD | Superoxide Dismutase |
| T2DM | Type 2 Diabetes Mellitus |
| TAS | Total Antioxidant Status |
| TC | Total Cholesterol |
| TG | Triglcyerides |
| TET | Ten-Eleven Translocation |
| TOS | Total Oxidant Status |
References
- American Diabetes Association Professional Practice Committee. 16. Diabetes Care in the Hospital: Standards of Care in Diabetes—2024. Diabetes Care 2024, 47, S295–S306. [Google Scholar] [CrossRef] [PubMed]
- Mannar, V.; Boro, H.; Patel, D.; Agstam, S.; Dalvi, M.; Bundela, V. Epigenetics of the Pathogenesis and Complications of Type 2 Diabetes Mellitus. touchREV Endocrinol. 2023, 19, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Sriraman, A.; Debnath, T.K.; Xhemalce, B.; Miller, K.M. Making It or Breaking It: DNA Methylation and Genome Integrity. Essays Biochem. 2020, 64, 687–703. [Google Scholar] [CrossRef]
- Kriebel, J.; Herder, C.; Rathmann, W.; Wahl, S.; Kunze, S.; Molnos, S.; Volkova, N.; Schramm, K.; Carstensen-Kirberg, M.; Waldenberger, M.; et al. Association between DNA Methylation in Whole Blood and Measures of Glucose Metabolism: KORA F4 Study. PLoS ONE 2016, 11, e0152314. [Google Scholar] [CrossRef]
- Pfeiffer, L.; Wahl, S.; Pilling, L.C.; Reischl, E.; Sandling, J.K.; Kunze, S.; Holdt, L.M.; Kretschmer, A.; Schramm, K.; Adamski, J.; et al. DNA Methylation of Lipid-Related Genes Affects Blood Lipid Levels. Circ. Cardiovasc. Genet. 2015, 8, 334–342. [Google Scholar] [CrossRef]
- Vekic, J.; Stromsnes, K.; Mazzalai, S.; Zeljkovic, A.; Rizzo, M.; Gambini, J. Oxidative Stress, Atherogenic Dyslipidemia, and Cardiovascular Risk. Biomedicines 2023, 11, 2897. [Google Scholar] [CrossRef] [PubMed]
- Pezone, A.; Olivieri, F.; Napoli, M.V.; Procopio, A.; Avvedimento, E.V.; Gabrielli, A. Inflammation and DNA Damage: Cause, Effect or Both. Nat. Rev. Rheumatol. 2023, 19, 200–211. [Google Scholar] [CrossRef]
- Yaribeygi, H.; Sathyapalan, T.; Atkin, S.L.; Sahebkar, A. Molecular Mechanisms Linking Oxidative Stress and Diabetes Mellitus. Oxid. Med. Cell. Longev. 2020, 2020, 8609213. [Google Scholar] [CrossRef]
- Vekic, J.; Vujcic, S.; Bufan, B.; Bojanin, D.; Al-Hashmi, K.; Al-Rasadi, K.; Stoian, A.P.; Zeljkovic, A.; Rizzo, M. The Role of Advanced Glycation End Products on Dyslipidemia. Metabolites 2023, 13, 77. [Google Scholar] [CrossRef]
- Islam, S.; Moinuddin; Mir, A.R.; Arfat, M.Y.; Alam, K.; Ali, A. Studies on Glycoxidatively Modified Human IgG: Implications in Immuno-Pathology of Type 2 Diabetes Mellitus. Int. J. Biol. Macromol. 2017, 104, 19–29. [Google Scholar] [CrossRef]
- Kowluru, R.A. Cross Talks between Oxidative Stress, Inflammation and Epigenetics in Diabetic Retinopathy. Cells 2023, 12, 300. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Mani, A.M.; Wu, Z.-H. DNA Damage-Induced Nuclear Factor-Kappa B Activation and Its Roles in Cancer Progression. J. Cancer Metastasis Treat. 2017, 3, 45–59. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA Hypermethylation in Disease: Mechanisms and Clinical Relevance. Epigenetics 2019, 14, 1141–1163. [Google Scholar] [CrossRef] [PubMed]
- Luttmer, R.; Spijkerman, A.M.; Kok, R.M.; Jakobs, C.; Blom, H.J.; Serne, E.H.; Dekker, J.M.; Smulders, Y.M. Metabolic Syndrome Components Are Associated with DNA Hypomethylation. Obes. Res. Clin. Pract. 2013, 7, e106–e115. [Google Scholar] [CrossRef]
- Pinzón-Cortés, J.A.; Perna-Chaux, A.; Rojas-Villamizar, N.S.; Díaz-Basabe, A.; Polanía-Villanueva, D.C.; Jácome, M.F.; Mendivil, C.O.; Groot, H.; López-Segura, V. Effect of Diabetes Status and Hyperglycemia on Global DNA Methylation and Hydroxymethylation. Endocr. Connect. 2017, 6, 708–725. [Google Scholar] [CrossRef]
- Nadiger, N.; Veed, J.K.; Chinya Nataraj, P.; Mukhopadhyay, A. DNA Methylation and Type 2 Diabetes: A Systematic Review. Clin. Epigenetics 2024, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Volkmar, M.; Dedeurwaerder, S.; Cunha, D.A.; Ndlovu, M.N.; Defrance, M.; Deplus, R.; Calonne, E.; Volkmar, U.; Igoillo-Esteve, M.; Naamane, N.; et al. DNA Methylation Profiling Identifies Epigenetic Dysregulation in Pancreatic Islets from Type 2 Diabetic Patients. EMBO J. 2012, 31, 1405–1426. [Google Scholar] [CrossRef]
- Thongsroy, J.; Mutirangura, A. Decreased Alu Methylation in Type 2 Diabetes Mellitus Patients Increases HbA1c Levels. J. Clin. Lab Anal. 2023, 37, e24966. [Google Scholar] [CrossRef]
- Muka, T.; Nano, J.; Voortman, T.; Braun, K.V.E.; Ligthart, S.; Stranges, S.; Bramer, W.M.; Troup, J.; Chowdhury, R.; Dehghan, A.; et al. The Role of Global and Regional DNA Methylation and Histone Modifications in Glycemic Traits and Type 2 Diabetes: A Systematic Review. Nutr. Metab. Cardiovasc. Dis. 2016, 26, 553–566. [Google Scholar] [CrossRef]
- Lisanti, S.; Omar, W.A.W.; Tomaszewski, B.; De Prins, S.; Jacobs, G.; Koppen, G.; Mathers, J.C.; Langie, S.A.S. Comparison of Methods for Quantification of Global DNA Methylation in Human Cells and Tissues. PLoS ONE 2013, 8, e79044. [Google Scholar] [CrossRef]
- Maghbooli, Z.; Hossein-nezhad, A.; Larijani, B.; Amini, M.; Keshtkar, A. Global DNA Methylation as a Possible Biomarker for Diabetic Retinopathy. Diabetes Metab. Res. 2015, 31, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-H.; Han, X.; Wang, M.; Hu, Q.; Li, S.; Wang, M.; Hu, J. The Association between Genomic DNA Methylation and Diabetic Peripheral Neuropathy in Patients with Type 2 Diabetes Mellitus. J. Diabetes Res. 2019, 2019, 2494057. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, Y.; Yamada, H.; Munetsuna, E.; Fujii, R.; Yamazaki, M.; Ando, Y.; Mizuno, G.; Ishikawa, H.; Ohashi, K.; Hashimoto, S.; et al. Global DNA Hypermethylation in Peripheral Blood Mononuclear Cells and Cardiovascular Disease Risk: A Population-Based Propensity Score-Matched Cohort Study. J. Epidemiol. Community Health 2021, 75, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Baccarelli, A.; Wright, R.; Bollati, V.; Litonjua, A.; Zanobetti, A.; Tarantini, L.; Sparrow, D.; Vokonas, P.; Schwartz, J. Ischemic Heart Disease and Stroke in Relation to Blood DNA Methylation. Epidemiology 2010, 21, 819–828. [Google Scholar] [CrossRef]
- Aavik, E.; Lumivuori, H.; Leppänen, O.; Wirth, T.; Häkkinen, S.-K.; Bräsen, J.-H.; Beschorner, U.; Zeller, T.; Braspenning, M.; van Criekinge, W.; et al. Global DNA Methylation Analysis of Human Atherosclerotic Plaques Reveals Extensive Genomic Hypomethylation and Reactivation at Imprinted Locus 14q32 Involving Induction of a miRNA Cluster. Eur. Heart J. 2015, 36, 993–1000. [Google Scholar] [CrossRef]
- Wang, C.; Ni, W.; Yao, Y.; Just, A.; Heiss, J.; Wei, Y.; Gao, X.; Coull, B.A.; Kosheleva, A.; Baccarelli, A.A.; et al. DNA Methylation-Based Biomarkers of Age Acceleration and All-Cause Death, Myocardial Infarction, Stroke, and Cancer in Two Cohorts: The NAS, and KORA F4. eBioMedicine 2021, 63, 103151. [Google Scholar] [CrossRef]
- Zuo, H.-P.; Guo, Y.-Y.; Che, L.; Wu, X.-Z. Hypomethylation of Interleukin-6 Promoter Is Associated with the Risk of Coronary Heart Disease. Arq. Bras. Cardiol. 2016, 107, 131–136. [Google Scholar] [CrossRef]
- Constâncio, V.; Nunes, S.P.; Henrique, R.; Jerónimo, C. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells 2020, 9, 624. [Google Scholar] [CrossRef]
- Lee, J.; Yun, J.-S.; Ko, S.-H. Advanced Glycation End Products and Their Effect on Vascular Complications in Type 2 Diabetes Mellitus. Nutrients 2022, 14, 3086. [Google Scholar] [CrossRef]
- Sun, X.; Gan, H.; Xia, Y. Changes of Serum Advanced Glycation End Products (AGEs), Matrix Metalloprotein-2 (MMP-2), and Urinary Microalbuminuria (mALB) in Diabetic Nephropathy and Their Predictive Value for Heart Failure. Transl. Androl. Urol. 2021, 10, 1279–1285. [Google Scholar] [CrossRef]
- Kuzan, A. Toxicity of Advanced Glycation End Products (Review). Biomed. Rep. 2021, 14, 46. [Google Scholar] [CrossRef] [PubMed]
- García-Guede, Á.; Vera, O.; Ibáñez-de-Caceres, I. When Oxidative Stress Meets Epigenetics: Implications in Cancer Development. Antioxidants 2020, 9, 468. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-F.; Wu, K.-J. Epigenetics, TET Proteins, and Hypoxia in Epithelial-Mesenchymal Transition and Tumorigenesis. Biomedicine 2016, 6, 1. [Google Scholar] [CrossRef]
- Rubio, K.; Hernández-Cruz, E.Y.; Rogel-Ayala, D.G.; Sarvari, P.; Isidoro, C.; Barreto, G.; Pedraza-Chaverri, J. Nutriepigenomics in Environmental-Associated Oxidative Stress. Antioxidants 2023, 12, 771. [Google Scholar] [CrossRef]
- Yi, Y.-S. Functional Interplay between Methyltransferases and Inflammasomes in Inflammatory Responses and Diseases. Int. J. Mol. Sci. 2021, 22, 7580. [Google Scholar] [CrossRef] [PubMed]
- Crosswhite, P.; Sun, Z. TNFα Induces DNA and Histone Hypomethylation and Pulmonary Artery Smooth Muscle Cell Proliferation Partly via Excessive Superoxide Formation. Antioxidants 2024, 13, 677. [Google Scholar] [CrossRef]
- Cucoreanu, C.; Tigu, A.-B.; Nistor, M.; Moldovan, R.-C.; Pralea, I.-E.; Iacobescu, M.; Iuga, C.-A.; Szabo, R.; Dindelegan, G.-C.; Ciuce, C. Epigenetic and Molecular Alterations in Obesity: Linking CRP and DNA Methylation to Systemic Inflammation. Curr. Issues Mol. Biol. 2024, 46, 7430–7446. [Google Scholar] [CrossRef]
- Sun, W.-D.; Zhu, X.-J.; Li, J.-J.; Mei, Y.-Z.; Li, W.-S.; Li, J.-H. Nicotinamide N-Methyltransferase (NNMT): A Novel Therapeutic Target for Metabolic Syndrome. Front. Pharmacol. 2024, 15, 1410479. [Google Scholar] [CrossRef]
- Campagna, R.; Mazzanti, L.; Pompei, V.; Alia, S.; Vignini, A.; Emanuelli, M. The Multifaceted Role of Endothelial Sirt1 in Vascular Aging: An Update. Cells 2024, 13, 1469. [Google Scholar] [CrossRef]
- Kim, H.C.; Mofarrahi, M.; Vassilakopoulos, T.; Maltais, F.; Sigala, I.; Debigare, R.; Bellenis, I.; Hussain, S.N.A. Expression and Functional Significance of Nicotinamide N-Methyl Transferase in Skeletal Muscles of Patients with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2010, 181, 797–805. [Google Scholar] [CrossRef]
- van Haren, M.J.; Gao, Y.; Buijs, N.; Campagna, R.; Sartini, D.; Emanuelli, M.; Mateuszuk, L.; Kij, A.; Chlopicki, S.; Escudé Martinez de Castilla, P.; et al. Esterase-Sensitive Prodrugs of a Potent Bisubstrate Inhibitor of Nicotinamide N-Methyltransferase (NNMT) Display Cellular Activity. Biomolecules 2021, 11, 1357. [Google Scholar] [CrossRef] [PubMed]
- van Haren, M.J.; Zhang, Y.; Thijssen, V.; Buijs, N.; Gao, Y.; Mateuszuk, L.; Fedak, F.A.; Kij, A.; Campagna, R.; Sartini, D.; et al. Macrocyclic Peptides as Allosteric Inhibitors of Nicotinamide N-Methyltransferase (NNMT). RSC Chem. Biol. 2021, 2, 1546–1555. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; van Haren, M.J.; Buijs, N.; Innocenti, P.; Zhang, Y.; Sartini, D.; Campagna, R.; Emanuelli, M.; Parsons, R.B.; Jespers, W.; et al. Potent Inhibition of Nicotinamide N-Methyltransferase by Alkene-Linked Bisubstrate Mimics Bearing Electron Deficient Aromatics. J. Med. Chem. 2021, 64, 12938–12963. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Sun, Y.; Nie, L.; Cui, A.; Zhao, P.; Leung, W.K.; Wang, Q. Metabolic Memory: Mechanisms and Diseases. Signal Transduct. Target. Ther. 2024, 9, 38. [Google Scholar] [CrossRef]
- Klisic, A.; Kavaric, N.; Ninic, A.; Kotur-Stevuljevic, J. Oxidative Stress and Cardiometabolic Biomarkers in Patients with Non-Alcoholic Fatty Liver Disease. Sci. Rep. 2021, 11, 18455. [Google Scholar] [CrossRef]
- Pia De La Maza, M.; Garrido, F.; Escalante, N.; Leiva, L.; Barrera, G.; Schnitzler, S.; Zanolli, M.; Verdaguer, J.; Hirsch, S.; Jara, N.; et al. Fluorescent Advanced Glycation End-Products (Ages) Detected by Spectro-Photofluorimetry, as a Screening Tool to Detect Diabetic Microvascular Complications. J. Diabetes Mellit. 2012, 02, 221–226. [Google Scholar] [CrossRef]
- Crescenti, A.; Solà, R.; Valls, R.M.; Caimari, A.; Del Bas, J.M.; Anguera, A.; Anglés, N.; Arola, L. Cocoa Consumption Alters the Global DNA Methylation of Peripheral Leukocytes in Humans with Cardiovascular Disease Risk Factors: A Randomized Controlled Trial. PLoS ONE 2013, 8, e65744. [Google Scholar] [CrossRef]
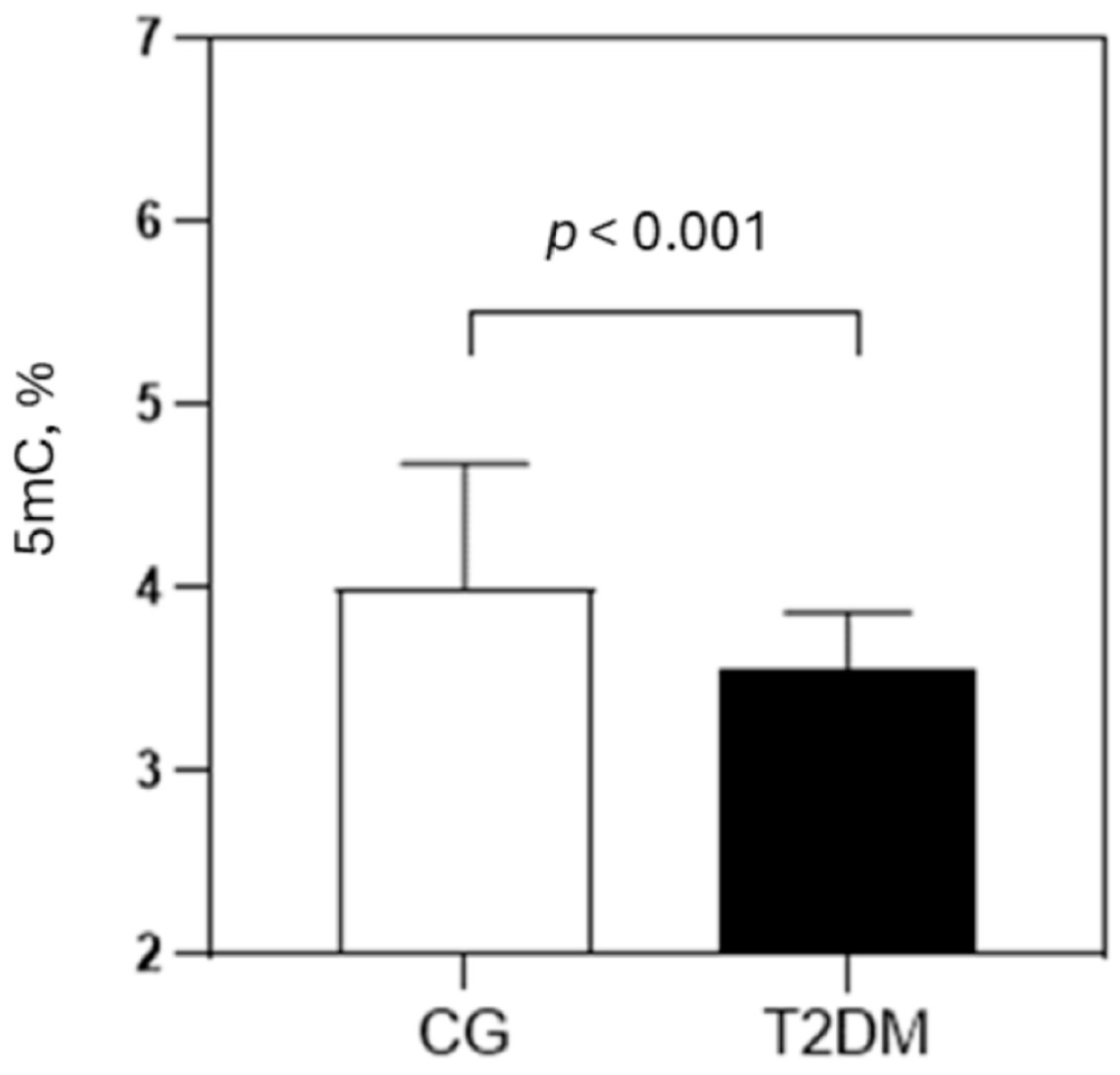
| Parameters | CG (N = 56) | T2DM (N = 107) | p |
|---|---|---|---|
| Age, years | 52.6 ± 9.3 (57) | 62.0 ± 11.3 (62.5) | <0.001 |
| Gender, m/f | 20/36 | 60/47 | 0.021 |
| BMI, kg/m2 | 25.6 ± 3.6 (25.9) | 31.5 ± 6.8 (29.6) | <0.001 |
| Obesity, % | 7.1 | 49.5 | <0.001 |
| Systolic blood pressure, mm Hg | 122.4 ± 20.3 (124) | 133.9 ± 20.7 (130) | 0.001 |
| Diastolic blood pressure, mm Hg | 80.5 ± 10.8 (85) | 80.5 ± 11.7 (80) | 0.990 |
| Hypertension, % | 35.7 | 88.7 | <0.001 |
| Smoking, % | 5.4 | 38.3 | <0.001 |
| Glucose, mmol/L | 5.23 ± 0.61 (5.20) | 10.60 ± 4.08 (10.05) | <0.001 |
| HbA1c, % | 3.4 ± 1.1 (3.2) | 9.9 ± 1.9 (10.0) | <0.001 |
| TC, mmol/L | 6.17 ± 1.14 (6.27) | 4.61 ± 1.21 (4.70) | <0.001 |
| LDL-C, mmol/L | 3.88 ± 1.01 (4.29) | 2.64 ± 0.99 (2.70) | <0.001 |
| HDL-C, mmol/L | 1.65 ± 0.39 (1.50) | 1.06 ± 0.39 (1.00) | <0.001 |
| TG, mmol/L # | 1.19 (0.98–1.54) | 1.75 (1.10–2.65) | <0.001 |
| Category of T2DM Patients | N | 5mC, % | p |
|---|---|---|---|
| Gender | |||
| Male | 60 | 3.56 ± 0.27 | 0.912 |
| Female | 47 | 3.56 ± 0.35 | |
| BMI | |||
| <25 kg/m2 | 54 | 3.53 ± 0.33 | 0.246 |
| ≥25 kg/m2 | 53 | 3.60 ± 0.29 | |
| Diabetes duration | |||
| <10 years | 69 | 3.62 ± 0.33 | 0.027 |
| ≥10 years | 38 | 3.48 ± 0.24 | |
| Smoking | |||
| No | 66 | 3.53 ± 0.28 | 0.090 |
| Yes | 41 | 3.64 ± 0.34 | |
| CVD | |||
| No | 59 | 3.58 ± 0.34 | 0.676 |
| Yes | 48 | 3.55 ± 0.27 | |
| Microvascular complications | |||
| No | 41 | 3.63 ± 0.36 | 0.060 |
| Yes | 66 | 3.51 ± 0.26 | |
| Neuropathy | |||
| No | 56 | 3.60 ± 0.36 | 0.236 |
| Yes | 51 | 3.52 ± 0.24 | |
| Retinopathy | |||
| No | 99 | 3.55 ± 0.32 | 0.324 |
| Yes | 8 | 3.67 ± 0.16 | |
| Nephropathy | |||
| No | 90 | 3.58 ± 0.31 | 0.097 |
| Yes | 17 | 3.45 ± 0.30 | |
| Diabetic foot ulcers | |||
| No | 82 | 3.60 ± 0.32 | 0.037 |
| Yes | 25 | 3.45 ± 0.26 |
| Biomarkers | CG (N = 56) | T2DM (N = 107) | p |
|---|---|---|---|
| Inflammatory biomarkers | |||
| hsCRP, mg/L | 0.30 (0.10–0.85) | 5.80 (1.85–13.55) | <0.001 |
| NLR | 1.95 (1.35–2.17) | 2.19 (1.63–2.93) | 0.013 |
| NWR | 0.60 (0.53–0.63) | 0.60 (0.54–0.67) | 0.407 |
| MLR | 0.16 (0.13–0.20) | 0.24 (0.20–0.31) | <0.001 |
| MWR | 0.05 (0.04–0.06) | 0.06 (0.05–0.08) | <0.001 |
| LWR | 0.31 (0.29–0.38) | 0.28 (0.23–0.34) | 0.001 |
| PLR | 106.90 (89.75–138.80) | 111.00 (91.10–150.70) | 0.326 |
| Pro-oxidant biomarkers | |||
| TOS, μmol/L | 4.5 (4.0–6.0) | 6.60 (4.7–8.0) | <0.001 |
| PAB, U/L | 74.24 (60.30–84.10) | 88.33 (67.60–127.20) | <0.001 |
| Oxidative damage biomarkers | |||
| IMA, AU | 0.28 (0.20–0.33) | 0.55 (0.47–0.62) | <0.001 |
| AOPP, μmol/L | 19.20 (18.25–27.35) | 48.90 (36.60–58.90) | <0.001 |
| Antioxidant defense biomarkers | |||
| TAS, μmol/L | 1290.0 (1217.0–1384.5) | 962.00 (714.0–1120.0) | <0.001 |
| SH-groups, mmol/L | 0.35 (0.31–0.39) | 0.25 (0.19–0.31) | <0.001 |
| SOD, U/L | 133 (124–141) | 136 (126–142) | 0.162 |
| 5mC, % | ||
|---|---|---|
| r | p | |
| Inflammatory biomarkers | ||
| hsCRP, mg/L | −0.388 | <0.001 |
| MLR | −0.174 | 0.035 |
| Redox biomarkers | ||
| PAB, U/L | −0.343 | <0.001 |
| IMA, AU | −0.412 | <0.001 |
| AOPP, μmol/L | −0.214 | 0.009 |
| TAS, μmol/L | 0.386 | <0.001 |
| SH-groups, mmol/L | 0.316 | <0.001 |
| Factors | Variables Included in the Factor | Loadings of the Variables | Factor Variability, % (Total Variance: 58%) |
|---|---|---|---|
| Proinflammatory factor | hsCRP, mg/L SOD, U/L NLR PAB, U/L IMA, AU | 0.774 −0.689 0.621 0.607 0.576 | 18 |
| Pro-oxidant factor | TOS, μmol/L AOPP, μmol/L BMI, kg/m2 | 0.708 0.643 0.599 | 12.5 |
| Ageing factor | Age, years Diabetes duration, years | 0.757 0.723 | 10.5 |
| Hyperglycemic factor | F-AGEs, U/mL Glucose, mmol/L Gender | 0.687 −0.544 0.525 | 9 |
| Antioxidant factor | SH-groups, mmol/L | 0.715 | 8 |
| Predictors | B (SE) | Wald Coefficient | OR (95% CI) | P |
|---|---|---|---|---|
| Proinflammatory factor | −0.777 (0.476) | 3.074 | 0.460 (0.193–1.096) | 0.080 |
| Prooxidant factor | 0.830 (0.377) | 4.863 | 2.294 (1.097–4.798) | 0.027 |
| Ageing factor | −0.120 (0.346) | 0.120 | 0.887 (0.450–1.749) | 0.730 |
| Hyperglycemic factor | 0.278 (0.410) | 0.460 | 1.321 (0.591–2.949) | 0.498 |
| Antioxidant factor | 0.173 (0.406) | 0.181 | 1.188 (0.537–2.631) | 0.671 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vujcic, S.; Kotur-Stevuljevic, J.; Vujcic, Z.; Stojanovic, S.; Beljic Zivkovic, T.; Vuksanovic, M.; Marjanovic Petkovic, M.; Perovic Blagojevic, I.; Koprivica-Uzelac, B.; Ilic-Mijailovic, S.; et al. Global DNA Methylation in Poorly Controlled Type 2 Diabetes Mellitus: Association with Redox and Inflammatory Biomarkers. Int. J. Mol. Sci. 2025, 26, 6716. https://doi.org/10.3390/ijms26146716
Vujcic S, Kotur-Stevuljevic J, Vujcic Z, Stojanovic S, Beljic Zivkovic T, Vuksanovic M, Marjanovic Petkovic M, Perovic Blagojevic I, Koprivica-Uzelac B, Ilic-Mijailovic S, et al. Global DNA Methylation in Poorly Controlled Type 2 Diabetes Mellitus: Association with Redox and Inflammatory Biomarkers. International Journal of Molecular Sciences. 2025; 26(14):6716. https://doi.org/10.3390/ijms26146716
Chicago/Turabian StyleVujcic, Sanja, Jelena Kotur-Stevuljevic, Zoran Vujcic, Sanja Stojanovic, Teodora Beljic Zivkovic, Miljanka Vuksanovic, Milica Marjanovic Petkovic, Iva Perovic Blagojevic, Branka Koprivica-Uzelac, Sanja Ilic-Mijailovic, and et al. 2025. "Global DNA Methylation in Poorly Controlled Type 2 Diabetes Mellitus: Association with Redox and Inflammatory Biomarkers" International Journal of Molecular Sciences 26, no. 14: 6716. https://doi.org/10.3390/ijms26146716
APA StyleVujcic, S., Kotur-Stevuljevic, J., Vujcic, Z., Stojanovic, S., Beljic Zivkovic, T., Vuksanovic, M., Marjanovic Petkovic, M., Perovic Blagojevic, I., Koprivica-Uzelac, B., Ilic-Mijailovic, S., Rizzo, M., Zeljkovic, A., Stefanovic, T., Bosic, S., & Vekic, J. (2025). Global DNA Methylation in Poorly Controlled Type 2 Diabetes Mellitus: Association with Redox and Inflammatory Biomarkers. International Journal of Molecular Sciences, 26(14), 6716. https://doi.org/10.3390/ijms26146716









