Genome-Wide Identification and Comprehensive Analysis of Ubiquitin-Specific Protease Gene Family in Soybean (Glycine max)
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of the UBP Genes in Soybean
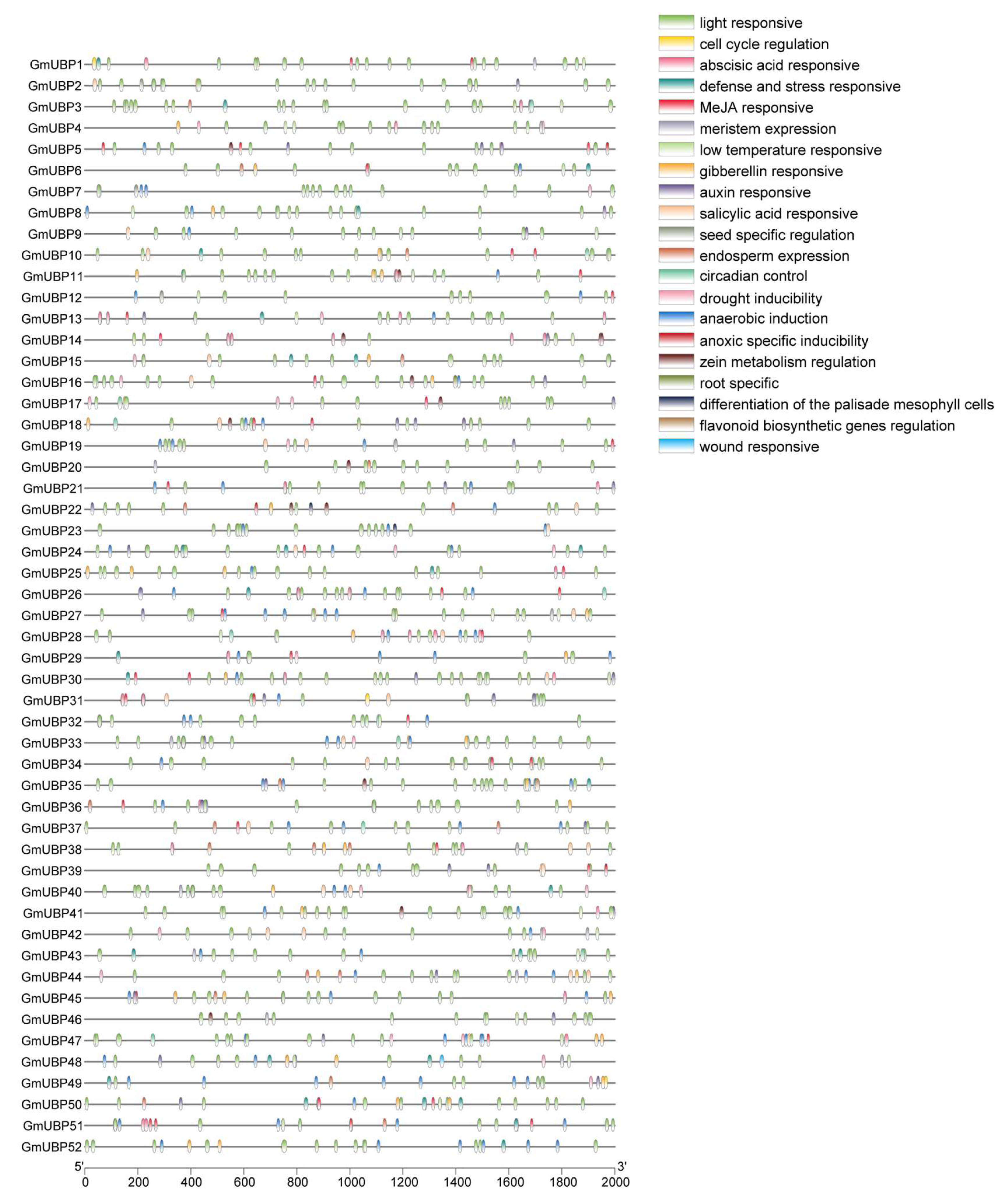
2.2. Gene Structures, Conserved Motifs, and cis-Acting Element Analysis of the GmUBP Genes
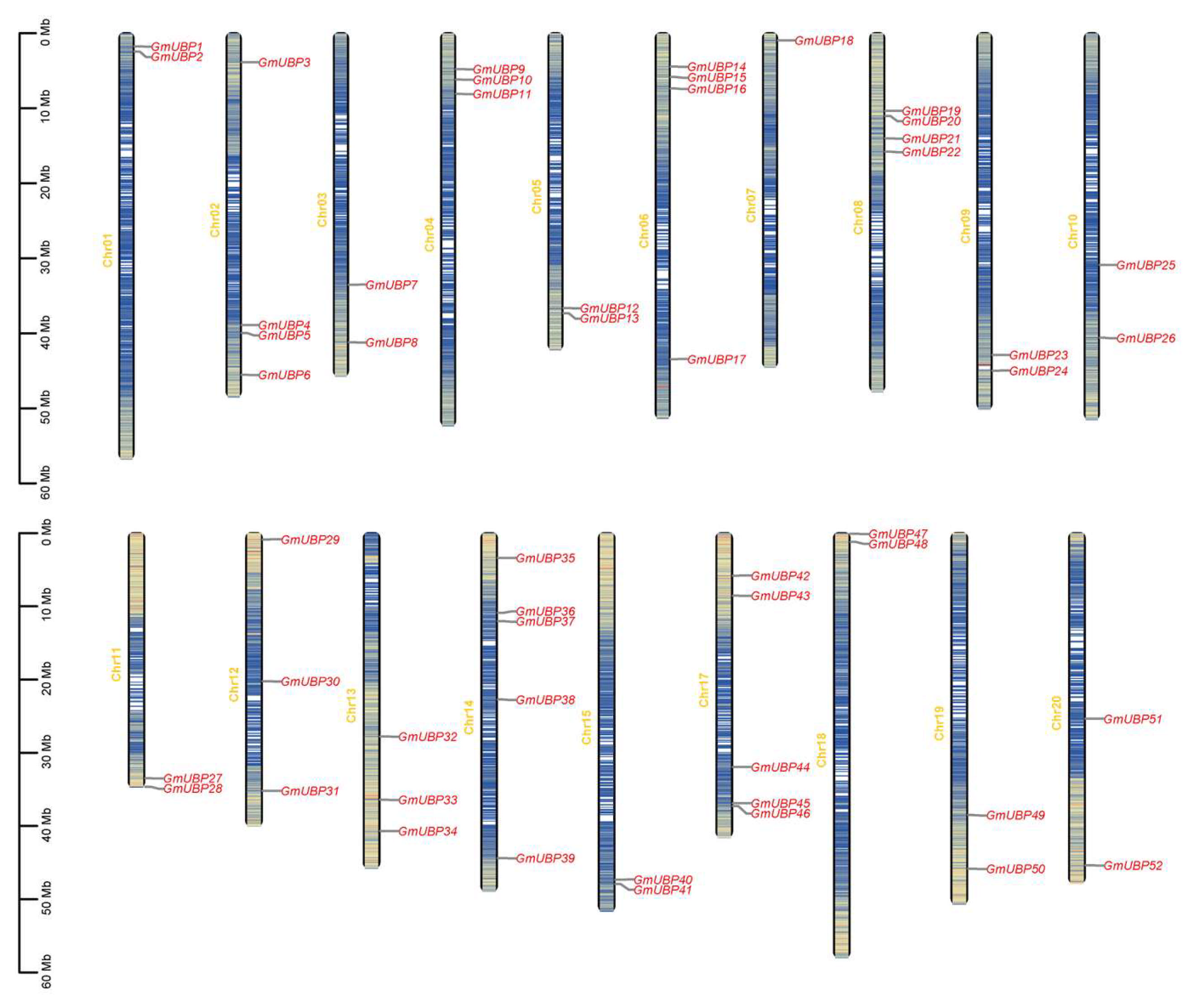
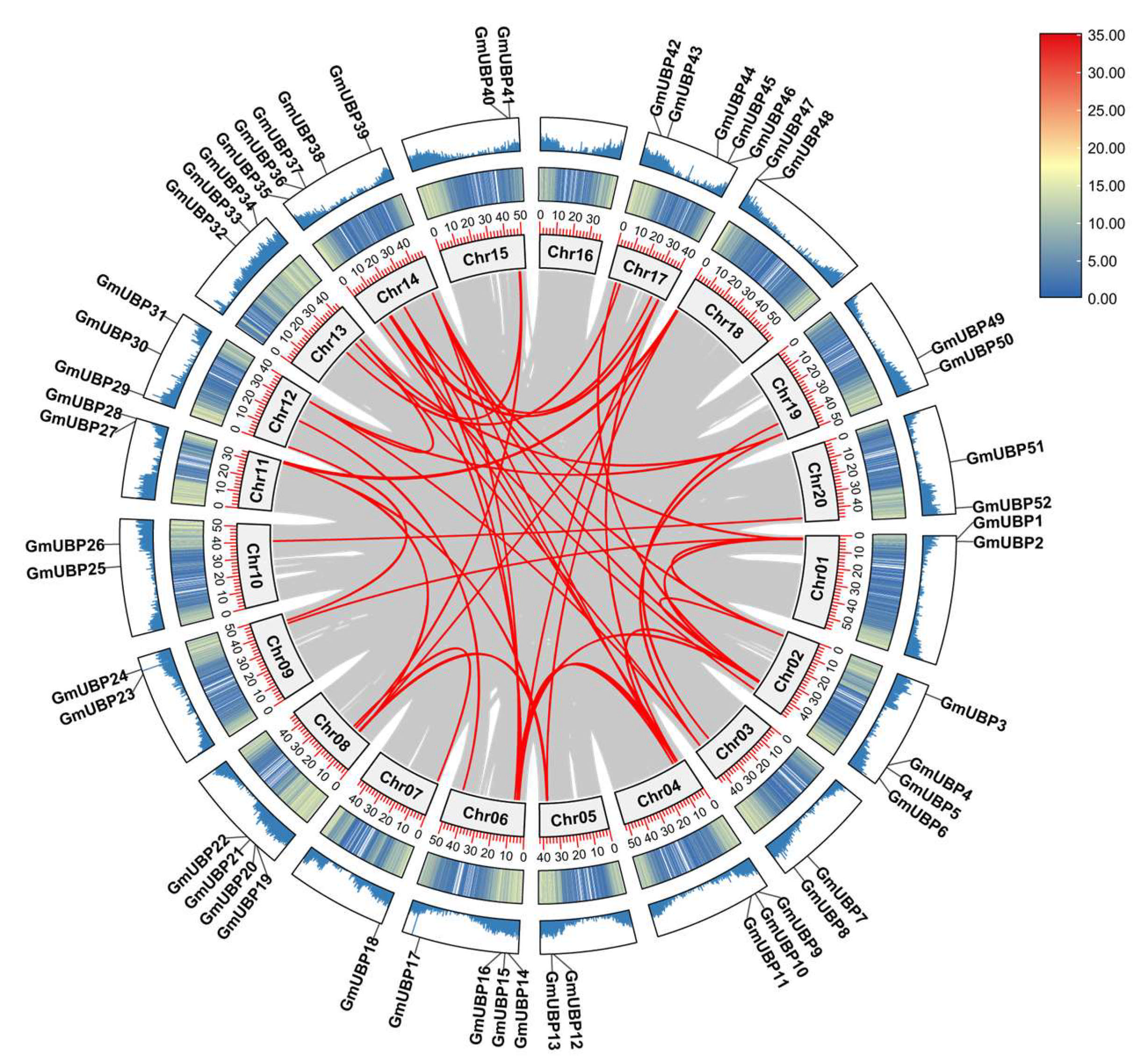
2.3. Visualization of Chromosomal Location and Duplication of GmUBPs
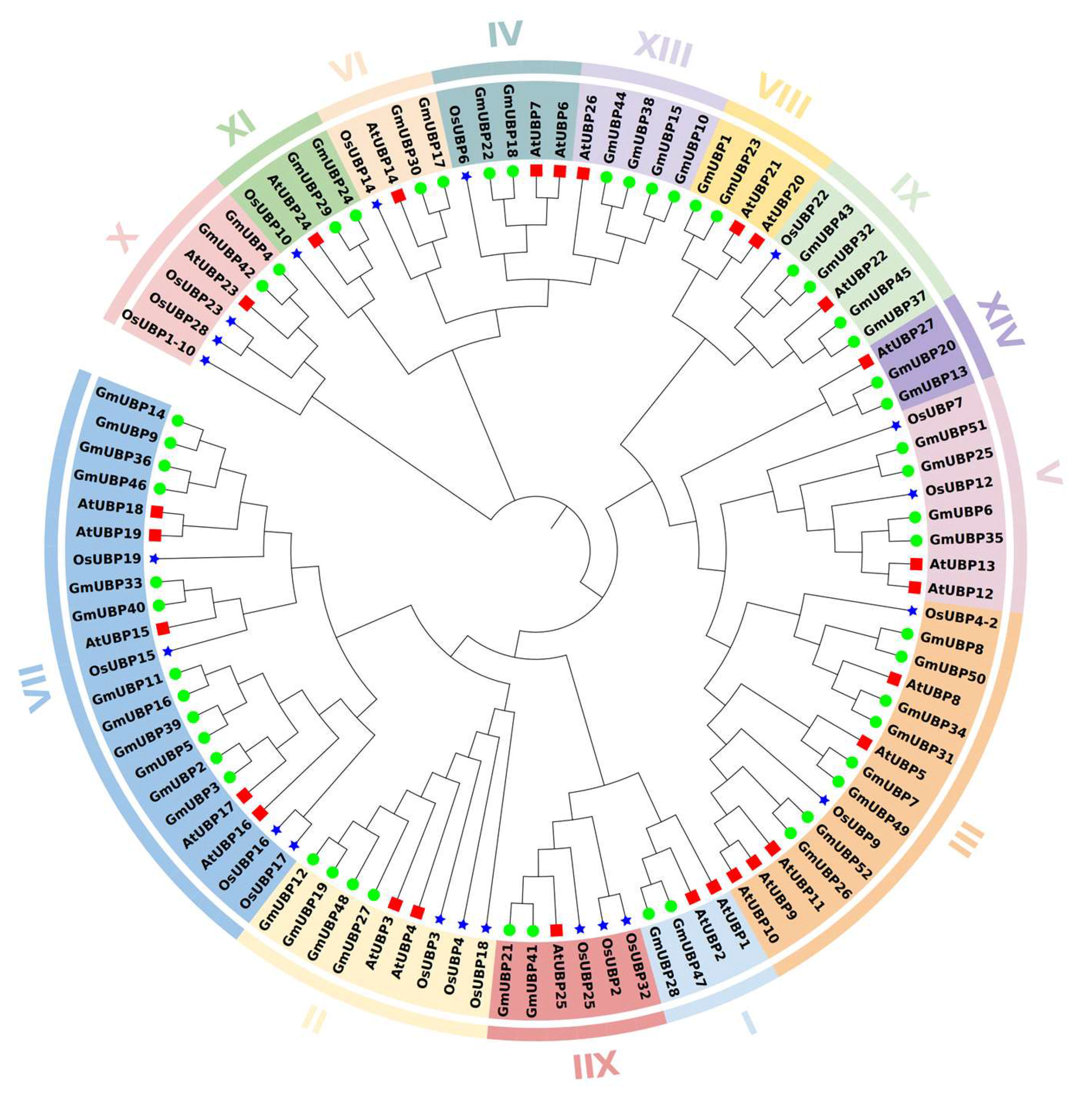
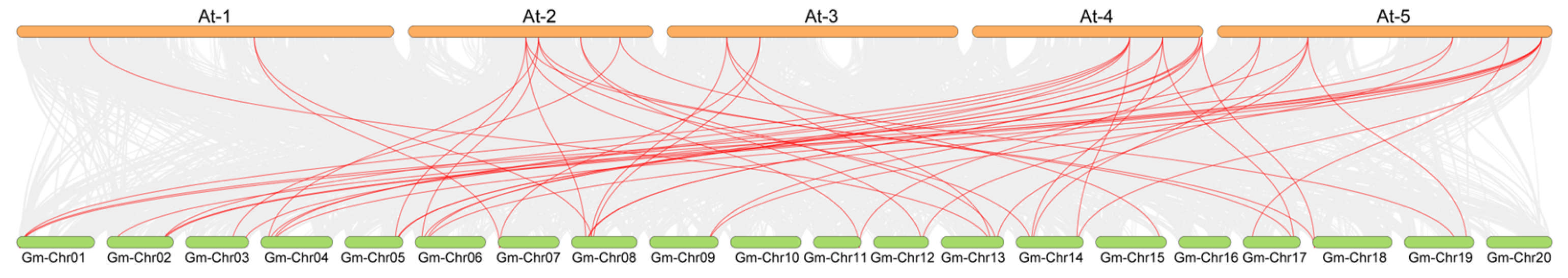
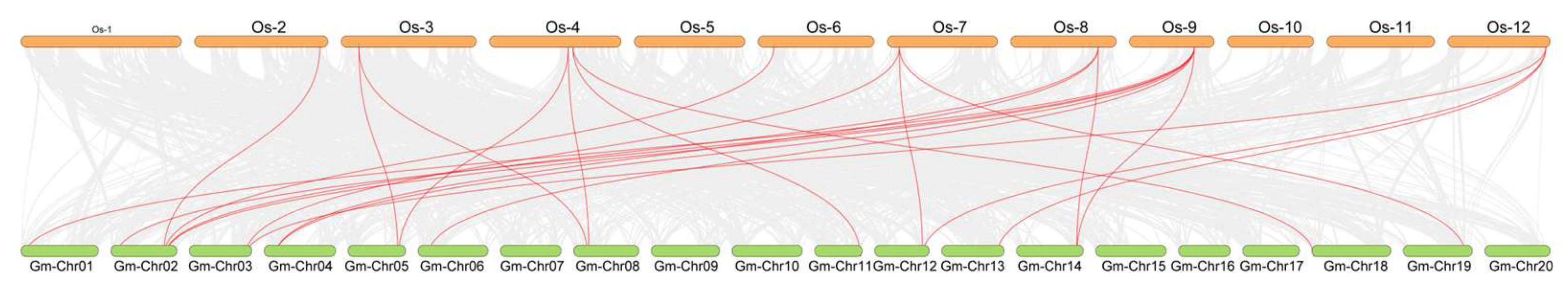
2.4. Phylogenetic Analysis and Collinearity Analysis of the UBP Genes Among Arabidopsis, Soybean, and Rice
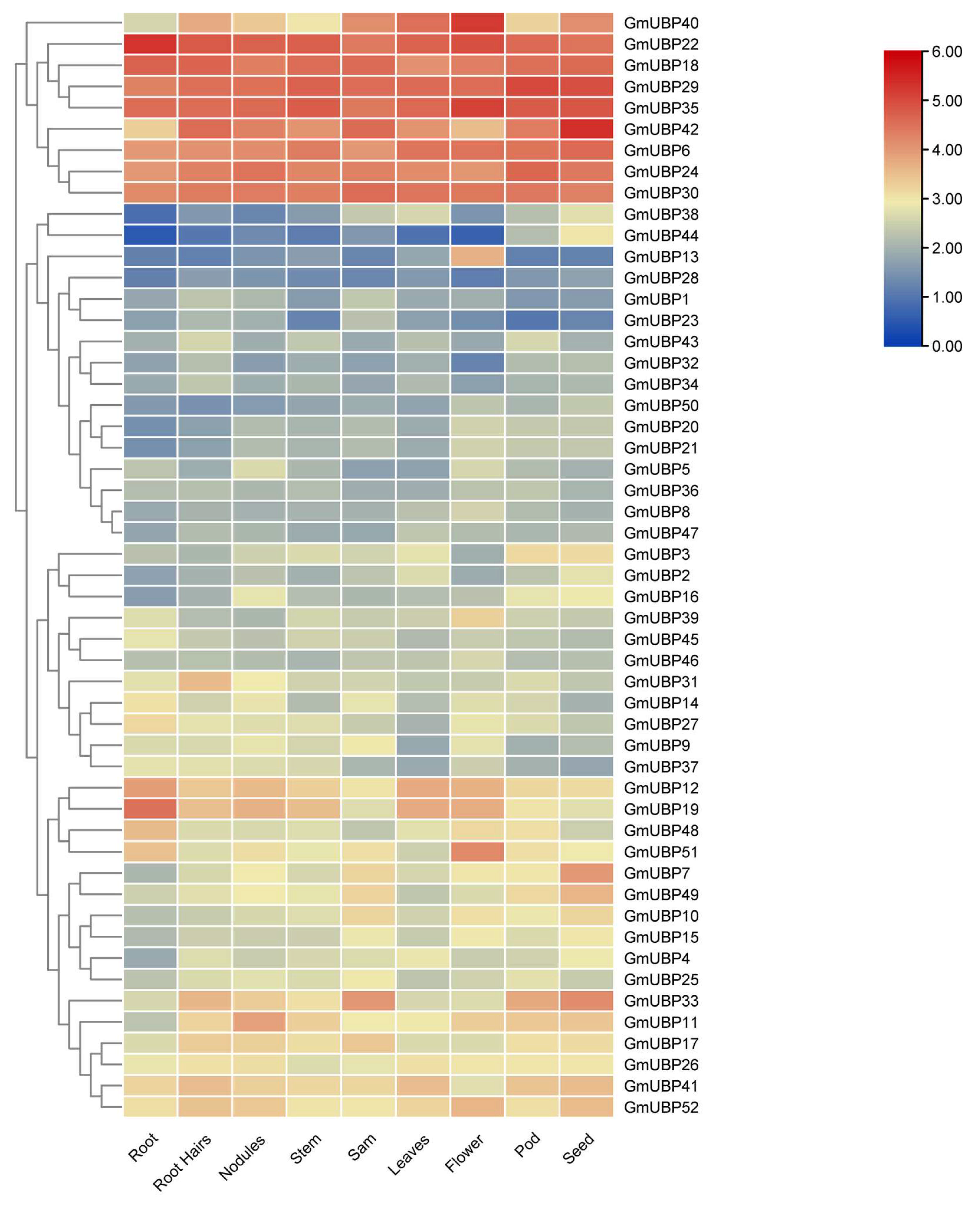
2.5. Expression Patterns of UBP Family in Different Tissues
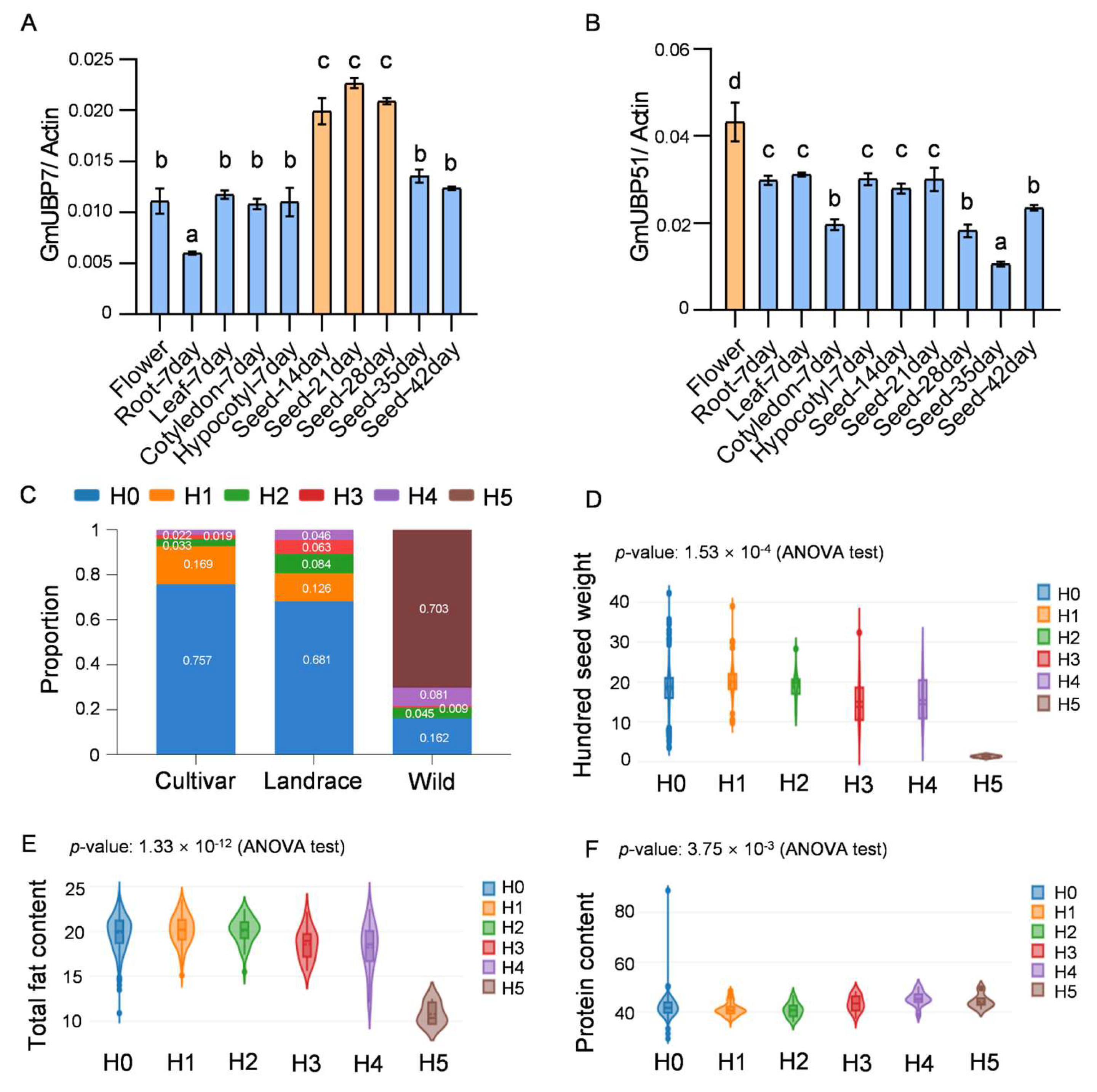
2.6. Functional Analysis of the GmUBP Family Genes
3. Discussion
4. Materials and Methods
4.1. Identification and Chromosomal Locations of Soybean UBP Genes
4.2. Chromosome Mapping and Phylogenetic Analysis
4.3. Gene Structure and Conserved Motif Analysis
4.4. Analysis of cis-Regulatory Elements of UBP Genes
4.5. Collinearity Analysis
4.6. Expression Analysis
4.7. Haplotype Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016, 33, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhao, J.; Cai, J.; Patil, S.B. UBIQUITIN-SPECIFIC PROTEASES function in plant development and stress responses. Plant Mol. Biol. 2017, 94, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Frappier, L.; Verrijzer, C.P. Gene expression control by protein deubiquitinases. Curr. Opin. Genet. Dev. 2011, 21, 207–213. [Google Scholar] [CrossRef]
- Zhang, Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003, 17, 2733–2740. [Google Scholar] [CrossRef]
- Atanassov, B.S.; Koutelou, E.; Dent, S.Y. The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett. 2011, 585, 2016–2023. [Google Scholar] [CrossRef]
- Neutzner, M.; Neutzner, A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012, 52, 37–50. [Google Scholar]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 2004, 1695, 189–207. [Google Scholar] [CrossRef]
- Wu, R.; Zheng, W.; Tan, J.; Sammer, R.; Du, L.; Lu, C. Protein partners of plant ubiquitin-specific proteases (UBPs). Plant Physiol. Biochem. 2019, 145, 227–236. [Google Scholar] [CrossRef]
- Yan, N.; Doelling, J.H.; Falbel, T.G.; Durski, A.M.; Vierstra, R.D. The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 2000, 124, 1828–1843. [Google Scholar] [CrossRef]
- Wang, D.H.; Song, W.; Wei, S.W.; Zheng, Y.F.; Chen, Z.S.; Han, J.D.; Zhang, H.T.; Luo, J.C.; Qin, Y.M.; Xu, Z.H.; et al. Characterization of the Ubiquitin C-Terminal Hydrolase and Ubiquitin-Specific Protease Families in Rice (Oryza sativa). Front. Plant Sci. 2018, 9, 1636. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Shi, Y.; Zhang, Q.; Zheng, W.; Chen, S.; Du, L.; Lu, C. Genome-Wide Identification and Characterization of the UBP Gene Family in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2019, 20, 4309. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Jin, P.; Liu, T.; Gao, S.; Zhang, T.; Zhang, F.; Han, X.; He, L.; Chen, J.; Yang, J. Genome-wide identification and characterization of UBP gene family in wheat (Triticum aestivum L.). PeerJ 2021, 9, e11594. [Google Scholar] [CrossRef]
- Fu, W.; Fan, D.; Liu, S.; Bu, Y. Genome-wide identification and expression analysis of Ubiquitin-specific protease gene family in maize (Zea mays L.). BMC Plant Biol. 2024, 24, 404. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, H.; Zhang, M.; Gao, Y.; Li, L.; Gao, Y.; Li, M.; Yang, Y.; Guo, Y.; Li, X. Ubiquitin-specific protease 24 negatively regulates abscisic acid signalling in Arabidopsis thaliana. Plant Cell Environ. 2016, 39, 427–440. [Google Scholar] [CrossRef]
- Liu, G.; Liang, J.; Lou, L.; Tian, M.; Zhang, X.; Liu, L.; Zhao, Q.; Xia, R.; Wu, Y.; Xie, Q.; et al. The deubiquitinases UBP12 and UBP13 integrate with the E3 ubiquitin ligase XBAT35.2 to modulate VPS23A stability in ABA signaling. Sci. Adv. 2022, 8, eabl5765. [Google Scholar] [CrossRef]
- Zhou, Y.; Park, S.H.; Chua, N.H. UBP12/UBP13-mediated deubiquitination of salicylic acid receptor NPR3 suppresses plant immunity. Mol. Plant 2023, 16, 232–244. [Google Scholar] [CrossRef]
- Zhou, H.; Zhao, J.; Yang, Y.; Chen, C.; Liu, Y.; Jin, X.; Chen, L.; Li, X.; Deng, X.W.; Schumaker, K.S.; et al. Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell 2012, 24, 5106–5122. [Google Scholar] [CrossRef]
- Jiang, R.; Zhou, S.; Da, X.; Chen, T.; Xu, J.; Yan, P.; Mo, X. Ubiquitin-Specific Protease 2 (OsUBP2) Negatively Regulates Cell Death and Disease Resistance in Rice. Plants 2022, 11, 2568. [Google Scholar] [CrossRef]
- Park, S.H.; Jeong, J.S.; Seo, J.S.; Park, B.S.; Chua, N.H. Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol. 2019, 223, 1447–1460. [Google Scholar] [CrossRef]
- An, Z.; Liu, Y.; Ou, Y.; Li, J.; Zhang, B.; Sun, D.; Sun, Y.; Tang, W. Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc. Natl. Acad. Sci. USA 2018, 115, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lu, F.; Li, Y.; Xue, Y.; Kang, Y.; Zhang, S.; Qiu, Q.; Cui, X.; Zheng, S.; Liu, B.; et al. Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 2013, 162, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, N.; Chen, L.; Xu, Y.; Li, Y.; Zhang, Y.; Li, C.; Li, Y. The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 2014, 26, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Ren, Y.; Liu, L.; Wang, F.; Zhang, H.; Tian, P.; Pan, T.; Wang, Y.; Jing, R.; Liu, T.; et al. Ubiquitin Specific Protease 15 Has an Important Role in Regulating Grain Width and Size in Rice. Plant Physiol. 2019, 180, 381–391. [Google Scholar] [CrossRef]
- Doelling, J.H.; Phillips, A.R.; Soyler-Ogretim, G.; Wise, J.; Chandler, J.; Callis, J.; Otegui, M.S.; Vierstra, R.D. The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 2007, 145, 801–813. [Google Scholar] [CrossRef]
- Doelling, J.H.; Yan, N.; Kurepa, J.; Walker, J.; Vierstra, R.D. The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 2001, 27, 393–405. [Google Scholar] [CrossRef]
- Luo, M.; Luo, M.Z.; Buzas, D.; Finnegan, J.; Helliwell, C.; Dennis, E.S.; Peacock, W.J.; Chaudhury, A. UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 2008, 180, 229–236. [Google Scholar] [CrossRef][Green Version]
- Sridhar, V.V.; Kapoor, A.; Zhang, K.; Zhu, J.; Zhou, T.; Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 2007, 447, 735–738. [Google Scholar] [CrossRef]
- Zou, T.; Li, G.; Liu, M.; Liu, R.; Yang, S.; Wang, K.; Lu, L.; Ye, Q.; Liu, J.; Liang, J.; et al. A ubiquitin-specific protease functions in regulating cell death and immune responses in rice. Plant Cell Environ. 2023, 46, 1312–1326. [Google Scholar] [CrossRef]
- Sun, J.; Song, W.; Chang, Y.; Wang, Y.; Lu, T.; Zhang, Z. OsLMP1, Encoding a Deubiquitinase, Regulates the Immune Response in Rice. Front. Plant Sci. 2021, 12, 814465. [Google Scholar] [CrossRef]
- Moon, Y.K.; Hong, J.P.; Cho, Y.C.; Yang, S.J.; An, G.; Kim, W.T. Structure and expression of OsUBP6, an ubiquitin-specific protease 6 homolog in rice (Oryza sativa L.). Mol. Cells 2009, 28, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Jin, J.; Dong, Q.; Qiu, J.; Li, Y.; Yang, Y.; Shi, Y.; Si, W.; Gu, L.; Yang, F.; et al. Maize factors ZmUBP15, ZmUBP16 and ZmUBP19 play important roles for plants to tolerance the cadmium stress and salt stress. Plant Sci. 2019, 280, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Duan, Z.; He, X.; Yang, X.; Yuan, Y.; Liang, Q.; Pan, Y.; Zhou, G.; Zhang, M.; Liu, S.; et al. Publisher Correction: Natural variation in GmSW17 controls seed size in soybean. Nat. Commun. 2024, 15, 8250. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Moore, B.M.; Panchy, N.L.; Meng, F.; Lehti-Shiu, M.D.; Shiu, S.H. Factors Influencing Gene Family Size Variation Among Related Species in a Plant Family, Solanaceae. Genome Biol. Evol. 2018, 10, 2596–2613. [Google Scholar] [CrossRef]
- Zheng, L.; Wu, H.; Qanmber, G.; Ali, F.; Wang, L.; Liu, Z.; Yu, D.; Wang, Q.; Xu, A.; Yang, Z. Genome-Wide Study of the GATL Gene Family in Gossypium hirsutum L. Reveals that GhGATL Genes Act on Pectin Synthesis to Regulate Plant Growth and Fiber Elongation. Genes 2020, 11, 64. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, F.; Zhang, H.; He, H.; Ma, L.; Deng, X.W. Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 2008, 55, 844–856. [Google Scholar] [CrossRef]
- March, E.; Farrona, S. Plant Deubiquitinases and Their Role in the Control of Gene Expression Through Modification of Histones. Front. Plant Sci. 2017, 8, 2274. [Google Scholar] [CrossRef]
- Ewan, R.; Pangestuti, R.; Thornber, S.; Craig, A.; Carr, C.; O’Donnell, L.; Zhang, C.; Sadanandom, A. Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 2011, 191, 92–106. [Google Scholar] [CrossRef]
- Godwin, J.; Govindasamy, M.; Nedounsejian, K.; March, E.; Halton, R.; Bourbousse, C.; Wolff, L.; Fort, A.; Krzyszton, M.; López Corrales, J.; et al. The UBP5 histone H2A deubiquitinase counteracts PRCs-mediated repression to regulate Arabidopsis development. Nat. Commun. 2024, 15, 667. [Google Scholar] [CrossRef]
- Walling, J.G.; Shoemaker, R.; Young, N.; Mudge, J.; Jackson, S. Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 2006, 172, 1893–1900. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Cannon, S.B.; Graham, M.M.; Grant, D.; Shoemaker, R.C. Changes in twelve homoeologous genomic regions in soybean following three rounds of polyploidy. Plant Cell 2011, 23, 3129–3136. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Shoemaker, R.C. Evolutionary and comparative analyses of the soybean genome. Breed. Sci. 2012, 61, 437–444. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Wang, W.; Jiang, W.; Liu, J.; Li, Y.; Gai, J.; Li, Y. Genome-wide characterization of the aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response. BMC Genom. 2017, 18, 518. [Google Scholar] [CrossRef]
- Casal, J.J.; Yanovsky, M.J. Regulation of gene expression by light. Int. J. Dev. Biol. 2005, 49, 501–511. [Google Scholar] [CrossRef]
- Mohanty, B.; Krishnan, S.P.; Swarup, S.; Bajic, V.B. Detection and preliminary analysis of motifs in promoters of anaerobically induced genes of different plant species. Ann. Bot. 2005, 96, 669–681. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176.e13. [Google Scholar] [CrossRef]
- Sedivy, E.J.; Wu, F.; Hanzawa, Y. Soybean domestication: The origin, genetic architecture and molecular bases. New Phytol. 2017, 214, 539–553. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Erratum: Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2016, 34, 441. [Google Scholar] [CrossRef]
- Aguilar-Hernández, V.; Guzmán, P. Spliceosomal introns in the 5′ untranslated region of plant BTL RING-H2 ubiquitin ligases are evolutionary conserved and required for gene expression. BMC Plant Biol. 2013, 13, 179. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.B.; Carter, A.; Korf, I.; Kojima, N. Intron sequences that stimulate gene expression in Arabidopsis. Plant Mol. Biol. 2016, 92, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, M.; Li, S.; Sun, L.; Wang, W.; Cai, C.; Dierking, E.C.; Ma, J. Plasticity and innovation of regulatory mechanisms underlying seed oil content mediated by duplicated genes in the palaeopolyploid soybean. Plant J. 2017, 90, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wei, W.; Li, Q.T.; Bian, X.H.; Lu, X.; Hu, Y.; Cheng, T.; Wang, Z.Y.; Jin, M.; Tao, J.J.; et al. A transcriptional regulatory module controls lipid accumulation in soybean. New Phytol. 2021, 231, 661–678. [Google Scholar] [CrossRef]
- O’Conner, S.; Zheng, W.; Qi, M.; Kandel, Y.; Fuller, R.; Whitham, S.A.; Li, L. GmNF-YC4-2 Increases Protein, Exhibits Broad Disease Resistance and Expedites Maturity in Soybean. Int. J. Mol. Sci. 2021, 22, 3586. [Google Scholar] [CrossRef]
- Zhang, M.; Zheng, H.; Jin, L.; Xing, L.; Zou, J.; Zhang, L.; Liu, C.; Chu, J.; Xu, M.; Wang, L. miR169o and ZmNF-YA13 act in concert to coordinate the expression of ZmYUC1 that determines seed size and weight in maize kernels. New Phytol. 2022, 235, 2270–2284. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, C.; Ban, D.; Li, H.; Wang, J.; Liu, B.; Zhang, C. Genome-Wide Identification and Comprehensive Analysis of Ubiquitin-Specific Protease Gene Family in Soybean (Glycine max). Int. J. Mol. Sci. 2025, 26, 6689. https://doi.org/10.3390/ijms26146689
Tan C, Ban D, Li H, Wang J, Liu B, Zhang C. Genome-Wide Identification and Comprehensive Analysis of Ubiquitin-Specific Protease Gene Family in Soybean (Glycine max). International Journal of Molecular Sciences. 2025; 26(14):6689. https://doi.org/10.3390/ijms26146689
Chicago/Turabian StyleTan, Cuirong, Dingyue Ban, Haiyang Li, Jinxing Wang, Baohui Liu, and Chunyu Zhang. 2025. "Genome-Wide Identification and Comprehensive Analysis of Ubiquitin-Specific Protease Gene Family in Soybean (Glycine max)" International Journal of Molecular Sciences 26, no. 14: 6689. https://doi.org/10.3390/ijms26146689
APA StyleTan, C., Ban, D., Li, H., Wang, J., Liu, B., & Zhang, C. (2025). Genome-Wide Identification and Comprehensive Analysis of Ubiquitin-Specific Protease Gene Family in Soybean (Glycine max). International Journal of Molecular Sciences, 26(14), 6689. https://doi.org/10.3390/ijms26146689





