Overexpression of Circular PRMT1 Transcripts in Colorectal Adenocarcinoma Predicts Recurrence and Poor Overall Survival
Abstract
1. Introduction
2. Results
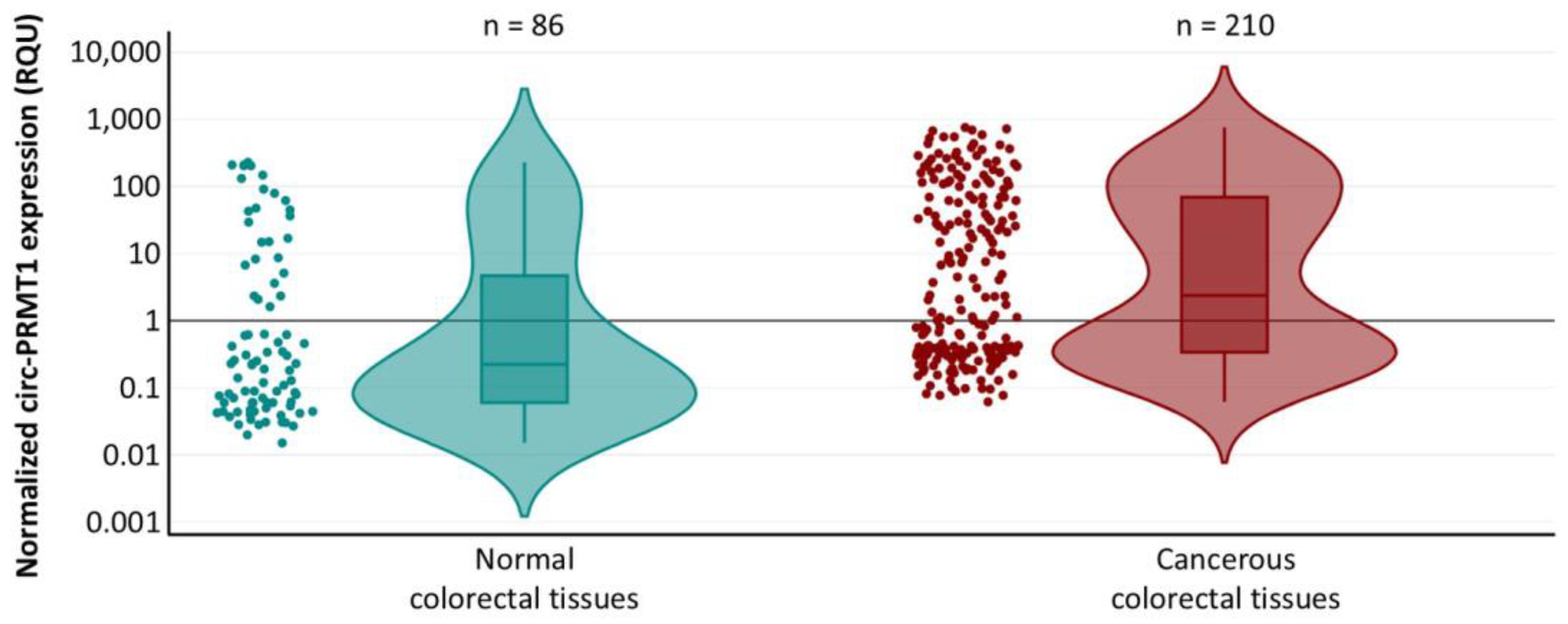
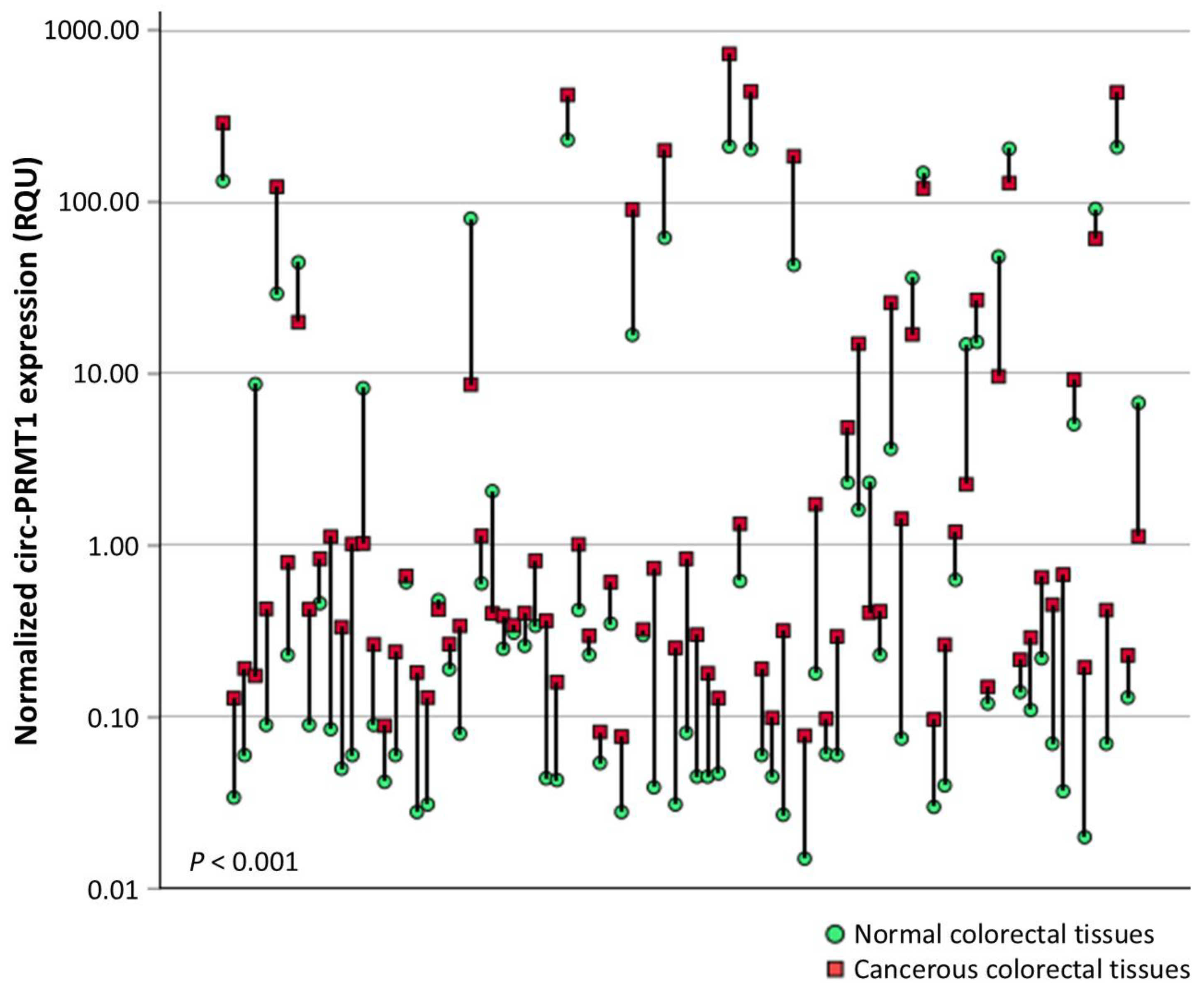
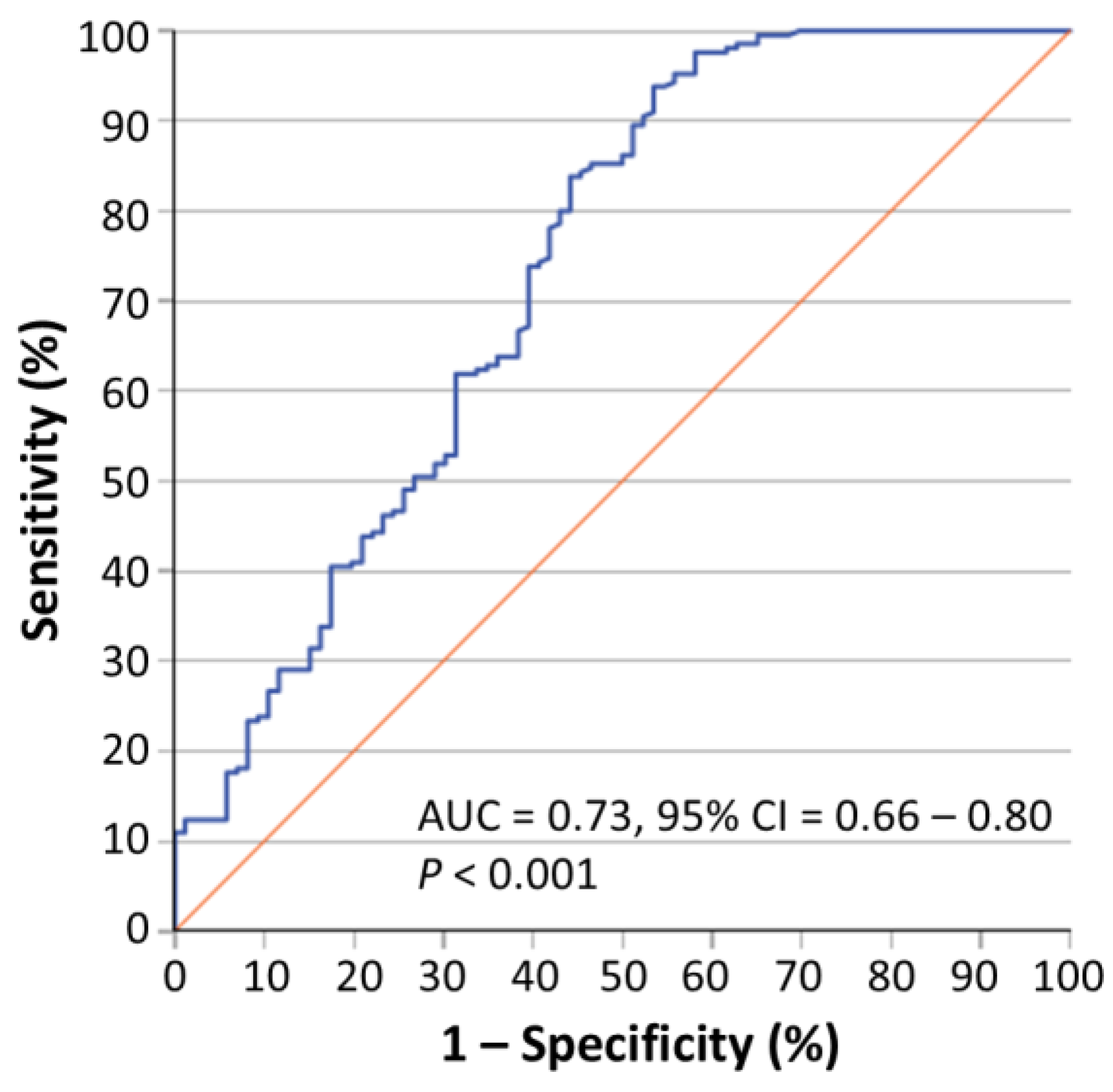
2.1. Circ-PRMT1 Expression Is Higher in Colorectal Adenocarcinoma Tissue Specimens than in Their Adjacent Non-Cancerous Tissues
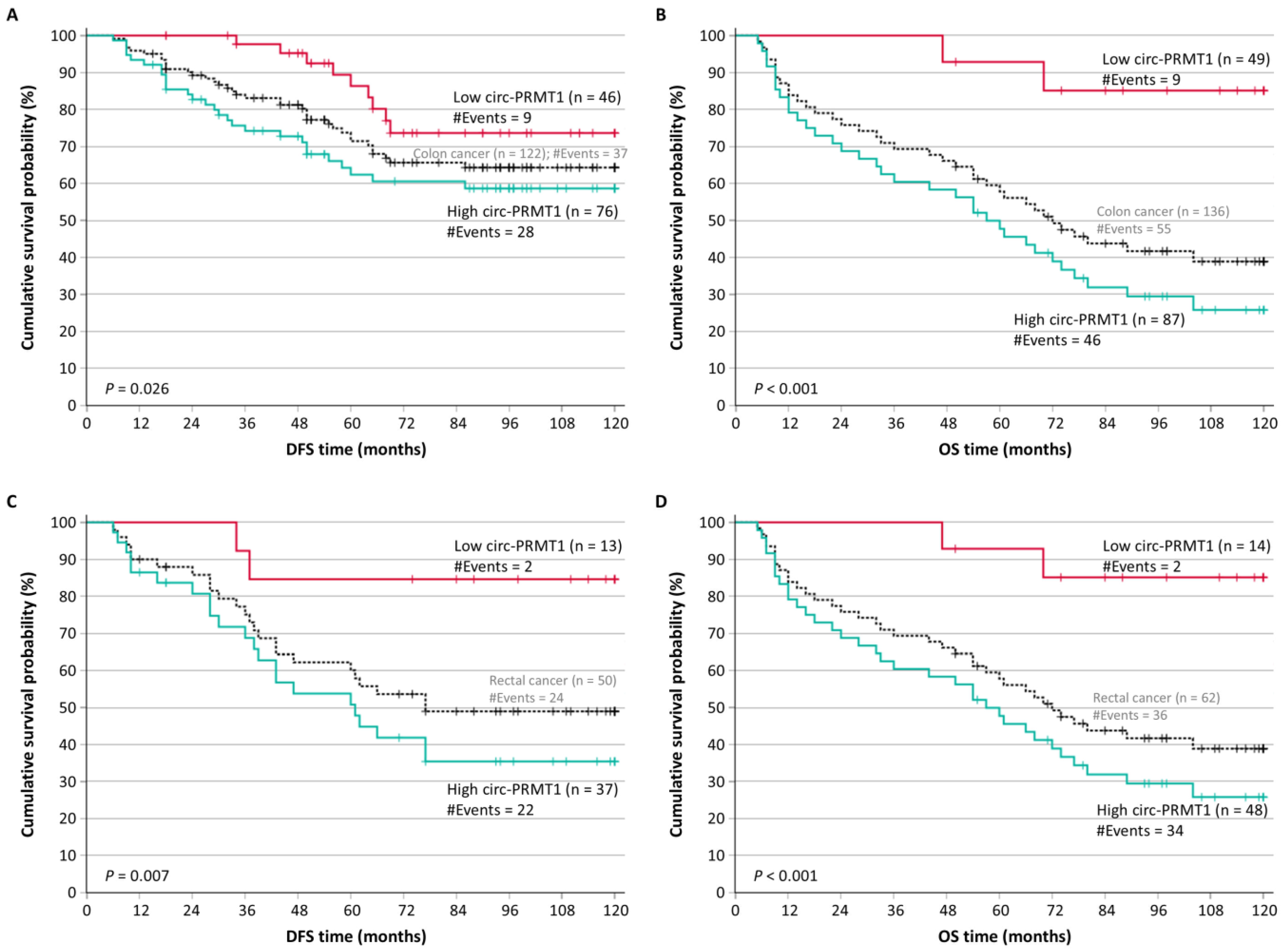
2.2. Circ-PRMT1 Overexpression Is an Unfavorable Prognosticator of Patients’ Outcome
2.3. Circ-PRMT1 Overexpression Retains Its Unfavorable Prognostic Significance Independently of Other Established Prognostic Factors
2.4. Circ-PRMT1 Is an Unfavorable Prognostic Marker in Distinct Subgroups of Colorectal Adenocarcinoma Patients
3. Discussion
4. Materials and Methods
4.1. Sample and Clinocopathological Data Collection
4.2. Total RNA Isolation and Reverse Transcription
4.3. Primer Designing and Real-Time Quantitative Polymerase Chain Reaction (qPCR)
4.4. Biostatistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of Colorectal Carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Argiles, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef] [PubMed]
- Glynne-Jones, R.; Wyrwicz, L.; Tiret, E.; Brown, G.; Rodel, C.; Cervantes, A.; Arnold, D.; Committee, E.G. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv263. [Google Scholar] [CrossRef]
- Cervantes, A.; Adam, R.; Rosello, S.; Arnold, D.; Normanno, N.; Taieb, J.; Seligmann, J.; De Baere, T.; Osterlund, P.; Yoshino, T.; et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 10–32. [Google Scholar] [CrossRef]
- Seligmann, J.F. Colorectal cancer staging-time for a re-think on TNM? Br. J. Surg. 2025, 112, znaf047. [Google Scholar] [CrossRef]
- Koncina, E.; Haan, S.; Rauh, S.; Letellier, E. Prognostic and Predictive Molecular Biomarkers for Colorectal Cancer: Updates and Challenges. Cancers 2020, 12, 319. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Malla, M.; Loree, J.M.; Kasi, P.M.; Parikh, A.R. Using Circulating Tumor DNA in Colorectal Cancer: Current and Evolving Practices. J. Clin. Oncol. 2022, 40, 2846–2857. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, M.; Liu, X.; Huang, Y.; Liu, C.; Zhu, J.; Fu, G.; Lei, Z.; Chu, X. Alternative splicing of mRNA in colorectal cancer: New strategies for tumor diagnosis and treatment. Cell Death Dis. 2021, 12, 752. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Kuo, H.C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Liu, H.S.; Wang, F.W.; Hu, T.; Liang, Z.X.; Lan, N.; He, X.W.; Zheng, X.B.; Wu, X.J.; Xie, D.; et al. circCAMSAP1 Promotes Tumor Growth in Colorectal Cancer via the miR-328-5p/E2F1 Axis. Mol. Ther. 2020, 28, 914–928. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; He, Y.; Xiao, Z.; Xin, L.; Deng, M.; Yao, M.; Huang, G. circEIF3I Promotes Colorectal Cancer Metastasis by Regulating the miR-328-3p/NCAPH Axis. Mol. Carcinog. 2025, 64, 450–462. [Google Scholar] [CrossRef]
- Fan, B.; Zheng, C.; Wang, N.; Chang, Z.; Liu, Y.; Wang, C.; Xiang, J.; Tao, Y.; Wang, G.; Zhang, Q. CircSTK3 drives the metastasis of colorectal cancer by regulating epithelial-mesenchymal transition. iScience 2023, 26, 106170. [Google Scholar] [CrossRef]
- Deng, J.; Liao, S.; Chen, C.; Han, F.; Lei, S.; Lai, X.; Ye, K.; Han, Q.; E, F.; Lu, C.; et al. Specific intracellular retention of circSKA3 promotes colorectal cancer metastasis by attenuating ubiquitination and degradation of SLUG. Cell Death Dis. 2023, 14, 750. [Google Scholar] [CrossRef]
- Shen, S.; Zhou, H.; Xiao, Z.; Zhan, S.; Tuo, Y.; Chen, D.; Pang, X.; Wang, Y.; Wang, J. PRMT1 in human neoplasm: Cancer biology and potential therapeutic target. Cell Commun. Signal. 2024, 22, 102. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Xu, P.; Li, X.; Lu, Y.; Jin, D.; Yin, X.; Jiang, H.; Huang, J.; Xiong, H.; et al. PRMT1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Theranostics 2021, 11, 5387–5403. [Google Scholar] [CrossRef]
- Altan, B.; Yokobori, T.; Ide, M.; Mochiki, E.; Toyomasu, Y.; Kogure, N.; Kimura, A.; Hara, K.; Bai, T.; Bao, P.; et al. Nuclear PRMT1 expression is associated with poor prognosis and chemosensitivity in gastric cancer patients. Gastric Cancer 2016, 19, 789–797. [Google Scholar] [CrossRef]
- Mathioudaki, K.; Scorilas, A.; Ardavanis, A.; Lymberi, P.; Tsiambas, E.; Devetzi, M.; Apostolaki, A.; Talieri, M. Clinical evaluation of PRMT1 gene expression in breast cancer. Tumour Biol. 2011, 32, 575–582. [Google Scholar] [CrossRef]
- Papadokostopoulou, A.; Mathioudaki, K.; Scorilas, A.; Xynopoulos, D.; Ardavanis, A.; Kouroumalis, E.; Talieri, M. Colon cancer and protein arginine methyltransferase 1 gene expression. Anticancer Res. 2009, 29, 1361–1366. Available online: https://ar.iiarjournals.org/content/29/4/1361.long (accessed on 7 July 2025). [PubMed]
- Wu, J.; Li, D.; Wang, L. Overview of PRMT1 modulators: Inhibitors and degraders. Eur. J. Med. Chem. 2024, 279, 116887. [Google Scholar] [CrossRef] [PubMed]
- Papatsirou, M.; Scorilas, A.; Sideris, D.C.; Kontos, C.K. Targeted nanopore sequencing for the identification of novel PRMT1 circRNAs unveils a diverse transcriptional profile of this gene in breast cancer cells. Genes Dis. 2024, 11, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Papatsirou, M.; Diamantopoulos, M.A.; Katsaraki, K.; Kletsas, D.; Kontos, C.K.; Scorilas, A. Identification of Novel Circular RNAs of the Human Protein Arginine Methyltransferase 1 (PRMT1) Gene, Expressed in Breast Cancer Cells. Genes 2022, 13, 1133. [Google Scholar] [CrossRef]
- Mathioudaki, K.; Papadokostopoulou, A.; Scorilas, A.; Xynopoulos, D.; Agnanti, N.; Talieri, M. The PRMT1 gene expression pattern in colon cancer. Br. J. Cancer 2008, 99, 2094–2099. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, J.; Yang, W.; Ye, W.C. CircRNAs in colorectal cancer: Potential biomarkers and therapeutic targets. Cell Death Dis. 2023, 14, 353. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Yan, M.; Li, H. Emerging Role of Circular RNAs in Cancer. Front. Oncol. 2020, 10, 663. [Google Scholar] [CrossRef]
- Ruan, H.; Wang, P.C.; Han, L. Characterization of circular RNAs with advanced sequencing technologies in human complex diseases. Wiley Interdiscip. Rev. RNA 2023, 14, e1759. [Google Scholar] [CrossRef]
- Xue, L.; Bao, L.; Roediger, J.; Su, Y.; Shi, B.; Shi, Y.B. Protein arginine methyltransferase 1 regulates cell proliferation and differentiation in adult mouse adult intestine. Cell Biosci. 2021, 11, 113. [Google Scholar] [CrossRef]
- Adamopoulos, P.G.; Mavrogiannis, A.V.; Kontos, C.K.; Scorilas, A. Novel alternative splice variants of the human protein arginine methyltransferase 1 (PRMT1) gene, discovered using next-generation sequencing. Gene 2019, 699, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.W.; Hsu, J.M.; Xia, W.; Wang, H.L.; Wang, Y.N.; Chang, W.C.; Arold, S.T.; Chou, C.K.; Tsou, P.H.; Yamaguchi, H.; et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J. Clin. Investig. 2015, 125, 4529–4543. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Gui, T.; Zeng, X.; Deng, Y.; Wang, Z.; Wang, Y.; Yang, D.; Li, Q.; Xu, P.; Hu, R.; et al. PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med. 2021, 13, 58. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, P. m6A-modified circXPO1 accelerates colorectal cancer progression via interaction with FMRP to promote WWC2 mRNA decay. J. Transl. Med. 2024, 22, 931. [Google Scholar] [CrossRef]
- Wang, X.; Lu, X.; Wang, P.; Chen, Q.; Xiong, L.; Tang, M.; Hong, C.; Lin, X.; Shi, K.; Liang, L.; et al. SRSF9 promotes colorectal cancer progression via stabilizing DSN1 mRNA in an m6A-related manner. J. Transl. Med. 2022, 20, 198. [Google Scholar] [CrossRef]
- Du, D.; Qin, M.; Shi, L.; Liu, C.; Jiang, J.; Liao, Z.; Wang, H.; Zhang, Z.; Sun, L.; Fan, H.; et al. RNA binding motif protein 45-mediated phosphorylation enhances protein stability of ASCT2 to promote hepatocellular carcinoma progression. Oncogene 2023, 42, 3127–3141. [Google Scholar] [CrossRef]
- Cazes, A.; Childers, B.G.; Esparza, E.; Lowy, A.M. The MST1R/RON Tyrosine Kinase in Cancer: Oncogenic Functions and Therapeutic Strategies. Cancers 2022, 14, 2037. [Google Scholar] [CrossRef]
- Brownmiller, T.; Caplen, N.J. The HNRNPF/H RNA binding proteins and disease. Wiley Interdiscip. Rev. RNA 2023, 14, e1788. [Google Scholar] [CrossRef]
- Luo, X.J.; Zhao, Q.; Liu, J.; Zheng, J.B.; Qiu, M.Z.; Ju, H.Q.; Xu, R.H. Novel Genetic and Epigenetic Biomarkers of Prognostic and Predictive Significance in Stage II/III Colorectal Cancer. Mol. Ther. 2021, 29, 587–596. [Google Scholar] [CrossRef]
- Yang, J.; Fan, Q.; Wang, Y.; Liu, Y.; Xu, X.; Liang, Y.; Xie, J.; Li, J.; Ai, F.; Cao, Y.; et al. CircRNAs in colorectal cancer: Potential roles, clinical applications, and natural product-based regulation. Front. Oncol. 2025, 15, 1525779. [Google Scholar] [CrossRef]
- Kalioraki, M.A.; Artemaki, P.I.; Sklirou, A.D.; Kontos, C.K.; Adamopoulos, P.G.; Papadopoulos, I.N.; Trougakos, I.P.; Scorilas, A. Heat shock protein beta 3 (HSPB3) is an unfavorable molecular biomarker in colorectal adenocarcinoma. Mol. Carcinog. 2020, 59, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Artemaki, P.I.; Sklirou, A.D.; Kontos, C.K.; Liosi, A.A.; Gianniou, D.D.; Papadopoulos, I.N.; Trougakos, I.P.; Scorilas, A. High clusterin (CLU) mRNA expression levels in tumors of colorectal cancer patients predict a poor prognostic outcome. Clin. Biochem. 2020, 75, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Karousi, P.; Kontos, C.K.; Nikou, S.T.; Carell, T.; Sideris, D.C.; Scorilas, A. Discovery of circular transcripts of the human BCL2-like 12 (BCL2L12) apoptosis-related gene, using targeted nanopore sequencing, provides new insights into circular RNA biology. Funct. Integr. Genom. 2025, 25, 66. [Google Scholar] [CrossRef]
- Alexopoulou, D.K.; Kontos, C.K.; Christodoulou, S.; Papadopoulos, I.N.; Scorilas, A. KLK11 mRNA expression predicts poor disease-free and overall survival in colorectal adenocarcinoma patients. Biomark. Med. 2014, 8, 671–685. [Google Scholar] [CrossRef]
- Christodoulou, S.; Alexopoulou, D.K.; Kontos, C.K.; Scorilas, A.; Papadopoulos, I.N. Kallikrein-related peptidase-6 (KLK6) mRNA expression is an independent prognostic tissue biomarker of poor disease-free and overall survival in colorectal adenocarcinoma. Tumour Biol. 2014, 35, 4673–4685. [Google Scholar] [CrossRef]
- Kontos, C.K.; Chantzis, D.; Papadopoulos, I.N.; Scorilas, A. Kallikrein-related peptidase 4 (KLK4) mRNA predicts short-term relapse in colorectal adenocarcinoma patients. Cancer Lett. 2013, 330, 106–112. [Google Scholar] [CrossRef]
- Kontos, C.K.; Papadopoulos, I.N.; Fragoulis, E.G.; Scorilas, A. Quantitative expression analysis and prognostic significance of L-DOPA decarboxylase in colorectal adenocarcinoma. Br. J. Cancer 2010, 102, 1384–1390. [Google Scholar] [CrossRef]
- Kontos, C.K.; Papadopoulos, I.N.; Scorilas, A. Quantitative expression analysis and prognostic significance of the novel apoptosis-related gene BCL2L12 in colon cancer. Biol. Chem. 2008, 389, 1467–1475. [Google Scholar] [CrossRef]
- Camp, R.L.; Dolled-Filhart, M.; Rimm, D.L. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004, 10, 7252–7259. [Google Scholar] [CrossRef]


| Variable | Number of Patients (%) |
|---|---|
| Gender | |
| Male | 110 (52.4%) |
| Female | 100 (47.6%) |
| Tumor location | |
| Colon | 143 (68.1%) |
| Rectum | 67 (31.9%) |
| Histological grade | |
| I | 19 (9.0%) |
| II | 160 (76.2%) |
| III | 31 (14.8%) |
| T (tumor invasion) | |
| T1 | 6 (2.9%) |
| T2 | 25 (11.9%) |
| T3 | 130 (61.9%) |
| T4 | 49 (23.3%) |
| N (nodal status) | |
| N0 | 118 (56.2%) |
| N1 | 54 (25.7%) |
| N2 | 38 (18.1%) |
| M (distant metastasis) | |
| M0 | 184 (87.6%) |
| M1 | 26 (12.4%) |
| TNM stage | |
| I | 28 (13.3%) |
| II | 85 (40.5%) |
| III | 71 (33.8%) |
| IV | 26 (12.4%) |
| Radiotherapy | |
| No | 166 (83.8%) |
| Yes | 32 (16.2%) |
| Chemotherapy | |
| No | 85 (42.9%) |
| Yes | 113 (57.1%) |
| Covariate | HR | 95% CI | p Value 1 | BCa 95% CI | Bootstrap p Value 1 | |
|---|---|---|---|---|---|---|
| Disease-free survival (DFS) | circ-PRMT1 expression | |||||
| Low | 1.00 | |||||
| High | 3.05 | 1.59–5.86 | <0.001 | 1.64–6.9 | 0.001 | |
| Tumor location | ||||||
| Colon | 1.00 | |||||
| Rectum | 1.64 | 0.98–2.74 | 0.059 | 1.02–2.6 | 0.052 | |
| Histological grade | ||||||
| I | 1.00 | |||||
| II | 1.73 | 0.62–4.81 | 0.29 | 0.66–2.6 × 104 | 0.24 | |
| III | 3.06 | 0.97–9.61 | 0.056 | 0.81–7.0 × 104 | 0.035 | |
| TNM stage | ||||||
| I | 1.00 | |||||
| II | 4.37 | 1.03–18.49 | 0.045 | 1.35–2.8 × 104 | 0.029 | |
| III | 9.95 | 2.39–41.47 | 0.002 | 3.21–7.6 × 104 | 0.001 | |
| Radiotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.78 | 0.99–3.19 | 0.052 | 0.97–3.04 | 0.044 | |
| Chemotherapy | ||||||
| No | 1.00 | |||||
| Yes | 2.32 | 1.32–4.06 | 0.003 | 1.31–4.35 | 0.004 | |
| Overall survival (OS) | circ-PRMT1 expression | |||||
| Low | 1.00 | |||||
| High | 4.39 | 2.33–8.24 | <0.001 | 2.52–9.56 | 0.001 | |
| Tumor location | ||||||
| Colon | 1.00 | |||||
| Rectum | 1.53 | 1.01–2.33 | 0.047 | 0.98–2.42 | 0.056 | |
| Histological grade | ||||||
| I | 1.00 | |||||
| II | 1.32 | 0.61–2.88 | 0.48 | 0.6–3.9 | 0.51 | |
| III | 2.72 | 1.14–6.48 | 0.024 | 1.13–8.05 | 0.023 | |
| TNM stage | ||||||
| I | 1.00 | |||||
| II | 2.60 | 0.91–7.48 | 0.075 | 0.83–3.6 × 104 | 0.062 | |
| III | 5.76 | 2.03–16.29 | <0.001 | 1.8–6.2 × 104 | 0.005 | |
| IV | 34.85 | 11.71–103.72 | <0.001 | 9.69–4.7 × 105 | 0.001 | |
| Radiotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.67 | 1.02–2.75 | 0.042 | 0.91–2.95 | 0.057 | |
| Chemotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.78 | 1.15–2.77 | 0.010 | 1.15–2.77 | 0.013 |
| Covariate | HR | 95% CI | p Value 1 | BCa 95% CI | Bootstrap p Value 1 | |
|---|---|---|---|---|---|---|
| Disease-Free Survival (DFS) | circ-PRMT1 expression | |||||
| Low | 1.00 | |||||
| High | 3.00 | 1.55–5.79 | 0.001 | 1.59–7.6 | 0.003 | |
| Tumor location | ||||||
| Colon | 1.00 | |||||
| Rectum | 2.56 | 1.26–5.18 | 0.009 | 1.11–8.23 | 0.029 | |
| Histological grade | ||||||
| I | 1.00 | |||||
| II | 1.07 | 0.37–3.11 | 0.90 | 0.28–2.3 × 104 | 0.87 | |
| III | 1.61 | 0.48–5.35 | 0.44 | 0.36–3.5 × 104 | 0.43 | |
| TNM stage | ||||||
| I | 1.00 | |||||
| II | 4.86 | 1.10–21.61 | 0.038 | 1.23–8.8 × 104 | 0.015 | |
| III | 9.74 | 2.09–45.39 | 0.004 | 2.09–1.9 × 105 | 0.002 | |
| Radiotherapy | ||||||
| No | 1.00 | |||||
| Yes | 0.48 | 0.21–1.09 | 0.080 | 0.19–0.98 | 0.086 | |
| Chemotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.37 | 0.72–2.63 | 0.34 | 0.61–3.03 | 0.38 | |
| Overall Survival (OS) | circ-PRMT1 expression | |||||
| Low | 1.00 | |||||
| High | 3.69 | 1.95–6.98 | <0.001 | 1.81–11.26 | 0.001 | |
| Tumor location | ||||||
| Colon | 1.00 | |||||
| Rectum | 1.42 | 0.83–2.44 | 0.20 | 0.73–2.62 | 0.22 | |
| Histological grade | ||||||
| I | 1.00 | |||||
| II | 0.78 | 0.34–1.79 | 0.56 | 0.29–2.27 | 0.62 | |
| III | 1.09 | 0.43–2.75 | 0.86 | 0.35–4.18 | 0.88 | |
| TNM stage | ||||||
| I | 1.00 | |||||
| II | 2.97 | 1.00–8.80 | 0.049 | 0.73–5.4 × 104 | 0.042 | |
| III | 6.98 | 2.26–21.58 | <0.001 | 1.7–2.2 × 105 | 0.001 | |
| IV | 36.97 | 11.31–120.83 | <0.001 | 8.18–1.3 × 106 | <0.001 | |
| Radiotherapy | ||||||
| No | 1.00 | |||||
| Yes | 0.96 | 0.50–1.83 | 0.90 | 0.47–1.89 | 0.90 | |
| Chemotherapy | ||||||
| No | 1.00 | |||||
| Yes | 0.78 | 0.46–1.32 | 0.35 | 0.41–1.43 | 0.36 |
| Assay | Target | Sequence (5′ → 3′) | Ta (°C) | Amplicon Length (bp) |
|---|---|---|---|---|
| Pre-amplification | PRMT1 circRNA | TGACTCCTACGCACACTTTGG | 60 | 401 |
| TCTTTGGATGTCATGTCCTCAGC | ||||
| GAPDH mRNA | CCACATCGCTCAGACACCAT | 60 | 223 | |
| TGACAAGCTTCCCGTTCTCA | ||||
| Real-time qPCR | PRMT1 circRNA | AGGTGGAGAGGTGCGG | 60 | 96 |
| TTGGGCTTCTCACTGCTTTCC | ||||
| GAPDH mRNA | ATGGGGAAGGTGAAGGTCG | 60 | 107 | |
| GGGTCATTGATGGCAACAATATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokoropoulos, P.; Christodoulou, S.; Tsiakanikas, P.; Vassiliu, P.; Kontos, C.K.; Arkadopoulos, N. Overexpression of Circular PRMT1 Transcripts in Colorectal Adenocarcinoma Predicts Recurrence and Poor Overall Survival. Int. J. Mol. Sci. 2025, 26, 6683. https://doi.org/10.3390/ijms26146683
Kokoropoulos P, Christodoulou S, Tsiakanikas P, Vassiliu P, Kontos CK, Arkadopoulos N. Overexpression of Circular PRMT1 Transcripts in Colorectal Adenocarcinoma Predicts Recurrence and Poor Overall Survival. International Journal of Molecular Sciences. 2025; 26(14):6683. https://doi.org/10.3390/ijms26146683
Chicago/Turabian StyleKokoropoulos, Panagiotis, Spyridon Christodoulou, Panagiotis Tsiakanikas, Panteleimon Vassiliu, Christos K. Kontos, and Nikolaos Arkadopoulos. 2025. "Overexpression of Circular PRMT1 Transcripts in Colorectal Adenocarcinoma Predicts Recurrence and Poor Overall Survival" International Journal of Molecular Sciences 26, no. 14: 6683. https://doi.org/10.3390/ijms26146683
APA StyleKokoropoulos, P., Christodoulou, S., Tsiakanikas, P., Vassiliu, P., Kontos, C. K., & Arkadopoulos, N. (2025). Overexpression of Circular PRMT1 Transcripts in Colorectal Adenocarcinoma Predicts Recurrence and Poor Overall Survival. International Journal of Molecular Sciences, 26(14), 6683. https://doi.org/10.3390/ijms26146683







