Identification and Characterization of the Complete Genome of the TGF-β Gene Family in Tupaia belangeri: Expression and Function of Adipose Tissue Under Cold Acclimation Conditions
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of TGF-β Family Genes
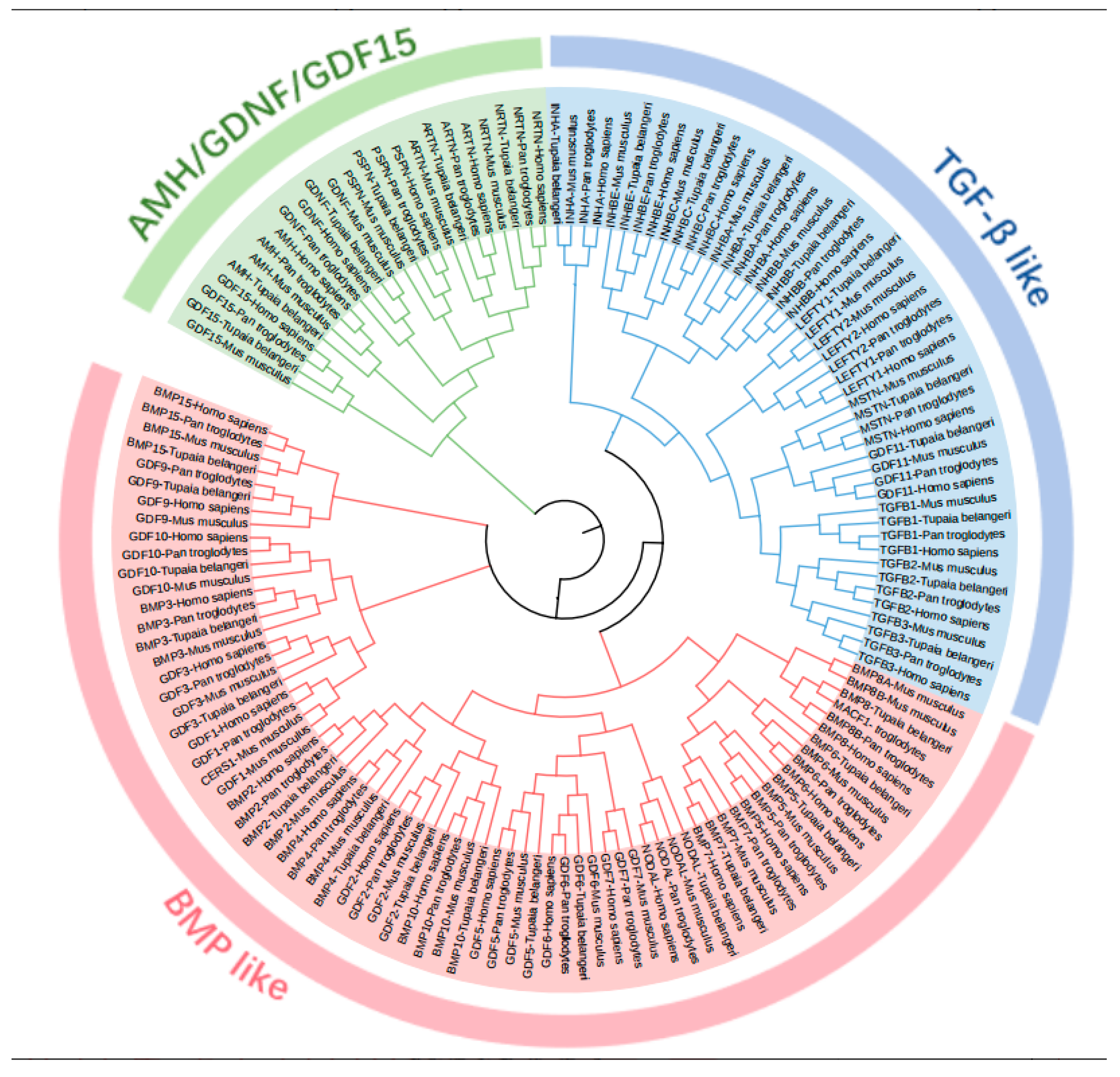
2.2. Phylogenetic Analysis
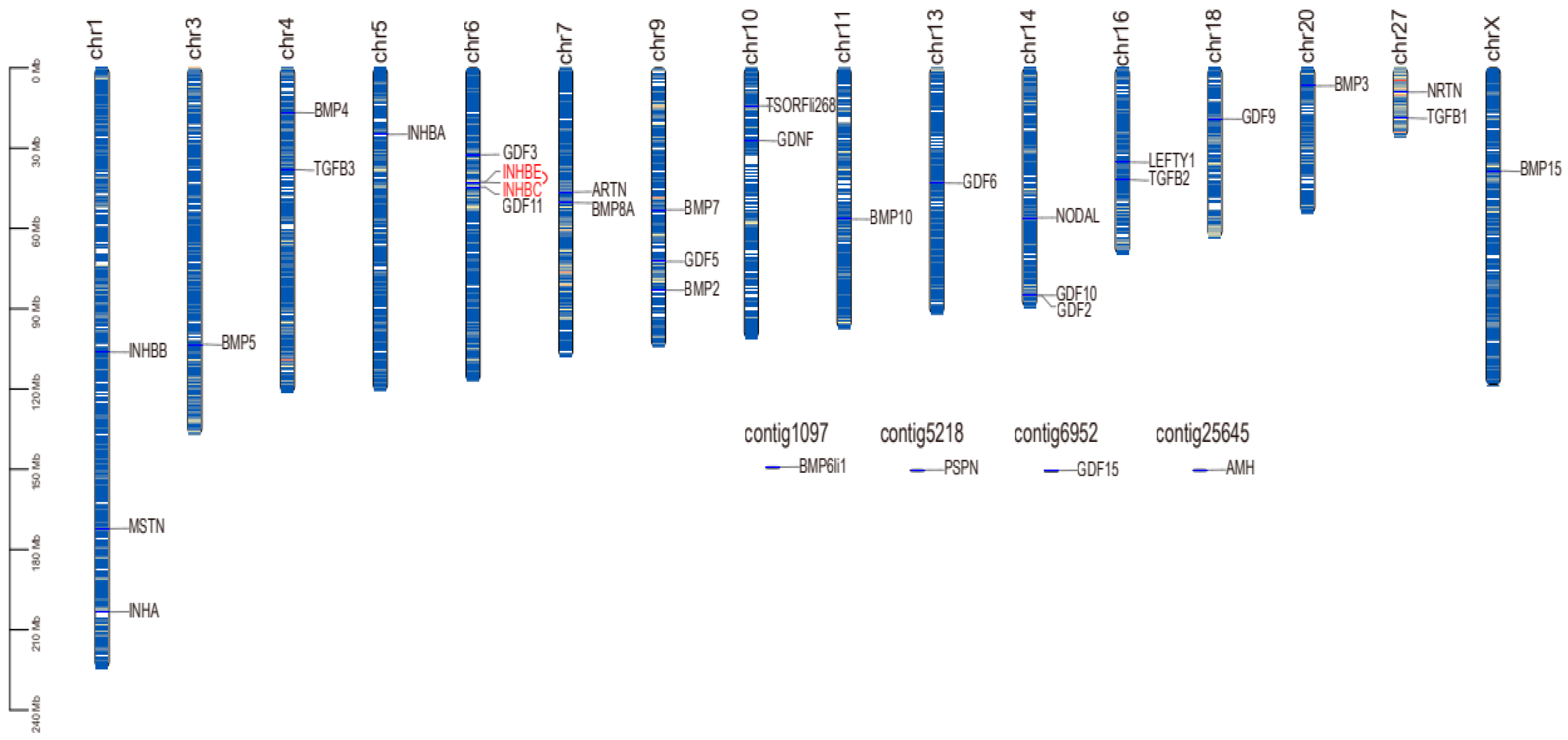
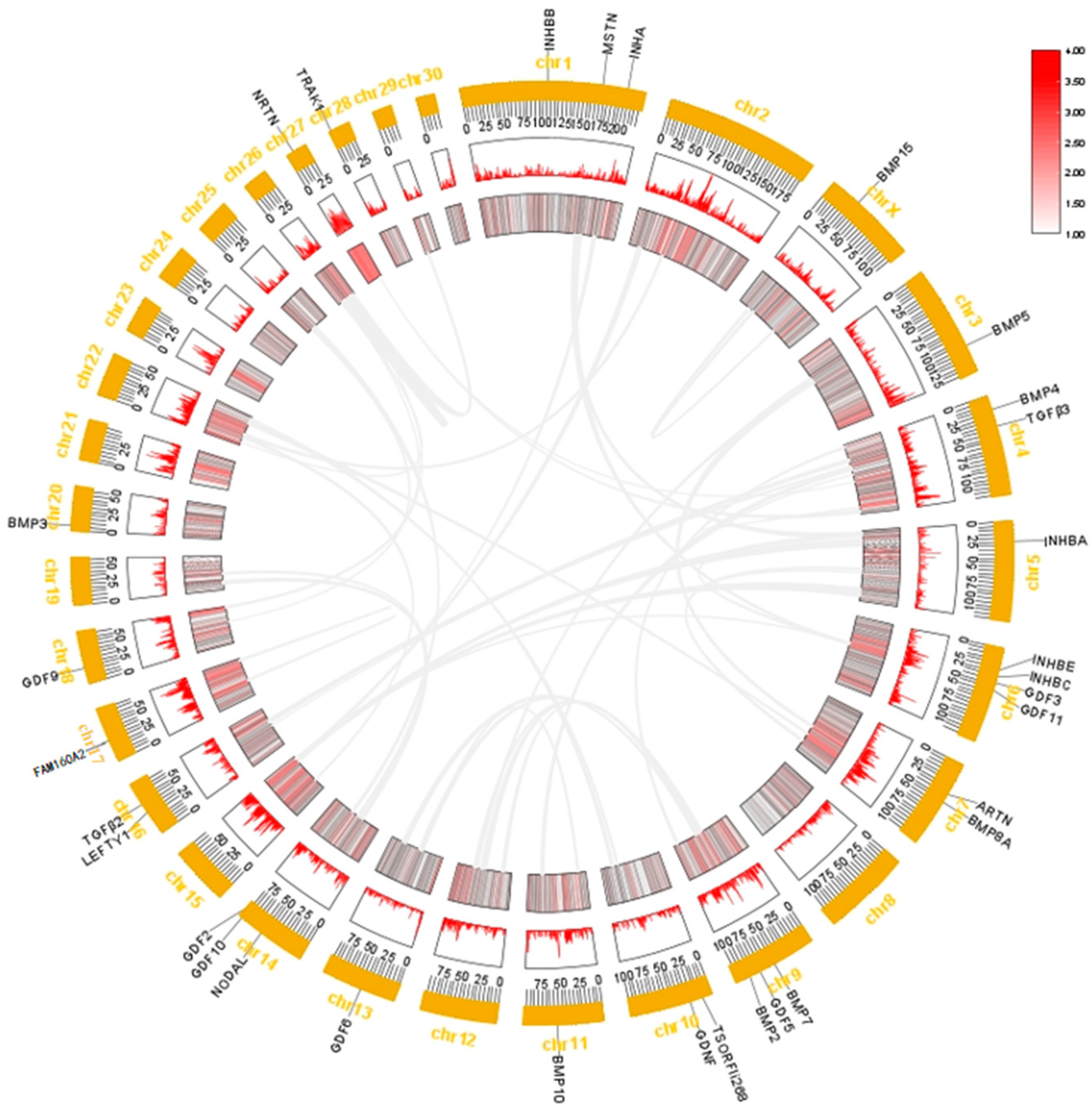
2.3. Chromosome Distribution and Collinearity Analysis
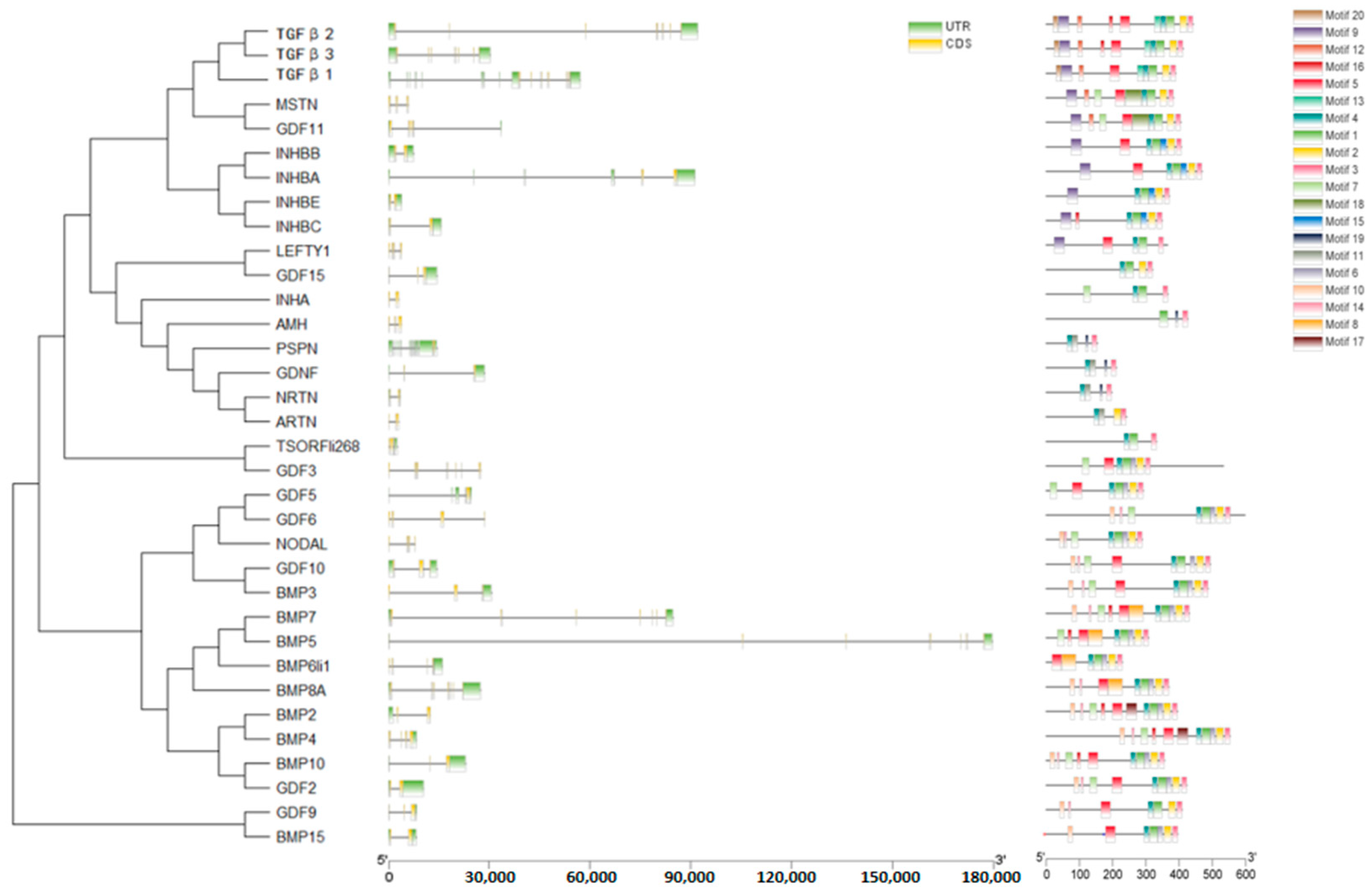
2.4. Gene Structure and Motif Analysis of TGF-β Gene Family
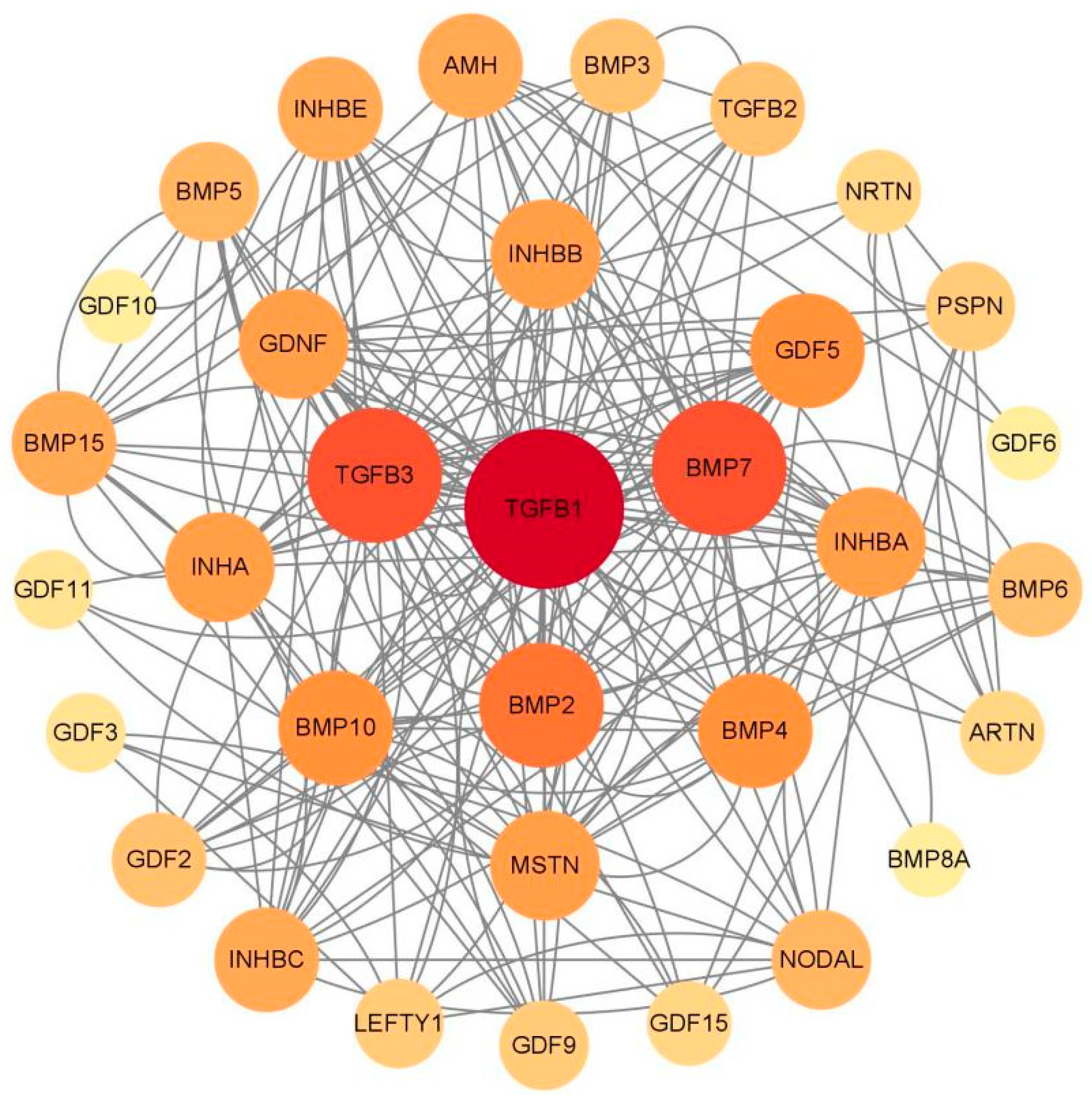
2.5. Protein Interaction Analysis Network of TGF-β Family Proteins
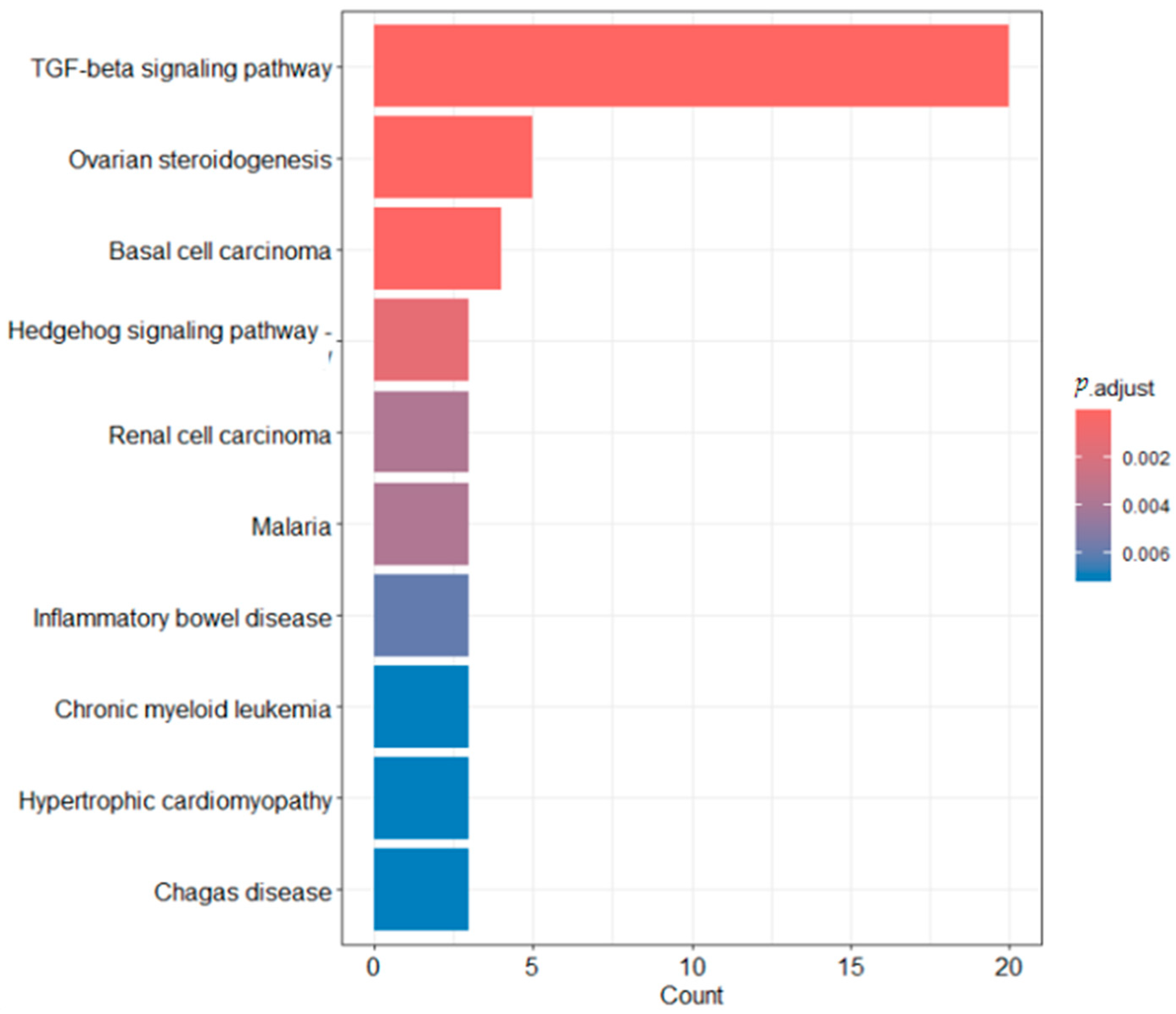
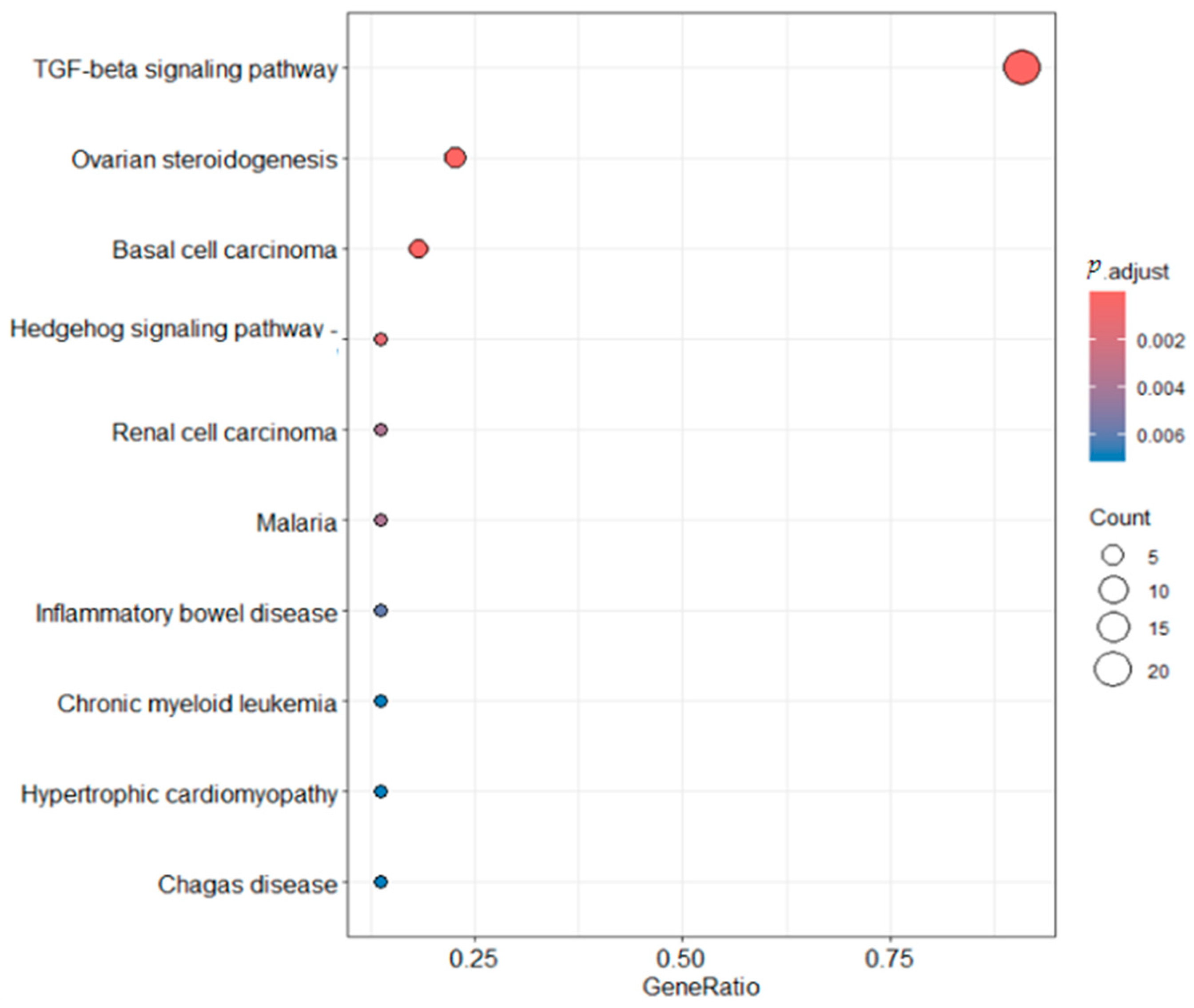
2.6. Functional Enrichment Analysis
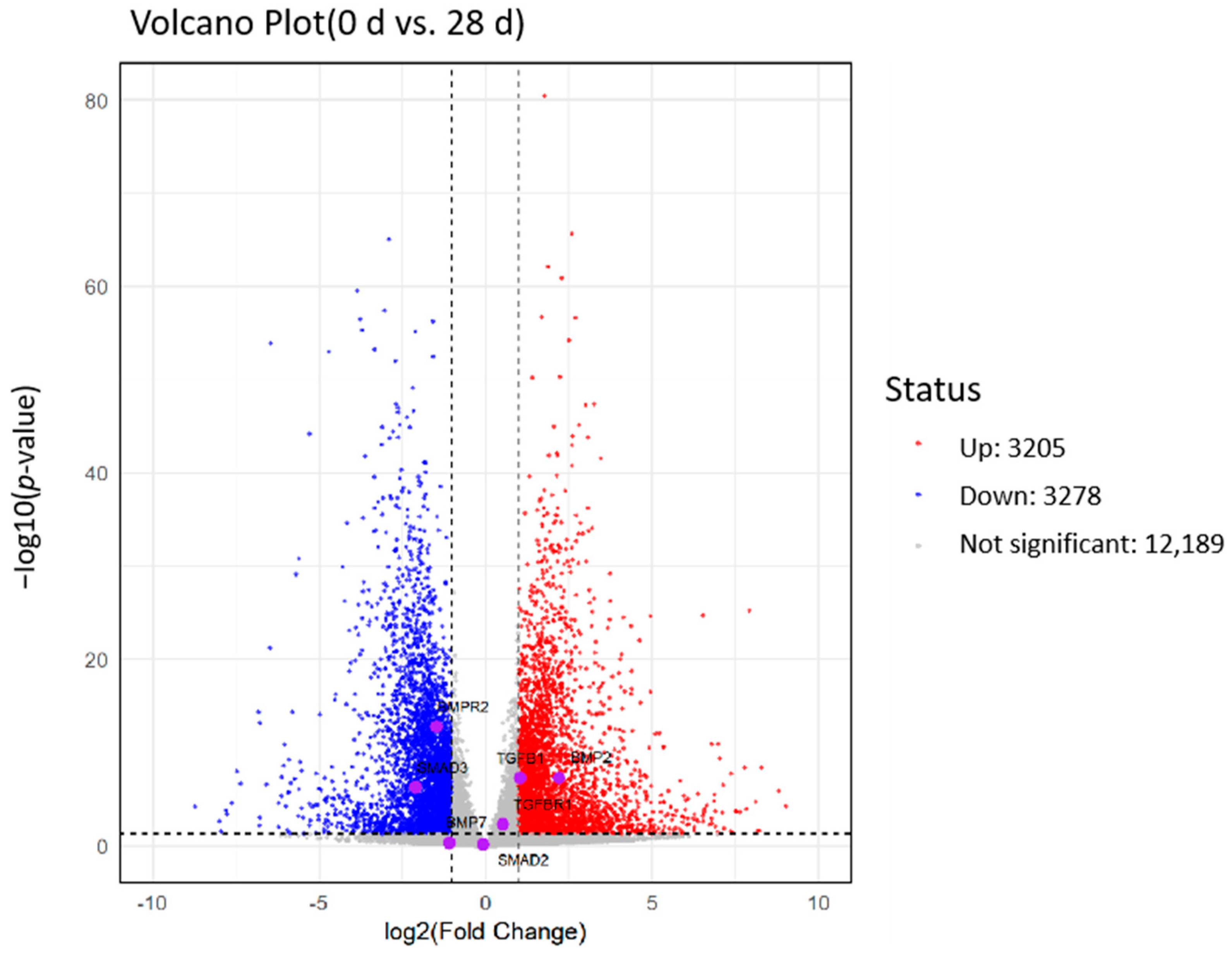
2.7. Analysis of Differentially Expressed Genes
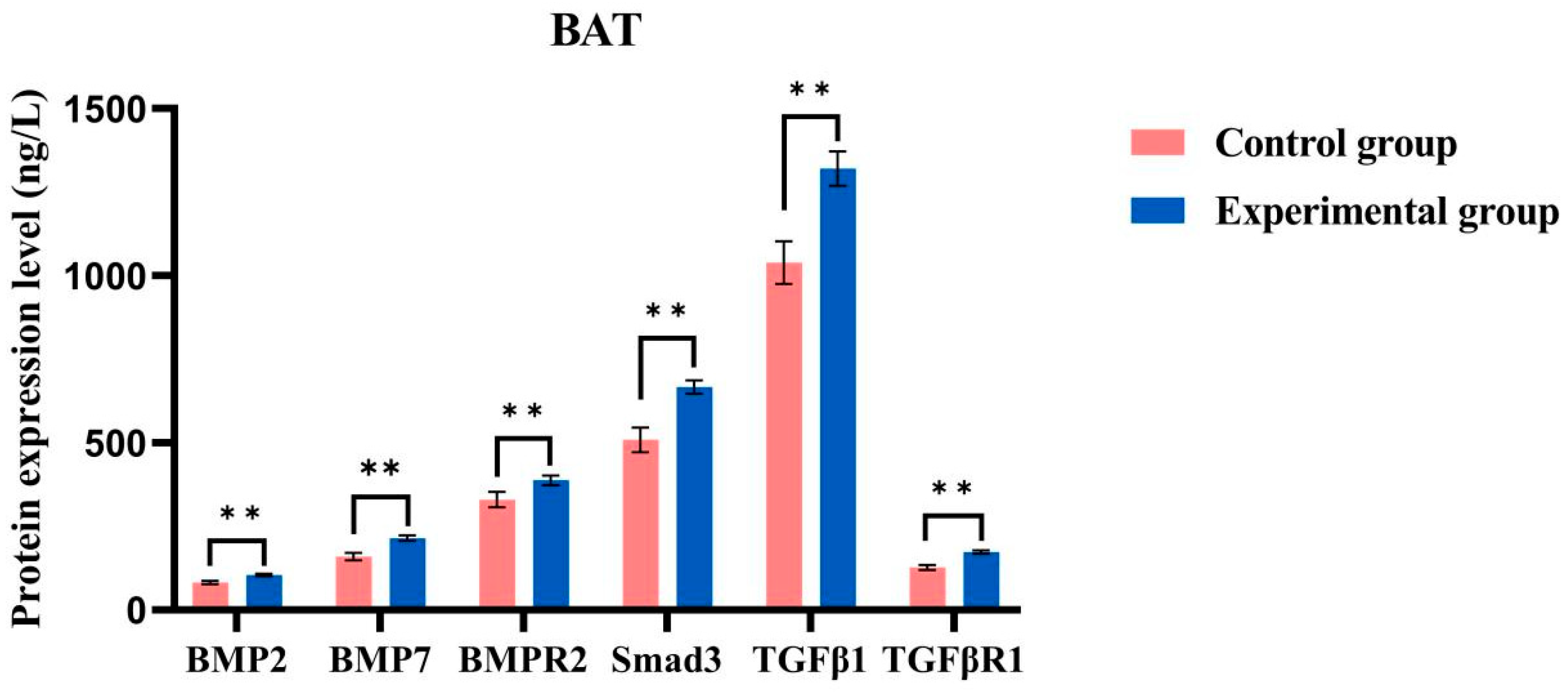
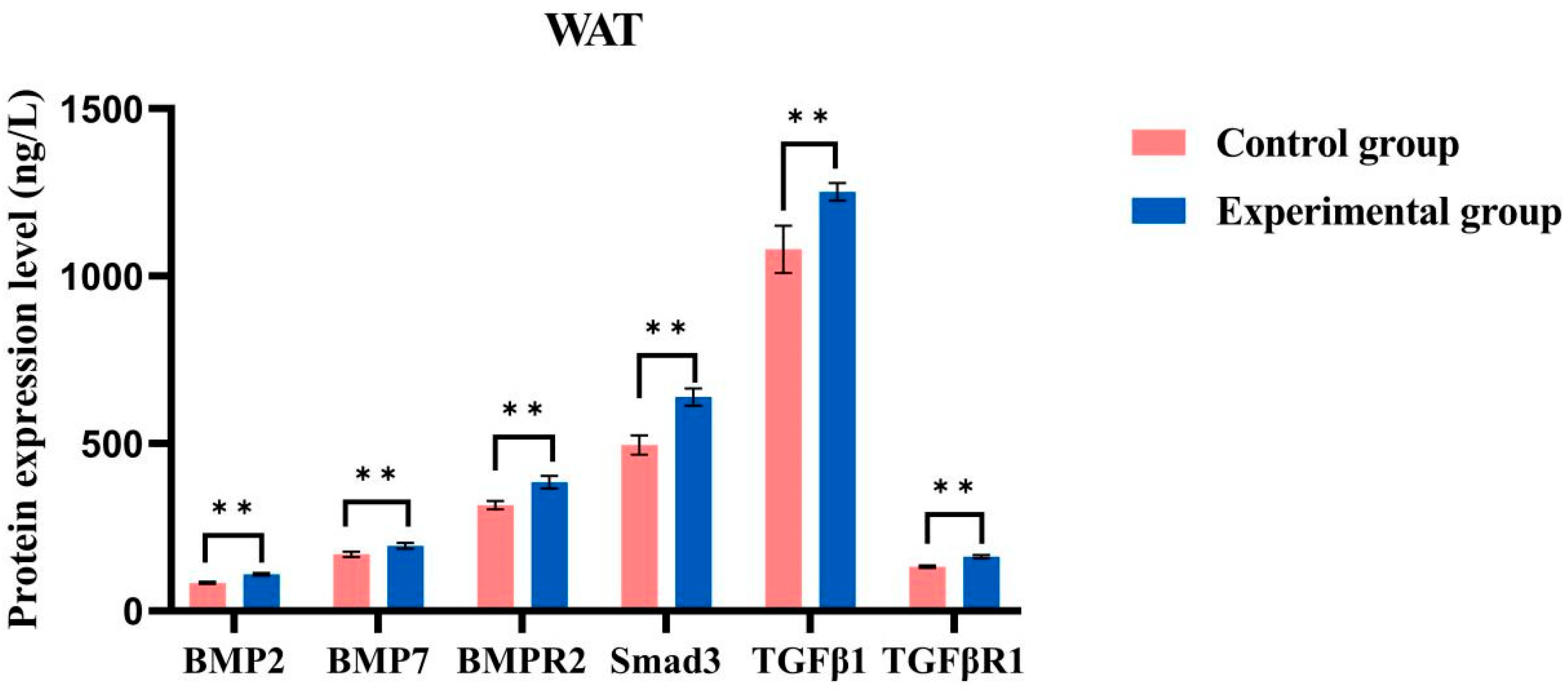
2.8. ELISA Detects Protein Content
3. Discussion
4. Material and Method
4.1. Identification and Characterization of TGF-β Family Genes
4.2. Phylogenetic Analysis
4.3. Chromosome Distribution and Collinearity Analysis
4.4. Gene Structure and Motif Analysis of TGF-β Gene Family
4.5. Protein Interaction Analysis Network of TGF-β Family Proteins
4.6. Functional Enrichment Analysis
4.7. Analysis of Differentially Expressed Genes
4.8. ELISA Detects Protein Content
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, D.; Zhu, W.; Gao, W.; Zhang, H.; Wang, Z. Progress and Prospect of Studies on Physiological and Behavioral Ecology in Tree Shrews, Tupaia belangeri. Bioprocess 2016, 6, 41–47. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, L.; Zhu, W.; Yang, S.; Yu, T.; Wang, Z. Thermoregulation and thermogenic characteristics of Tupaia belangeri from kunming and luquan district, yunnan province. Sichuan J. Zool. 2013, 32, 670–675. [Google Scholar]
- Gao, W.; Zhu, W. Oriental endemic species-progress and prospect of studies on physiological ecology in tree shrews, Tupaia belangeri. Bioprocess 2014, 4, 82–89. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, S.; Wu, Q.; Li, X.; Zeng, W.; Dong, F.; An, W.; Qin, F.; Lei, L.; Zhao, C. Tree shrews as a new animal model for systemic sclerosis research. Front. Immunol. 2024, 15, 1315198. [Google Scholar] [CrossRef]
- Hinck, A.P.; Mueller, T.D.; Springer, T.A. Structural Biology and Evolution of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2016, 8, a022103. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Han, C.; Huang, J.; Li, M.; Yang, J.; Li, G.; Lin, H.; Li, S.; Zhang, Y. Genome-wide identification, evolution and expression of TGF-β signaling pathway members in mandarin fish (Siniperca chuatsi). Int. J. Biol. Macromol. 2023, 253, 126949. [Google Scholar] [CrossRef]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, a021873. [Google Scholar] [CrossRef]
- Liu, S.; Guo, J.; Cheng, X.; Li, W.; Lyu, S.; Chen, X.; Li, Q.; Wang, H. Molecular evolution of transforming growth factor-β (TGF-β) gene family and the functional characterization of lamprey TGF-β2. Front. Immunol. 2022, 13, 836226. [Google Scholar] [CrossRef]
- Andreou, T.; Williams, J.; Brownlie, R.J.; Salmond, R.J.; Watson, E.; Shaw, G.; Melcher, A.; Wurdak, H.; Short, S.C.; Lorger, M. Hematopoietic stem cell gene therapy targeting TGFβ enhances the efficacy of irradiation therapy in a preclinical glioblastoma model. J. Immunother. Cancer 2021, 9, e001143. [Google Scholar] [CrossRef]
- Apostolou, E.; Blau, H.; Chien, K.; Lancaster, M.A.; Tata, P.R.; Trompouki, E.; Watt, F.M.; Zeng, Y.A.; Zernicka-Goetz, M. Progress and challenges in stem cell biology. Nat. Cell Biol. 2023, 25, 203–206. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, F.; Wang, Z.; Zhu, W. Role of thermal physiology and bioenergetics on adaptation in tree shrew (Tupaia belangeri): The experiment test. Sci. Rep. 2017, 7, 41352. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zou, D.; Zhang, X.; Wang, D. Huddling behavior regulate adaptive thermogenesis in Brandt’s voles (Lasiopodomys brandtii). Cell Biosci. 2025, 15, 51. [Google Scholar] [CrossRef]
- Shi, Y.; Massagué, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Zi, Z.; Chapnick, D.A.; Liu, X. Dynamics of TGF-β/Smad signaling. FEBS Lett. 2012, 586, 1921–1928. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; He, Y.; Cheng, L.; Alghazzawi, D.; Yang, Y.; Zhan, X. Machine learning for predicting protein properties: A comprehensive review. Neurocomputing 2024, 597, 128103. [Google Scholar] [CrossRef]
- Teng, Y.; Appiah, B.; Andrieux, G.; Schrempp, M.; Rose, K.; Hofmann, A.S.; Ku, M.; Beyes, S.; Boerries, M.; Hecht, A. TGF-β signaling redirects Sox11 gene regulatory activity to promote partial EMT and collective invasion of oncogenically transformed intestinal organoids. Oncogenesis 2025, 14, 17. [Google Scholar] [CrossRef]
- Kalluri, A.M.; Weinberg, R.A. Diverse Cellular Responses to TGF-β: Context-Dependent Signaling. Nat. Rev. Mol. Cell Biol. 2023, 24, 361–375. [Google Scholar]
- Pask, A.J.; Miller, R.J.; Duchêne, D.A.; Springer, M.S.; Gatesy, J.A. Genomic Insights into the Origins and Evolution of Placental Mammals. Mol. Biol. Evol. 2023, 40, 1234–1248. [Google Scholar]
- Lo, W.S.; Roca, M.; Dardiry, M.; Mackie, M.; Eberhardt, G.; Witte, H.; Hong, R.; Sommer, R.J.; Lightfoot, J.W. Evolution and Diversity of TGF-β Pathways are Linked with Novel Developmental and Behavioral Traits. Mol. Biol. Evol. 2022, 39, msac252. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Zhang, L. Comparative genomics and evolutionary analysis of the transforming growth factor-beta (TGF-β) superfamily in vertebrates. PLoS ONE 2014, 9, e85290. [Google Scholar]
- Hattori, M. Regulatory mechanism of Reelin activity: A platform for exploiting Reelin as a therapeutic agent. Front. Mol. Neurosci. 2025, 18, 1546083. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, H.R.; Wang, M. DeeperBind: Enhancing Prediction of Sequence Specificities of DNA Binding Proteins. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 178–183. [Google Scholar]
- Ghosh, S.; Saha, T. Central Role of Ubiquitination in Genome Maintenance: DNA Replication and Damage Repair. ISRN Mol. Biol. 2012, 2012, 146748. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Sheppard, D. TGF-β signaling in health and disease. Cell 2023, 186, 4007–4037. [Google Scholar] [CrossRef]
- Zhong, Y.; Ding, J.; Xia, F. DNA methylation and its impact on ovarian steroidogenesis in women with polycystic ovary syndrome: Insights from human and animal models. Arch. Gynecol. Obstet. 2025, 401, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Aktary, Z.; Raymond, J.; Pouteaux, M.; Delmas, V.; Petit, V.; Larue, L. Derivation and use of cell lines from mouse models of melanoma. J. Investig. Dermatol. 2023, 143, 538–544.e2. [Google Scholar] [CrossRef]
- Miyazono, K.; Kusanagi, K.; Inoue, H. Divergence and convergence of TGF-beta/BMP signaling. J. Cell. Physiol. 2001, 187, 265–276. [Google Scholar] [CrossRef]
- Yoneshiro, T.; Aita, S.; Matsushita, M.; Kameya, T.; Nakada, K.; Kawai, Y.; Saito, M. Brown adipose tissue, whole-body energy expenditure, and thermogenesis in healthy adult men. Obesity 2011, 19, 13–16. [Google Scholar] [CrossRef]
- Nedergaard, J.; Cannon, B. The changed metabolic world with human brown adipose tissue: Therapeutic visions. Cell Metab. 2018, 27, 738–752. [Google Scholar] [CrossRef]
- Cypess, A.M.; Cannon, B.; Nedergaard, J.; Kazak, L.; Chang, D.C.; Krakoff, J.; Tseng, Y.H.; Schéele, C.; Boucher, J.; Petrovic, N.; et al. Emerging debates and resolutions in brown adipose tissue research. Cell Metab. 2025, 37, 12–33. [Google Scholar] [CrossRef]
- Yang, K.; Wang, J.; Lu, W. Bone morphogenetic protein signalling in pulmonary hypertension: Advances and therapeutic implications. Exp. Physiol. 2017, 102, 1083–1089. [Google Scholar] [CrossRef]
- Walsh, D.W.; Godson, C.; Brazil, D.P.; Martin, F. Extracellular bmp-antagonist regulation in development and disease: Tied up in knots. Trends Cell Biol. 2010, 20, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Graja, A.; Schulz, T.J. Mechanisms of aging-related impairment of brown adipocyte development and function. Gerontology 2014, 61, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Surisaeng, T.; Wisitrasameewong, W.; Champaiboon, C.; Sa-Ard-Iam, N.; Chanamuangkon, T.; Thongnuek, P.; Tam, Y.K.; Muramatsu, H.; Weissman, D.; Pardi, N.; et al. BMP-2 mRNA-transfected BMSCs promote superior calvarial bone regeneration. Sci. Rep. 2025, 15, 15022. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Kusminski, C.M.; Scherer, P.E. Adipose tissue remodeling and obesity. J. Clin. Investig. 2011, 121, 2094–2101. [Google Scholar] [CrossRef]
- Dewida, B.; Meyer, C.; Dooley, S.; Meindl-Beinker, A.N. TGF-β in Hepatic Stellate Cell Activation and Liver Fibrogenesis-Updated 2019. Cells 2019, 8, 1419. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, R.; Wang, D.; Li, P.; Qu, Y. Genome-wide characterization of the TGF-β gene family and their expression in different tissues during tail regeneration in the Schlegel’s Japanese gecko Gekko japonicus. Int. J. Biol. Macromol. 2024, 255, 128127. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.C.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. S2), W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Rustici, G.; Kuhn, M.; Wyder, S.; Jensen, L.J. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Roncaglia, P.; van Dam, T.J.P.; Christie, K.R.; Nacheva, L.; Toedt, G.; Huynen, M.A.; Huntley, R.P.; Gibson, T.J.; Lomax, J. The Gene Ontology of eukaryotic cilia and flagella. Cilia 2017, 6, 10. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.; Han, Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Karin, J.; Mintz, R.; Raveh, B.; Nitzan, M. Interpreting single-cell and spatial omics data using deep neural network training dynamics. Nat. Comput. Sci. 2024, 4, 941–954. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]

| Gene Name | Protein Serial Number | Number of Amino Acids | Relative Molecular Weight of Protein/kDa | Isoelectric Point | Instability Coefficient | Overall Average Hydrophilicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| TGFB2 | TSDBTID.10930.1 | 442 | 50,504.96 | 8.74 | 52.37 | −0.393 | P; N |
| TGFB3 | TSDBTID.22079.2 | 412 | 47,353.42 | 8.56 | 49.70 | −0.477 | PM; P; G |
| TGFB1 | TSDBTID.1119.1 | 390 | 44,505.37 | 8.92 | 53.05 | −0.357 | P; PM |
| MSTN | TSDBTID.4722.1 | 383 | 43,653.21 | 6.67 | 40.99 | −0.399 | C;P; M |
| GDF11 | TSDBTID.24154.1 | 405 | 44,964.62 | 8.18 | 61.51 | −0.299 | PM; ER; P |
| INHBB | TSDBTID.4539.1 | 407 | 45,061.52 | 8.35 | 58.22 | −0.288 | P; PM |
| INHBA | TSDBTID.22580.1 | 469 | 52,597.32 | 9.04 | 64.26 | −0.598 | N; C; P |
| INHBE | TSDBTID.24129.1 | 371 | 40,672.71 | 9.28 | 56.76 | −0.295 | PM; N |
| INHBC | TSDBTID.24130.1 | 349 | 37,820.47 | 8.28 | 49.41 | 0.010 | P; PM |
| LEFTY1 | TSDBTID.10569.1 | 365 | 40,842.85 | 8.61 | 54.99 | −0.302 | P; PM |
| GDF15 | TSDBTID.27506.2 | 320 | 35,836.12 | 9.72 | 55.59 | −0.361 | P; N; PM |
| INHA | TSDBTID.4074.1 | 366 | 39,189.08 | 8.02 | 64.25 | −0.063 | P; PM |
| AMH | TSDBTID.26794.1 | 426 | 46,391.31 | 9.00 | 53.59 | −0.299 | N; C |
| PSPN | TSDBTID.2357.1 | 154 | 16,218.64 | 9.23 | 42.57 | −0.106 | PM |
| GDNF | TSDBTID.6036.1 | 211 | 23,722.16 | 9.25 | 63.19 | −0.560 | P |
| NRTN | TSDBTID.18517.1 | 197 | 22,393.00 | 11.12 | 72.88 | −0.436 | P |
| ARTN | TSDBTID.25276.1 | 242 | 25,936.37 | 11.51 | 80.63 | −0.846 | N |
| TSORFli2 | TSDBTID.5994.1 | 332 | 37,772.67 | 6.21 | 58.98 | −0.051 | P |
| GDF3 | TSDBTID.1893.1 | 533 | 60,304.47 | 9.36 | 51.68 | −0.282 | P |
| GDF5 | TSDBTID.5249.1 | 292 | 33,188.20 | 9.35 | 43.17 | −0.461 | N; C |
| GDF6 | TSDBTID.8166.1 | 598 | 65,620.63 | 9.43 | 51.31 | −0.585 | N; C; M |
| NODAL | TSDBTID.8844.1 | 289 | 33,063.76 | 6.55 | 60.36 | −0.401 | P; PERO |
| GDF10 | TSDBTID.8961.2 | 494 | 54,332.31 | 9.44 | 55.13 | −0.455 | P; ER |
| BMP3 | TSDBTID.14860.1 | 487 | 55,013.28 | 9.72 | 69.46 | −0.567 | P; M |
| BMP7 | TSDBTID.5163.1 | 431 | 49,340.96 | 7.35 | 54.47 | −0.403 | P; M |
| BMP5 | TSDBTID.20495.2 | 308 | 35,007.68 | 9.04 | 52.41 | −0.511 | N; C |
| BMP6 | TSDBTID.26352.1 | 229 | 25,826.29 | 8.47 | 53.87 | −0.313 | P |
| BMP8A | TSDBTID.24686.2 | 369 | 40,367.58 | 9.49 | 69.48 | −0.386 | P |
| BMP2 | TSDBTID.5339.1 | 395 | 44,645.81 | 9.12 | 54.95 | −0.495 | P; ER |
| BMP4 | TSDBTID.21977.2 | 553 | 62,888.18 | 9.94 | 70.04 | −0.820 | N |
| BMP10 | TSDBTID.6702.2 | 356 | 40,694.94 | 5.17 | 47.32 | −0.546 | N; C |
| GDF2 | TSDBTID.8962.1 | 423 | 46,732.97 | 7.18 | 49.32 | −0.362 | P |
| GDF9 | TSDBTID.12441.1 | 409 | 46,828.30 | 8.97 | 57.75 | −0.512 | N |
| BMP15 | TSDBTID.19556.1 | 396 | 45,786.83 | 9.27 | 55.64 | −0.393 | PM; PERO |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, L.; Zhu, W.; Zhang, L. Identification and Characterization of the Complete Genome of the TGF-β Gene Family in Tupaia belangeri: Expression and Function of Adipose Tissue Under Cold Acclimation Conditions. Int. J. Mol. Sci. 2025, 26, 6681. https://doi.org/10.3390/ijms26146681
Du L, Zhu W, Zhang L. Identification and Characterization of the Complete Genome of the TGF-β Gene Family in Tupaia belangeri: Expression and Function of Adipose Tissue Under Cold Acclimation Conditions. International Journal of Molecular Sciences. 2025; 26(14):6681. https://doi.org/10.3390/ijms26146681
Chicago/Turabian StyleDu, Lijie, Wanlong Zhu, and Lin Zhang. 2025. "Identification and Characterization of the Complete Genome of the TGF-β Gene Family in Tupaia belangeri: Expression and Function of Adipose Tissue Under Cold Acclimation Conditions" International Journal of Molecular Sciences 26, no. 14: 6681. https://doi.org/10.3390/ijms26146681
APA StyleDu, L., Zhu, W., & Zhang, L. (2025). Identification and Characterization of the Complete Genome of the TGF-β Gene Family in Tupaia belangeri: Expression and Function of Adipose Tissue Under Cold Acclimation Conditions. International Journal of Molecular Sciences, 26(14), 6681. https://doi.org/10.3390/ijms26146681







