Abstract
Research on structural variations in the field of crop genetics has expanded with the rapid development of genome sequencing technologies. As an important aspect of genomic variations, structural variations have a profound impact on the genetic characteristics of crops and significantly affect their key agronomic traits, such as yield, quality, and disease and stress resistance—by changing the gene arrangement order, copy number, and the positions of regulatory elements. Compared with single-nucleotide polymorphisms, structural variations present a diverse range of types, including deletions, duplications, inversions, and translocations, and their impacts are more extensive and profound. However, research on structural variations in crops still faces many challenges, for example those relating to different ploidy levels, genome repetitiveness, and their associations with phenotypes. Nevertheless, breakthroughs in long-read sequencing technologies and the integration of multi-omics data offer hope for solving these problems. A deep understanding of the impact of structural variations on crops is of great significance for accurately analyzing the evolutionary history of crops and guiding modern crop breeding, and is expected to provide strong support for global food security and the sustainable development of agriculture.
1. Introduction
As the source of biological diversity, genetic variations are an indispensable element for the exploration and development of modern genetics. Our understanding of genetic variation is continuously deepening with the advancement of sequencing technologies and data analysis methods, thereby providing momentum for innovation and discovery across various fields. Genetic variations encompass several key types that contribute to the diversity within a species, including single-nucleotide polymorphisms (SNPs), insertions/deletions (InDels), simple sequence repeat polymorphisms, and structural variations. Among these, SNPs and small InDels have frequently been investigated and used to dissect the genetic basis of key agronomic traits and guide breeding practices [1,2]. Recently, advances in sequencing and computational algorithms have enabled the genome-wide identification of complex structural variations, generally defined as large-fragment deletions or insertions, copy number variations, inversions, and translocations between chromosomes of different individuals [3]. These variations can significantly impact key traits as they can lead to gene loss, duplication, and generation.
Early research on structural variations mainly focused on the human genome, and a series of structural variations associated with human diseases were detected [4,5]. However, the identification of structural variations in plant genomes is challenging due to their size and complexity [6]. Although the efficiency, accuracy, and cost of identification regarding structural variations are all being improved, there are still many urgent problems to be solved in the field of crop research. For example, the genomic structures of different crops vary greatly, making it difficult to widely apply universal methods for identifying structural variations. Even though structural variations have been widely identified in crops, their mechanisms of impact on the specific physiological processes and agronomic traits of crops are difficult to clarify. The study of the frequency and distribution patterns of structural variations at the crop population level has also progressed slowly due to factors such as large sample sizes and complex genetic backgrounds [7]. Despite these challenges, the prospects for progress in crop structural variations are promising. Recent breakthroughs in long-read sequencing technologies, such as PacBio sequencing [8] and Nanopore sequencing [9], offer higher-resolution insights into crop genomes. These technologies can span large repetitive regions that are difficult to sequence with short-read methods, greatly facilitating the detection of complex structural variations. The integration of multi-omics data, including transcriptomics, proteomics, and metabolomics, has also emerged as a powerful approach to better understand the functional consequences of structural variations in crops.
A deep and comprehensive understanding of the impact of structural variations on crops, the accurate identification of structural variations closely related to beneficial traits, and, on this basis, the full use of advanced technologies such as molecular marker-assisted selection and gene editing can greatly accelerate the breeding process of varieties. This can significantly increase crop yields to meet the growing global food demand. This review summarizes the research progress on the impact of structural variations on crops, deeply analyzes the key problems faced in current research, and looks ahead to the promising research directions in the future.
2. The Types and Detection of Structural Variations
Structural variations are defined as alterations in the length, copy number, orientation, or chromosomal location of DNA segments among different individuals within the same species. Structural variations mainly encompass six basic types (deletions, insertions, duplications, copy number variations, inversions, and translocations) and involve DNA segments typically larger than 50 base pairs (Figure 1) [10]. Such changes have the potential to impact gene functions, expression levels, or regulatory elements, which may consequently result in phenotypic differences or diseases. Insertions involve the addition of nucleotides into a DNA sequence, while deletions involve the removal of nucleotides. Small insertions or deletions, which are typically less than 50 base pairs in length, occur within coding regions or promoters. They can cause frameshift mutations or the abnormal expression of genes [11]. In contrast, larger insertions or deletions—which may encompass entire genes or chromosomal segments—may have more profound impacts on genome structure and function [12]. Duplication involves the replication of specific DNA segments in the genome, leading to an increase in their copy numbers. They can be categorized into two types: tandem duplications and dispersed duplications. To some extent, copy number variation can be regarded as a type of duplication. It can cover a relatively large genomic region, ranging from changes involving a few genes to those encompassing multiple gene clusters or even chromosomal segments. It therefore has the potential to significantly impact the functions of multiple genes, which has been confirmed in plants [13] and humans [14]. Inversion refers to a structural change in which a segment of DNA on a chromosome is reversed in orientation. Inversions can be classified into two types depending on whether the inverted region includes the centromere: pericentric inversions and paracentric inversions. Translocation refers to the movement of a chromosomal segment from one position to another. This movement can occur within the same chromosome (intra-chromosomal translocation) or between different chromosomes (inter-chromosomal translocation). Translocation may have significant impacts on gene function, chromosomal stability, and the phenotype of an organism.
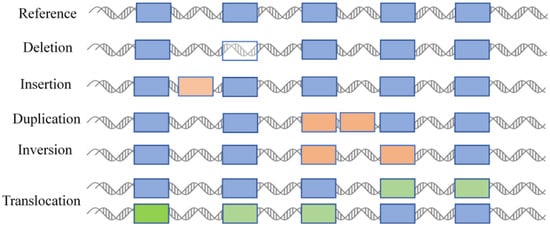
Figure 1.
Types of structural variations. Different colored boxes represent different fragments or genes in the genome. >50 bp is used as an operational demarcation.
Due to the limitations of early technologies, structural variations were defined mainly as large-fragment genomic alterations of >3 Mb that could be detected at the microscopic level [4]. About 20 years ago, the advent of hybridization-based microarray approaches and SNP arrays significantly enhanced the feasibility and efficiency of detecting structural variations [15,16]. These methods are now mostly used in fields such as the diagnosis of genetic diseases, genetic analysis of tumors, and population genetics research, and they have aided the discovery of genomic structural variations associated with human diseases. Minute deletions and duplications, which may lead to various congenital diseases, were detected in the fetal genome using hybridization-based microarray approaches [17]. With the advances in detection methods in recent years, especially DNA sequencing technologies and algorithms, whole-genome analysis has become viable for a wide range of plant species, and an increasing number of structural variations have been identified with higher efficiency. Initially, single-end and paired-end reads were employed for DNA sequencing [18]. However, owing to the limitations of these technologies, the sequencing reads were so short that structural variations could not be accurately detected. The latest breakthroughs in long-read sequencing and high-throughput chromosome conformation capture technologies present viable solutions to certain challenges related to short sequence reads [19]. The lengths of sequences obtained using long-read sequencing technologies, such as synthetic long-read sequencing and single-molecule long-read sequencing, can reach 10 to >100 kb [20,21,22]. These technologies have greatly improved the detection efficiency of structural variations. Particularly in recent years, with the development of pan-genomics and telomere-to-telomere (T2T) genomics, the genomes of many crops have been sequenced, like rice, maize, soybean, sorghum, and wheat. Based on these data, numerous structural variations have been detected and confirmed to be associated with traits [23,24]. Wei et al. sequenced two sorghum T2T genomes using ultra-long reads and 52 large inversions (>50 kb), and 68 translocations were detected between two sorghum T2T genomes [25]. The high-quality genomes of 17 representative wheat varieties from more than 5000 cultivars bred in China were assembled, and nearly 250,000 structural variations were identified. Among these, structural variations in the VRN-A1 and Ha genes were found to promote the adaptability and grain hardness of wheat, respectively [26].
3. The Naturally Occurring Causes of Structural Variations in Plants
Structural variations can be generated through multiple mechanisms, including non-allelic homologous recombination (NAHR), non-homologous end joining (NHEJ), fork stalling and template switching (FoSTeS), and transposable elements (TEs). NAHR is a biological mechanism for repairing broken chromosomes, which results in gross genomic rearrangements. This mechanism involves recombination between non-allelic homologous sequences. These homologous sequences, which are often repetitive elements such as low-copy repeats or transposable elements, can misalign during meiosis. When recombination occurs between these misaligned sequences, it can lead to the deletion or duplication of the DNA segment between the recombination sites [27]. The size of the deletion or duplication is largely dependent on the positioning of misaligned repeats and even translocations and chromosome fusions [28].
NHEJ is one of the important DNA repair methods for joining double-strand breaks in DNA. Under normal circumstances, it can connect broken DNA ends to maintain the integrity of the genome [29]. DNA double-strand breaks are induced when cells are exposed to physical factors like radiation, chemical factors such as specific mutagens, or biological factors, including viral infections. The NHEJ mechanism is likely to be activated and connect the broken ends. However, if the mechanism is abnormal, it may connect DNA ends incorrectly; for instance, during the repair of multiple DNA double-strand breaks, NHEJ may wrongly connect the broken ends of different chromosomes, thus leading to structural variations such as chromosomal translocations [30].
FoSTeS was proposed by Zhang F in 2009 to explain the genomic rearrangements associated with human diseases [31]. During DNA replication, the replication fork can stall due to various obstacles, such as DNA damage, tightly bound proteins, or sequences that are difficult to replicate. When the replication fork stalls, the replication machinery may switch to a different, nearby DNA template to continue replication. Depending on the location and nature of the template switch, this behavior can lead to the duplication or deletion of DNA segments.
TEs are DNA sequences in the genome that can move autonomously, and they are classified into two major categories based on their movement mechanisms: DNA transposons and retrotransposons. DNA transposons move via a “Cut-and-Paste” mechanism, relying on transposase to recognize and cleave the inverted repeat sequences at both ends of the transposon, excising it from the original locus and inserting it into a new genomic site. In contrast, retrotransposons mobilize via an RNA intermediate. Insertion of the element near or within a gene may disrupt the gene or lead to gene duplication. The transposable elements are widespread in plants and were also first discovered in them [32,33]. They result in the lineage-specific genome expansion and chromosome rearrangements after the split of two plant relatives [34].
4. The Impacts of Structural Variations
4.1. Crop Growth and Development
In rice, the “Green Revolution” gene SD1 encodes a key enzyme in the gibberellin synthesis pathway (Table 1). When a 383 bp deletion occurs in SD1, the synthesis of gibberellin in rice plants decreases, resulting in plant dwarfism [35]. Dwarf rice varieties have stronger lodging resistance, which is beneficial for increasing yields and has been widely used in rice breeding. An insertion of Ty1/Copia-like retrotransposon leads to the mutation of PH13 in soybean, which affects the gibberellin content, ultimately resulting in a reduction in plant height [36]. A major gene controlling plant height in upland cotton, GhPH1, was cloned and encodes gibberellin 2-oxidase 1A, a 1133 bp structural variation located approximately 16 kb upstream of GhPH1 that influences the expression of GhPH1, thereby affecting plant height [37]. The wheat dwarfing gene Rht-D1c leads to a reduction in plant height due to an increase in the copy number, and its dwarfing ability is more than three times that of the single copy gene [38].

Table 1.
Recent structural variations studies in crops.
Some research has reported that structural variations can impact the development of roots or nodulation. Zhang reported that the insertion of GmSINE1 transposons results in the loss of function of a specific R gene [45]. This change promotes the recognition of native soil rhizobia by soybeans and enhances the nitrogenase activity of nodules as well as the aboveground biomass. The type-II chalcone isomerase genes, unique to leguminous plants, have experienced substantial structural differentiation, which is likely the result of tandem duplication events. This genetic diversification has played a significant role in enhancing the nodulation process in soybeans and Medicago truncatul [46]. Transposable elements were found in maize and have influenced many traits. Many allelic variations related to flowering time are caused by transposon insertions [65]. The transposons of ZmCCT9 and ZmCCT10 arose sequentially following domestication and were targeted by selection as maize spread from the tropics to higher latitudes [53]. The insertion of a Ty1/copia-like retrotransposon into the E4 gene led to a decrease in the expression level of the E4 gene. As a result, soybeans became insensitive to long day conditions [47]. The deletion of the E1 gene, a core gene associated with flowering time, could significantly advance the flowering time of soybeans [48,49].
4.2. Crop Yield
Structural variations are not only associated with agronomic traits but also contribute to crop yield. For example, the gain number is an important trait for rice yield. Structural variations in Gn1 disrupt its function, resulting in cytokinin accumulation in inflorescence meristems and an increase in the number of reproductive organs [39]. A 1.2 kb transposon-containing insertion 60 Kb downstream of KRN4 regulated the level of its expression, resulting in kernel row number in maize [54]. A large number of structural variations in tomato were identified through super-pangenome analyses. Among them, a 244 bp deletion detected in exon of cytochrome P450 may affect plant architecture and yield [61].
Moreover, structural variations have the potential to exert an influence on the diverse alternative splicing patterns of AUXIN RESPONSE FACTOR 2 transcripts, subsequently contributing to the processes of pod development and size selection in peanut [60]. The size and shape of tomato fruits are important appearance quality traits. Through research on the tomato genome, it has been found that multiple structural variations are related to fruit size and shape. For example, a 102 kb inversion at the SUN locus changes the gene expression regulation pattern in this region, causing the tomato fruit to change from round to oval, which significantly affects the appearance quality [62]. An increase in the copy number of genes related to cell division and expansion has been discovered in large-fruited tomato varieties. These structural variations promote the division and expansion of fruit cells, thereby increasing the size of the fruits [66]. In rice, a two-haplotype deletion in the promoter of GSE5 reduced the expression of GSE5, which caused wide and heavy grain. Conversely, the overexpression of GES5 resulted in narrow grains [43].
4.3. Crop Quality
Crop quality is important in many fields such as human life, agricultural production, and the ecological environment. Taking the nutritional quality of crops as an example, many studies have revealed the crucial roles played by structural variations. Glucosinolates—an important class of secondary metabolites in rapeseed—not only affect the disease and pest resistance of rapeseed but also are closely related to the quality of rapeseed oil. By integrating population transcriptome data, structural variations can regulate the expression of genes related to the synthesis and transport of glucosinolates in rapeseed [64]. A 1454 bp insertion downstream of BnaA03.MAMf regulated the level of its expression, resulting in lower 4C:(4C + 5C) or higher 5C:(4C + 5C) ratios of glucosinolate. Meanwhile, accessions carrying a 41.6 kb insertion in the BnaA09.MYB28 exhibited significantly higher glucosinolate content compared to those without the insertion [64]. The low-molecular-weight glutenin subunit is an essential part of the storage protein in wheat, and the composition, copy number, and expression of core genes all influence wheat quality traits [67,68].
4.4. Crop Stress Resistance
Although plants are highly adaptable to these environments, transient changes in their growth environment still pose stress to them. Structural variations enable plants to adapt to environmental changes, and environmental changes can also give rise to the occurrence of structural variations. Some studies have found that environmental stress can activate transposons. For example, the transcriptional activity of TE-related genes in rice under stress and normal conditions was detected, and it was found that transcriptional activity of TE-related genes was significantly upregulated under stress conditions [69]. Tto1 and Tnt1 retrotransposons in tobacco and oat were activated by physical stresses like cutting and stab wounding, elicitors of plant defense responses, and pathogen infection [70,71].
In barley, an increase in the copy number of the boron transporter gene Bot1 enables the barley variety Sahara to tolerate boron toxicity [58]. CBF (C-repeat binding factor) is located at the frost resistance locus 2 (FR-2) in wheat and barley, and its copy number variations are associated with low-temperature tolerance. The copy number of CBF14 in winter wheat is higher than that in spring wheat. Deletions of large genes (including CBF-B12, CBF-B14, and CBF-B15) at the FR-2 locus in tetraploid durum wheat and hexaploid bread wheat lead to reduced cold tolerance [56,72,73,74]. In rice, cold can induce the expression of the LIP19 gene, the high expression level of which is significantly associated with cold tolerance. Interestingly, the 150 bp insertion in the promoter of LIP19 resulted in significantly higher LIP19 than normal, which promoted cold tolerance in Oryza sativa japonica (with a 150 bp insertion) compared to Oryza sativa indica without a 150 bp insertion [41]. He et al. conducted a large-scale screening of the genomic inversion in cultivated and wild rice accessions and found that genomic inversion widely exists [42]. An inversion cluster including MADS56 showed significantly higher expression levels and stronger heat-resistance compared to the non-inversion cluster. In wheat, an increase in the copy number of Vrn-A1 prolongs the vernalization time and results in delayed flowering, while a high copy number of HvFT1 in barley accelerates the flowering time [57,59]. A compelling association was unearthed between the expression of genes implicated in seed oil content and genomic structural variations within the quantitative trait loci of Brassica napus. Specifically, an insertion of 6313 bp in the promoter region of the BnaA09g48250D gene had a significant connection with the seed oil content [63]. Transposon-mediated inverted repeats (21.4 kb) in maize were found though bioinformatics, genetics, and molecular biology, resulting in environmental adaptation and yield balance [55].
4.5. Domestication and Evolution
Structural variations have an interactive and close relationship with crop domestication and evolution, providing an important genetic basis and driving force. In turn, crop domestication and evolution influence the direction and frequency of structural variations and, together, these mutual influences drive the development of crops from a wild state to one that is better adapted to human needs and environmental changes. Structural variations have the capacity to engender novel genetic combinations at the genomic scale. This process serves as a wellspring of rich raw materials, fueling the domestication and evolution of crops. During soybean domestication, the seed coat color shifted from wild soybean black to the most cultivated soybean yellow, and this process was influenced by structural variations. The classical I locus drives this change. Certain structural variation events formed different I locus haplotypes, which are genomic level structural changes. These alter gene expression, diversifying seed coat colors. Different haplotypes, linked to different domestication stages, show that structural variation-induced genetic differences underpin the evolution of soybean seed coat color traits. Humans, based on appearance needs, chose yellow coated soybeans, spurring their evolution into cultivars [47,50,51].
In wheat, copy number variations in the vernalization gene VRN-A1 play a crucial role in the evolution of spring-type and winter-type wheat. It was found, through the analysis of high-quality genomes of 17 representative wheat varieties, that there is a large block copy number variation of approximately 0.5 Mb in the interval where the VRN-A1 gene is located. This variation includes structural changes such as duplications and deletions, which directly affect the gene expression level. In cold regions, winter-type varieties with high copy numbers are retained due to their frost resistance advantages, while in warm regions, spring-type varieties with low copy numbers are preferred [26]. Maize was directly domesticated from teosinte (Zea mays ssp. parviglumis). Before it was confirmed that teosinte was the domesticated ancestor of maize, Tripsacum was thought to be the origin of maize domestication. Through genomic comparative analysis, this study identified the largest (3.2 Mb) variant fragment (named Region A) between the genomes of two inbred maize lines, B73 and Mo17. In the natural population, this fragment showed a high degree of sequence integrity, existing only in two forms: complete presence and complete absence. Through in-depth research on the sequence of this fragment, it was found that there are long terminal repeat retrotransposons specific to Tripsacum within it, indicating that the Tripsacum genome has contributed to the formation of this region in the maize genome. Signals of gene flow between maize and Tripsacum were also detected within this fragment, suggesting that Tripsacum may have directly participated in the speciation process of maize [75].
Some studies have revealed that structural variations have an important effect on the domestication and evolution of rice. Qin selected 35 rice materials and constructed a pan-genome using third-generation sequencing technology. Based on these sequences, 171,072 structural variations, 125,889 InDels, 627 inversions, and 4346 translocations were detected. Of them, a 66.6 kb deletion in japonica rice materials and a 43.3 kb deletion in Indica rice materials both contain the complete sequence of the known negative regulatory gene for blast resistance, OsWAK112d. Evidently, these two independent deletions contribute to the environmental adaptation of rice by enhancing blast resistance [40].
5. How to Apply Structural Variations to Crop Breeding
5.1. Molecular Marker-Assisted Breeding
Based on closely linking the molecular marker and target traits, molecular marker-assisted breeding enables accurate selection in the early stages of crop breeding, which helps to accelerate the development of superior varieties and enhances the quality and adaptability of the final varieties. Compared to SNPs, structural variations can be more easily developed into molecular markers and efficiently used for material detection. The soybean cyst nematode is estimated to be the pest that causes the most yield loss in soybean. The copy number of the Rhg1 gene mediated resistance to soybean cyst nematode. Lee et al. have developed an efficient method for measuring the copy number variation, and have applied this method to improve cyst nematode resistance in soybean [52]. In rice, the copy number variation in GL7 regulating longitudinal cell elongation was associated with grain size diversity and selected for and used in breeding [44]. Moreover, structural variations can be used to understand the genetic basis of complex agronomic traits. By analyzing the distribution of structural variations in different crop populations, we can gain insights into the evolutionary processes and genetic diversity of crops. This knowledge can guide us to more precisely combine different genetic resources, creating new crop varieties with beneficial, comprehensive traits and better adaptation to various environmental challenges in the future. Shading can cause exaggerated stem elongation in soybean, leading to lodging and reduced yields when planted at a high density. A Ty1/Copia-like retrotransposon insertion in the PH13 gene results in a truncated PH13 protein, which causes reduced plant height and enables high-density planting [36]. Molecular markers based on retrotransposon insertion in the PH13 gene were developed, followed by introgression of the insertion-type ph13 allele into wild-type materials, enabling the rapid genotypic selection of progeny (Figure 2A).
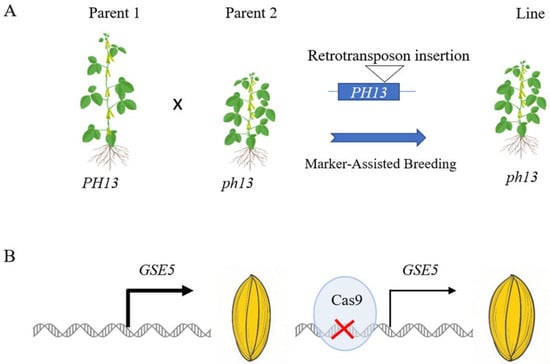
Figure 2.
Application of structural variations for crop breeding. (A) in a molecular-marker-assisted breeding approach, Parent 1 carrying the wild-type PH13 allele was crossed with the parent harboring the retrotransposon insertion ph13 allele. Subsequently, a specific molecular marker was developed and employed to screen the genotypes of the breeding progeny with ph13 allele. (B) using a gene editing-based breeding approach, cultivars carrying the wild-type GSE5 were employed to edit the promoter of GSE5, which reduced the expression of GSE5 and resulted in wider and heavier seeds.
5.2. Gene Editing Technology
With the development of gene editing technologies, it is now possible to perform precise modifications at specific locations within the genome, including introducing, repairing, or deleting structural variations. By precisely editing the structural variation regions in crops, researchers can determine the specific impacts of these structural variations on target traits and thereby accurately screen out those with breeding value.
Based on the crop breeding target, researchers can design and create novel structural variations. They can simultaneously edit multiple genes associated with important traits and introduce them into a single crop variety to aggregate beneficial traits. In 2024, researchers introduced a 10 bp heat shock element into tomato and rice through a new editing system, which increased the yield in a heat environment [76]. GSE5 regulated grain size in rice, and deletion in the promoter of GSE5 reduced expression and then caused wide grain and increased yield [43]. Gene editing was used to knock out the promoter of GSE5, thereby reducing its expression level and rapidly improving grain size (Figure 2B). With the combination of gene editing and structural variation, researchers can rapidly improve the target traits and increase grain yield.
6. Opportunities and Challenges
In the field of crop genetics and breeding, structural variations are gradually becoming the focus of research. Structural variations are an important source of genetic diversity in crops. Traditional genetic variations mainly focus on single-nucleotide polymorphisms, while structural variations can generate more complex and extensive genetic changes. These variations can create new gene combinations and alleles, providing rich genetic materials for crop breeding. With the help of modern molecular biology techniques, such as high-throughput sequencing and gene-editing technology, we can quickly and accurately detect and identify structural variations in crops. This enables us to precisely select target traits at the early stage of breeding, greatly shortening the breeding cycle. However, research on structural variations also faces some challenges, for example in terms of their detection and identification. Structural variations typically involve large genomic fragments, and their detection and identification require high-precision technical means. Although great progress has been made in high-throughput sequencing technology, there are still problems of inaccurate detection or missed detection for some complex structural variations, such as hidden inversions and translocations. Moreover, different sequencing platforms and analysis methods may lead to differences in results, which poses a significant challenge to the accurate identification of structural variations. Determining the biological functions of structural variations is an extremely challenging task. Since structural variations may affect multiple genes and their regulatory networks, their impacts on crop traits often exhibit complex pleiotropy. A single structural variation can influence multiple traits simultaneously, and different genetic backgrounds and environmental conditions can also affect its functions. Therefore, to accurately decipher the functions of structural variations, it is necessary to comprehensively apply multidisciplinary methods such as genetics, molecular biology, and bioinformatics and conduct a large number of experimental verifications.
Author Contributions
X.W., C.L., X.S., G.S., C.Z., Y.Q., Y.B. and W.L. drafted the manuscript. F.K., H.L. and Y.W. conceived the article and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the National Soybean Industry Technology System (CARS-04-CES09), Heilongjiang Provincial Department of Finance (CZKYF2025-1-A001), and Agricultural Science and Technology Innovation Leaping Project in Heilongjiang Province (Grant No. CX23ZD04).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Shi, J.; Lai, J. Patterns of genomic changes with crop domestication and breeding. Curr. Opin. Plant Biol. 2015, 24, 47–53. [Google Scholar] [CrossRef]
- Huang, X.; Huang, S.; Han, B.; Li, J. The integrated genomics of crop domestication and breeding. Cell 2022, 185, 2828–2839. [Google Scholar] [CrossRef] [PubMed]
- Escaramís, G.; Docampo, E.; Rabionet, R. A decade of structural variants: Description, history and methods to detect structural variation. Brief Funct. Genom. 2015, 14, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Feuk, L.; Carson, A.; Scherer, S. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Scott, A.J.; Davis, J.R.; Tsang, E.K.; Li, X.; Kim, Y.; Hadzic, T.; Damani, F.N.; Ganel, L.; Montgomery, S.B.; et al. The impact of structural variation on human gene expression. Nat. Genet. 2017, 49, 692–699. [Google Scholar] [CrossRef]
- Meyers, L.A.; Levin, D.A. On the abundance of polyploids in flowering plants. Evolution 2006, 60, 1198–1206. [Google Scholar]
- Yuan, Y.; Bayer, P.E.; Batley, J.; Edwards, D. Current status of structural variation studies in plants. Plant Biotechnol. J. 2021, 19, 2153–2163. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef]
- Schiessl, S.V.; Katche, E.; Ihien, E.; Chawla, H.S.; Mason, A.S. The role of genomic structural variation in the genetic improvement of polyploid crops. Crop J. 2019, 7, 127–140. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Chia, J.M.; Song, C.; Bradbury, P.J.; Costich, D.; de Leon, N.; Doebley, J.; Elshire, R.J.; Gaut, B.; Geller, L.; Glaubitz, J.C.; et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 2012, 44, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Iovene, M.; Zhang, T.; Lou, Q.; Buell, C.R.; Jiang, J. Copy number variation in potato an asexually propagated autotetraploid species. Plant J. 2013, 75, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Zarrei, M.; MacDonald, J.R.; Merico, D.; Scherer, S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015, 16, 172–183. [Google Scholar] [CrossRef]
- Solinas-Toldo, S.; Lampel, S.; Stilgenbauer, S.; Nickolenko, J.; Benner, A.; Döhner, H.; Cremer, T.; Lichter, P. Matrix-based comparative genomic hybridization: Biochips to screen for genomic imbalances. Genes Chromosomes Cancer 1997, 20, 399–407. [Google Scholar] [CrossRef]
- Pinkel, D.; Segraves, R.; Sudar, D.; Clark, S.; Poole, I.; Kowbel, D.; Collins, C.; Kuo, W.L.; Chen, C.; Zhai, Y.; et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998, 20, 207–211. [Google Scholar] [CrossRef]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010, 86, 749–764. [Google Scholar] [CrossRef]
- Alkan, C.; Coe, B.P.; Eichler, E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011, 12, 363–376. [Google Scholar] [CrossRef]
- Harewood, L.; Kishore, K.; Eldridge, M.D.; Wingett, S.; Pearson, D.; Schoenfelder, S.; Collins, V.P.; Fraser, P. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biology 2017, 18, 125. [Google Scholar] [CrossRef]
- Huddleston, J.; Chaisson, M.J.P.; Steinberg, K.M.; Warren, W.; Hoekzema, K.; Gordon, D.; Graves-Lindsay, T.A.; Munson, K.M.; Kronenberg, Z.N.; Vives, L.; et al. Corrigendum: Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2018, 28, 144. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Heller, D.; Vingron, M. SVIM: Structural variant identification using mapped long reads. Bioinformatics 2019, 35, 2907–2915. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 2019, 12, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Della Coletta, R.; Qiu, Y.; Ou, S.; Hufford, M.B.; Hirsch, C.N. How the pan-genome is changing crop genomics and improvement. Genome Biol. 2021, 22, 3. [Google Scholar] [CrossRef]
- Wei, C.; Gao, L.; Xiao, R.; Wang, Y.; Chen, B.; Zou, W.; Li, J.; Mace, E.; Jordan, D.; Tao, Y. Complete telomere-to-telomere assemblies of two sorghum genomes to guide biological discovery. Imeta 2024, 3, e193. [Google Scholar] [CrossRef]
- Jiao, C.; Xie, X.; Hao, C.; Chen, L.; Xie, Y.; Garg, V.; Zhao, L.; Wang, Z.; Zhang, Y.; Li, T.; et al. Pan-genome bridges wheat structural variations with habitat and breeding. Nature 2025, 637, 384–393. [Google Scholar] [CrossRef]
- Parks, M.M.; Lawrence, C.E.; Raphael, B.J. Detecting non-allelic homologous recombination from high-throughput sequencing data. Genome Biol. 2015, 16, 72. [Google Scholar] [CrossRef]
- Robberecht, C.; Voet, T.; Zamani Esteki, M.; Nowakowska, B.A.; Vermeesch, J.R. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res. 2013, 23, 411–418. [Google Scholar] [CrossRef]
- Nenarokova, A.; Záhonová, K.; Krasilnikova, M.; Gahura, O.; McCulloch, R.; Zíková, A.; Yurchenko, V.; Lukeš, J. Causes and effects of loss of classical nonhomologous end joining pathway in Parasitic Eukaryotes. Microbiol. Biol. 2019, 10, e01541-19. [Google Scholar] [CrossRef]
- Ferguson, D.O.; Sekiguchi, J.M.; Chang, S.; Frank, K.M.; Gao, Y.; DePinho, R.A.; Alt, F.W. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA 2000, 97, 6630–6633. [Google Scholar] [CrossRef]
- Zhang, F.; Khajavi, M.; Connolly, A.M.; Towne, C.F.; Batish, S.D.; Lupski, J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009, 41, 849–853. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Liu, J.; Zhang, T.; Su, T.; Li, S.; Cheng, Q.; Kong, L.; Li, X.; Bu, T.; Li, H.; et al. A recent retrotransposon insertion of J caused E6 locus facilitating soybean adaptation into low latitude. J. Integr. Plant Biol. 2021, 63, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, J.; Cai, Z.; Huang, Y.; Lv, M.; Du, H.; Gao, Q.; Zuo, Y.; Dong, Z.; Huang, W.; et al. Evolution and domestication footprints uncovered from the genomes of Coix. Mol. Plant 2020, 13, 295–308. [Google Scholar] [CrossRef]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Qin, C.; Li, Y.H.; Li, D.; Zhang, X.; Kong, L.; Zhou, Y.; Lyu, X.; Ji, R.; Wei, X.; Cheng, Q.; et al. PH13 improves soybean shade traits and enhances yield for high-density planting at high latitudes. Nat. Commun. 2023, 14, 6813. [Google Scholar] [CrossRef]
- Tian, Z.; Chen, B.; Li, H.; Pei, X.; Sun, Y.; Sun, G.; Pan, Z.; Dai, P.; Gao, X.; Geng, X.; et al. Strigolactone-gibberellin crosstalk mediated by a distant silencer fine-tunes plant height in upland cotton. Mol. Plant 2024, 17, 1539–1557. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, J.; Wu, J.; Duan, J.; Liu, Y.; Ye, X.; Zhang, X.; Guo, X.; Gu, Y.; Zhang, L.; et al. A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 2012, 196, 282–291. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Qin, P.; Lu, H.; Du, H.; Wang, H.; Chen, W.; Chen, Z.; He, Q.; Ou, S.; Zhang, H.; Li, X.; et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 2021, 184, 3542–3558. [Google Scholar] [CrossRef]
- Li, X.; Dai, X.; He, H.; Lv, Y.; Yang, L.; He, W.; Liu, C.; Wei, H.; Liu, X.; Yuan, Q.; et al. A pan-TE map highlights transposable elements underlying domestication and agronomic traits in Asian rice. Natl. Sci. Rev. 2024, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- He, W.; He, H.; Yuan, Q.; Zhang, H.; Li, X.; Wang, T.; Yang, Y.; Yang, L.; Yang, Y.; Liu, X.; et al. Widespread inversions shape the genetic and phenotypic diversity in rice. Sci. Bull. 2024, 69, 593–596. [Google Scholar] [CrossRef]
- Duan, P.; Xu, J.; Zeng, D.; Zhang, B.; Geng, M.; Zhang, G.; Huang, K.; Huang, L.; Xu, R.; Ge, S.; et al. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 2017, 10, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, M.; Sun, Y.; Zhao, P.; Liu, C.; Qing, K.; Hu, X.; Zhong, Z.; Cheng, J.; Wang, H.; et al. Glycine max NNL1 restricts symbiotic compatibility with widely distributed bradyrhizobia via root hair infection. Nat. Plants 2021, 7, 73–86. [Google Scholar] [CrossRef]
- Liu, T.; Liu, H.; Xian, W.; Liu, Z.; Yuan, Y.; Fan, J.; Xiang, S.; Yang, X.; Liu, Y.; Liu, S.; et al. Duplication and sub-functionalization of flavonoid biosynthesis genes plays important role in Leguminosae root nodule symbiosis evolution. J. Integr. Plant Biol. 2024, 66, 2191–2207. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-genome of wild and cultivated soybeans. Cell 2020, 182, 162–176. [Google Scholar] [CrossRef]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lü, S.; et al. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef]
- Zhai, H.; Lü, S.; Wu, H.; Zhang, Y.; Zhang, X.; Yang, J.; Wang, Y.; Yang, G.; Qiu, H.; Cui, T.; et al. Diurnal expression pattern, allelic variation, and association analysis reveal functional features of the E1 gene in control of photoperiodic flowering in soybean. PLoS ONE 2015, 10, e0135909. [Google Scholar] [CrossRef]
- Tuteja, J.H.; Clough, S.J.; Chan, W.C.; Vodkin, L.O. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 2004, 16, 819–835. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.G.; Diers, B.W.; Hudson, M.E. An efficient method for measuring copy number variation applied to improvement of nematode resistance in soybean. Plant J. 2016, 88, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Sun, H.; Xu, D.; Chen, Q.; Liang, Y.; Wang, X.; Xu, G.; Tian, J.; Wang, C.; Li, D.; et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 2018, 115, E334–E341. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Du, Y.; Shen, X.; Li, M.; Sun, W.; Huang, J.; Liu, Z.; Tao, Y.; Zheng, Y.; Yan, J.; et al. KRN4 controls quantitative variation in maize kernel row number. PLoS Genet. 2015, 11, e1005670. [Google Scholar] [CrossRef]
- Sun, X.; Xiang, Y.; Dou, N.; Zhang, H.; Pei, S.; Franco, A.V.; Menon, M.; Monier, B.; Ferebee, T.; Liu, T.; et al. The role of transposon inverted repeats in balancing drought tolerance and yield-related traits in maize. Nat. Biotechnol. 2023, 41, 120–127. [Google Scholar] [CrossRef]
- Francia, E.; Morcia, C.; Pasquariello, M.; Mazzamurro, V.; Milc, J.A.; Rizza, F.; Terzi, V.; Pecchioni, N. Copy number variation at the HvCBF4-HvCBF2 genomic segment is a major component of frost resistance in barley. Plant Mol. Biol. Rep. 2016, 92, 161–175. [Google Scholar] [CrossRef]
- Armour, J.A.; Sismani, C.; Patsalis, P.C.; Cross, G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000, 28, 605–609. [Google Scholar] [CrossRef]
- Sutton, T.; Baumann, U.; Hayes, J.; Collins, N.C.; Shi, B.J.; Schnurbusch, T.; Hay, A.; Mayo, G.; Pallotta, M.; Tester, M.; et al. Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 2007, 318, 1446–1449. [Google Scholar] [CrossRef]
- Nitcher, R.; Distelfeld, A.; Tan, C.; Yan, L.; Dubcovsky, J. Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol. Plant Genom. 2013, 288, 261–275. [Google Scholar] [CrossRef]
- Yin, D.; Ji, C.; Song, Q.; Zhang, W.; Zhang, X.; Zhao, K.; Chen, C.Y.; Wang, C.; He, G.; Liang, Z.; et al. Comparison of Arachis monticola with diploid and cultivated tetraploid genomes reveals asymmetric subgenome evolution and improvement of peanut. Adv. Sci. 2019, 7, 1901672. [Google Scholar] [CrossRef]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.; Huang, S.; Yang, T.; Tang, Y.; Yang, S.; Aisimutuola, P.; et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, L.; Li, H.; He, J.; Chao, H.; Yan, S.; Yin, Y.; Zhao, W.; Li, M. 3D genome structural variations play important roles in regulating seed oil content of Brassica napus. Plant Commun. 2024, 5, 100666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Z.; He, Y.; Liu, D.; Liu, Y.; Liang, C.; Xie, M.; Jia, Y.; Ke, Q.; Zhou, Y.; et al. Structural variation reshapes population gene expression and trait variation in 2105 Brassica napus accessions. Nat. Genet. 2024, 56, 2538–2550. [Google Scholar] [CrossRef]
- Su, H.; Cao, L.; Ren, Z.; Sun, W.; Zhu, B.; Ma, S.; Sun, C.; Zhang, D.; Liu, Z.; Zeng, H.; et al. ZmELF6-ZmPRR37 module regulates maize flowering and salt response. Plant Biotechnol. J. 2024, 22, 929–945. [Google Scholar] [CrossRef]
- Alonge, M.; Wang, X.; Benoit, M.; Soyk, S.; Pereira, L.; Zhang, L.; Suresh, H.; Ramakrishnan, S.; Maumus, F.; Ciren, D.; et al. Major impacts of widespread structural variation on gene expression and crop improvement in Tomato. Cell 2020, 182, 145–161.e23. [Google Scholar] [CrossRef]
- Harberd, N.P.; Bartels, D.; Thompson, R.D. Analysis of the gliadin multigene loci in bread wheat using nullisomic-tetrasomic lines. Mol. Gen. Genet. 1985, 198, 234–242. [Google Scholar] [CrossRef]
- Beom, H.R.; Kim, J.S.; Jang, Y.R.; Lim, S.H.; Kim, C.K.; Lee, C.K.; Lee, J.Y. Proteomic analysis of low-molecular-weight glutenin subunits and relationship with their genes in a common wheat variety. 3 Biotech 2018, 8, 56. [Google Scholar] [CrossRef]
- Jiao, Y.; Deng, X.W. A genome-wide transcriptional activity survey of rice transposable element-related genes. Genome Biol. 2007, 8, R28. [Google Scholar] [CrossRef]
- Yosuke, K.; Yukio, T.; Saori, S.; Ryohei, S.; Motoaki, K.; Tetsuo, S.; Shigeyuki, B.; Yukiko, E.; Hitoshi, N.; Shigeyuki, M. OARE-1, a Ty1-copia retrotransposon in oat activated by abiotic and biotic stresses. Plant Cell Physiol. 2001, 42, 1345–1354. [Google Scholar]
- Takeda, S.; Sugimoto, K.; Otsuki, H.; Hirochika, H. A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 1999, 18, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Knox, A.K.; Dhillon, T.; Cheng, H.; Tondelli, A.; Pecchioni, N.; Stockinger, E.J. CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor. Appl. Genet. 2010, 121, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.; Zhu, J.; Boldizsár, Á.; Vágújfalvi, A.; Burke, A.; Garland-Campbell, K.; Galiba, G.; Dubcovsky, J. Large deletions in the CBF gene cluster at the Fr-B2 locus are associated with reduced frost tolerance in wheat. Theor. Appl. Genet. 2013, 126, 2683–2697. [Google Scholar] [CrossRef] [PubMed]
- Díaz, A.; Zikhali, M.; Turner, A.S.; Isaac, P.; Laurie, D.A. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE 2012, 7, e33234. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, W.; Meng, Z.; Braz, G.T.; Li, Y.; Wang, K.; Wang, H.; Lai, J.; Jiang, J.; Dong, Z.; et al. Megabase-scale presence-absence variation with Tripsacum origin was under selection during maize domestication and adaptation. Genome Biol. 2021, 22, 237. [Google Scholar] [CrossRef]
- Lou, H.; Li, S.; Shi, Z.; Zou, Y.; Zhang, Y.; Huang, X.; Yang, D.; Yang, Y.; Li, Z.; Xu, C. Engineering source-sink relations by prime editing confers heat-stress resilience in tomato and rice. Cell 2025, 188, 530–549. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).