Disclosing Pathogenic Variant Effects on the Structural Dynamics of the VAPB MSP Domain Causing Familial ALS
Abstract
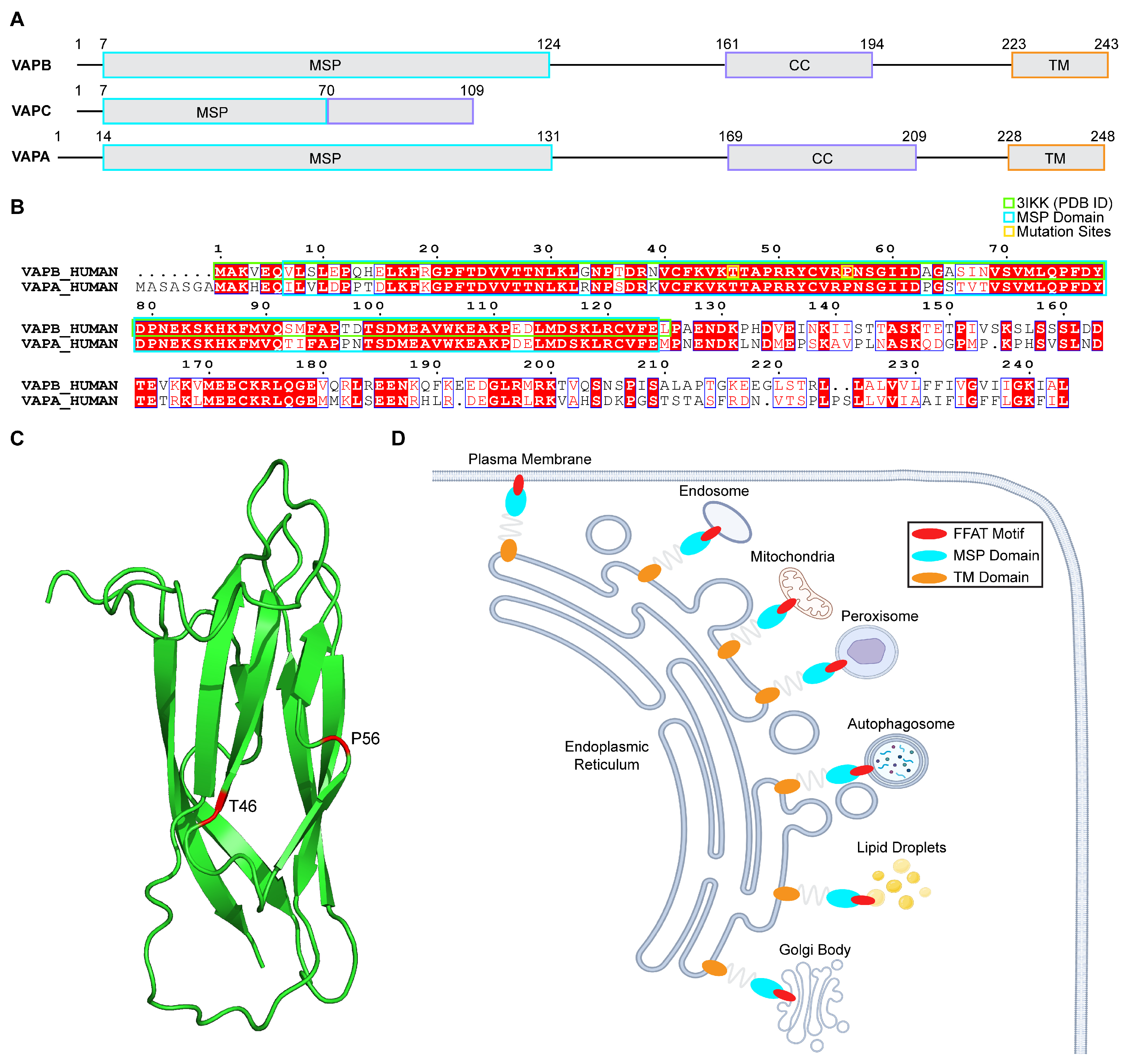
1. Introduction
2. Results
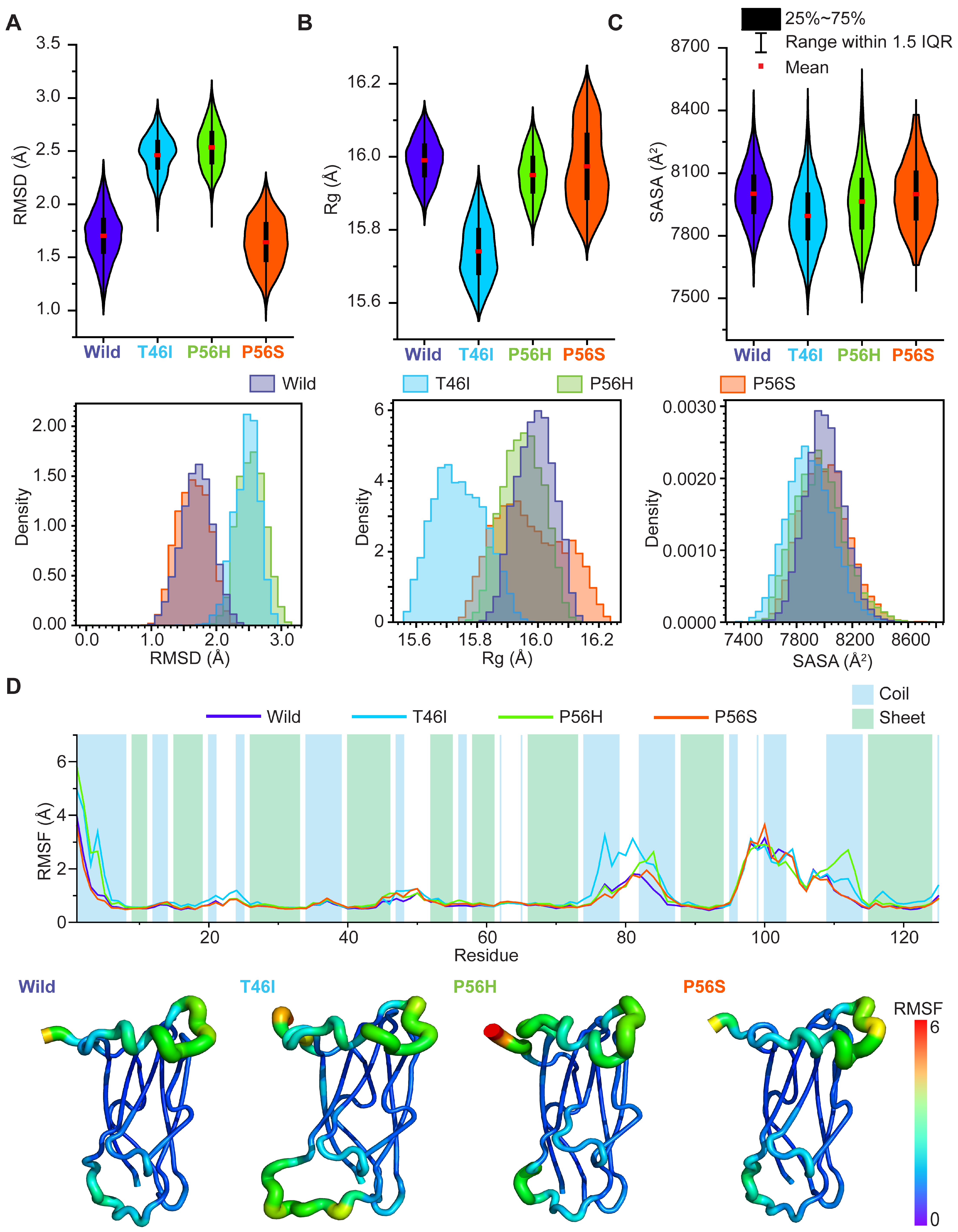
2.1. Pathogenic Variants Induce Conformational Alterations in the VAPB MSP Domain
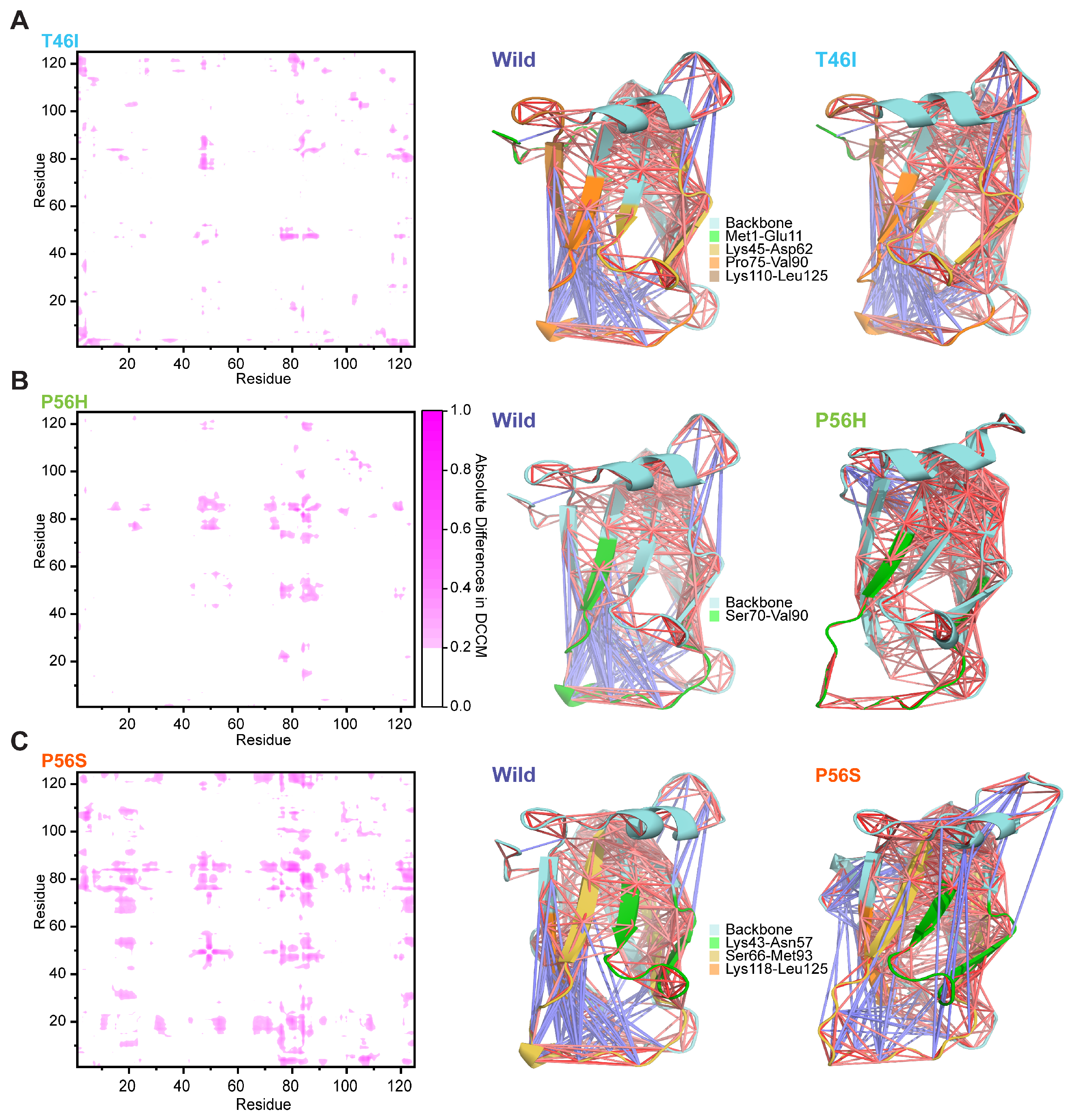
2.2. Pathogenic Variants Alter the Dynamic Correlated and Anticorrelated Motions of the VAPB MSP Domain
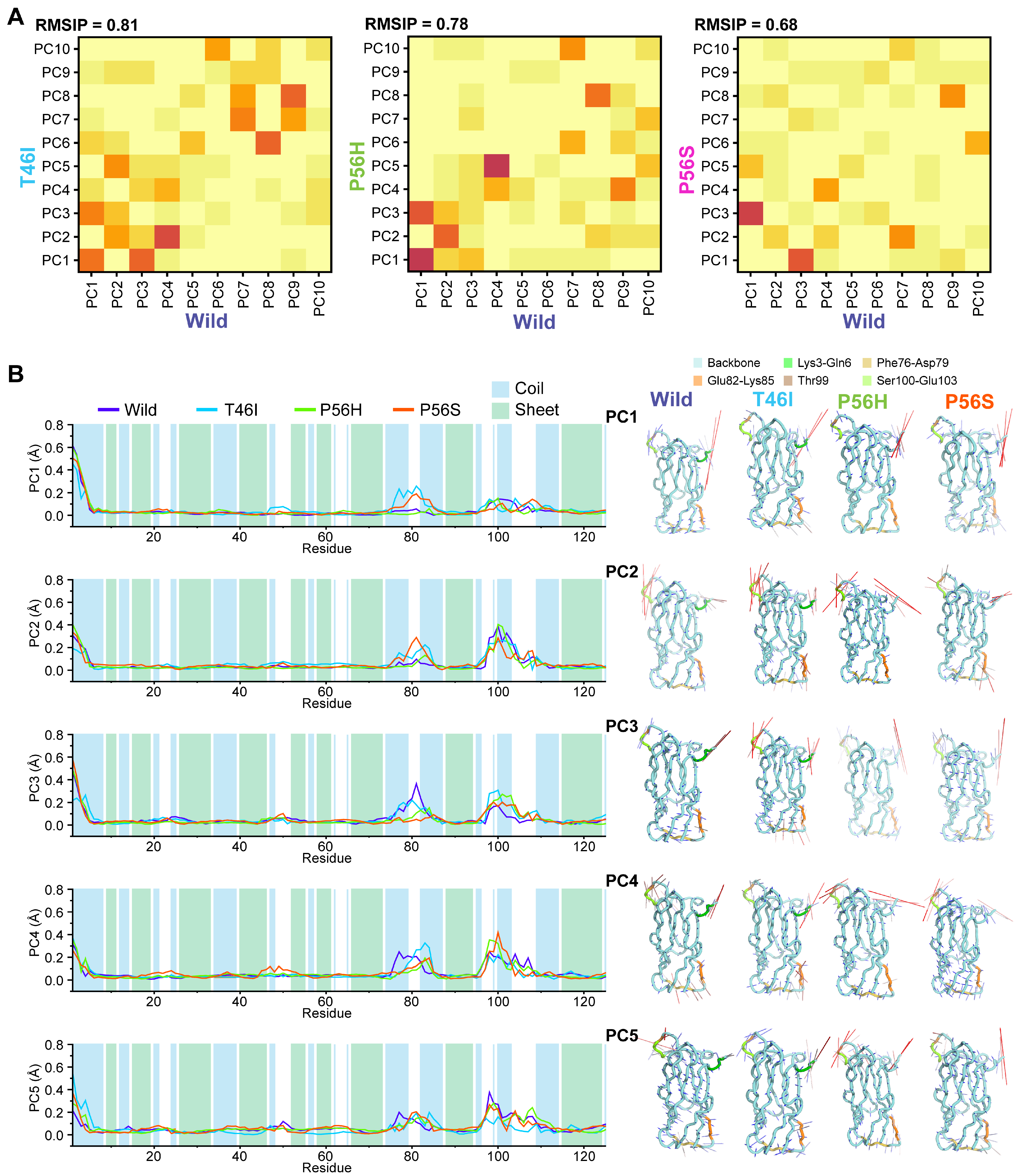
2.3. Pathogenic Variants Alter the Residual Conformational Motions
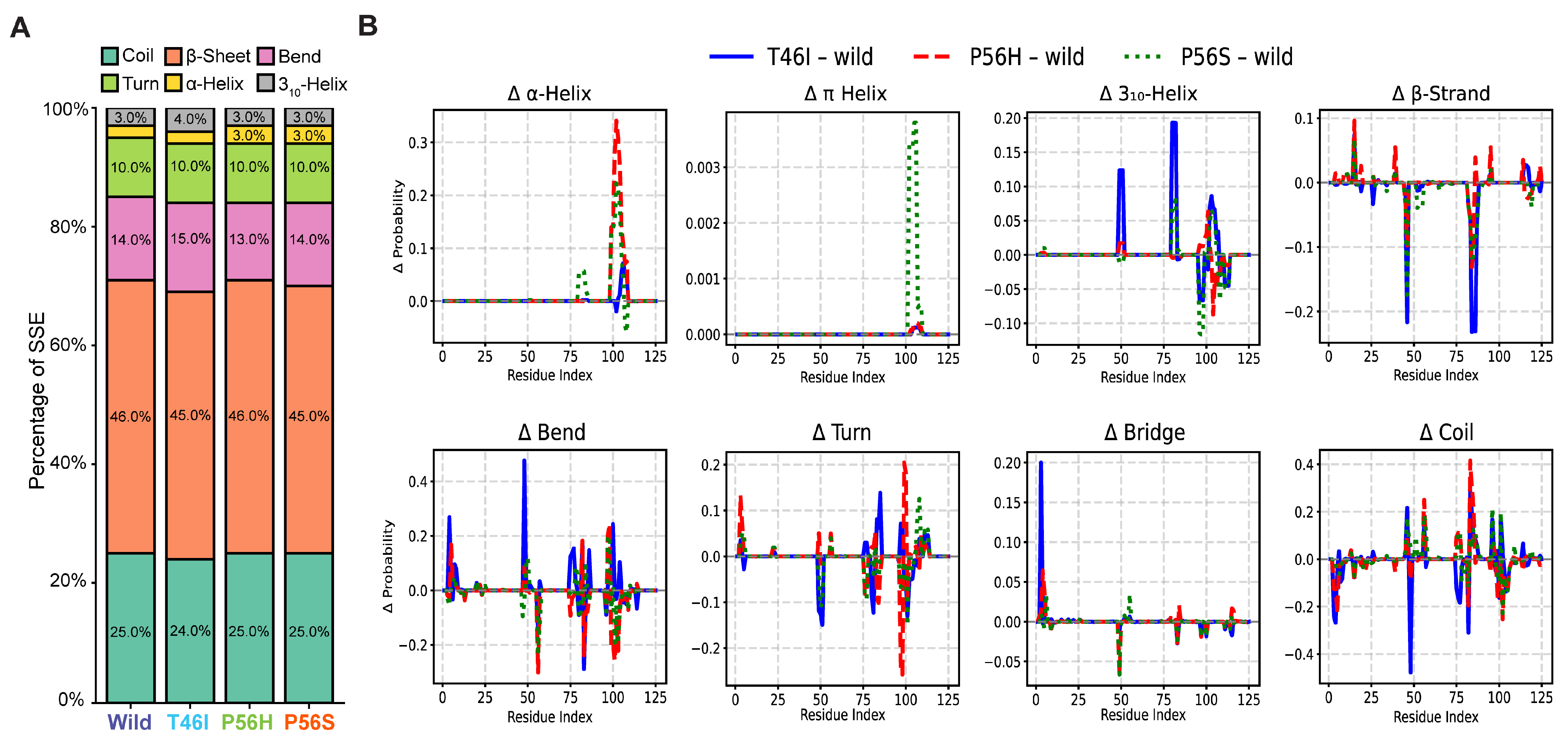
2.4. Pathogenic Variants Alter the Organization of Secondary Structure Formation in the VAPB MSP Domain
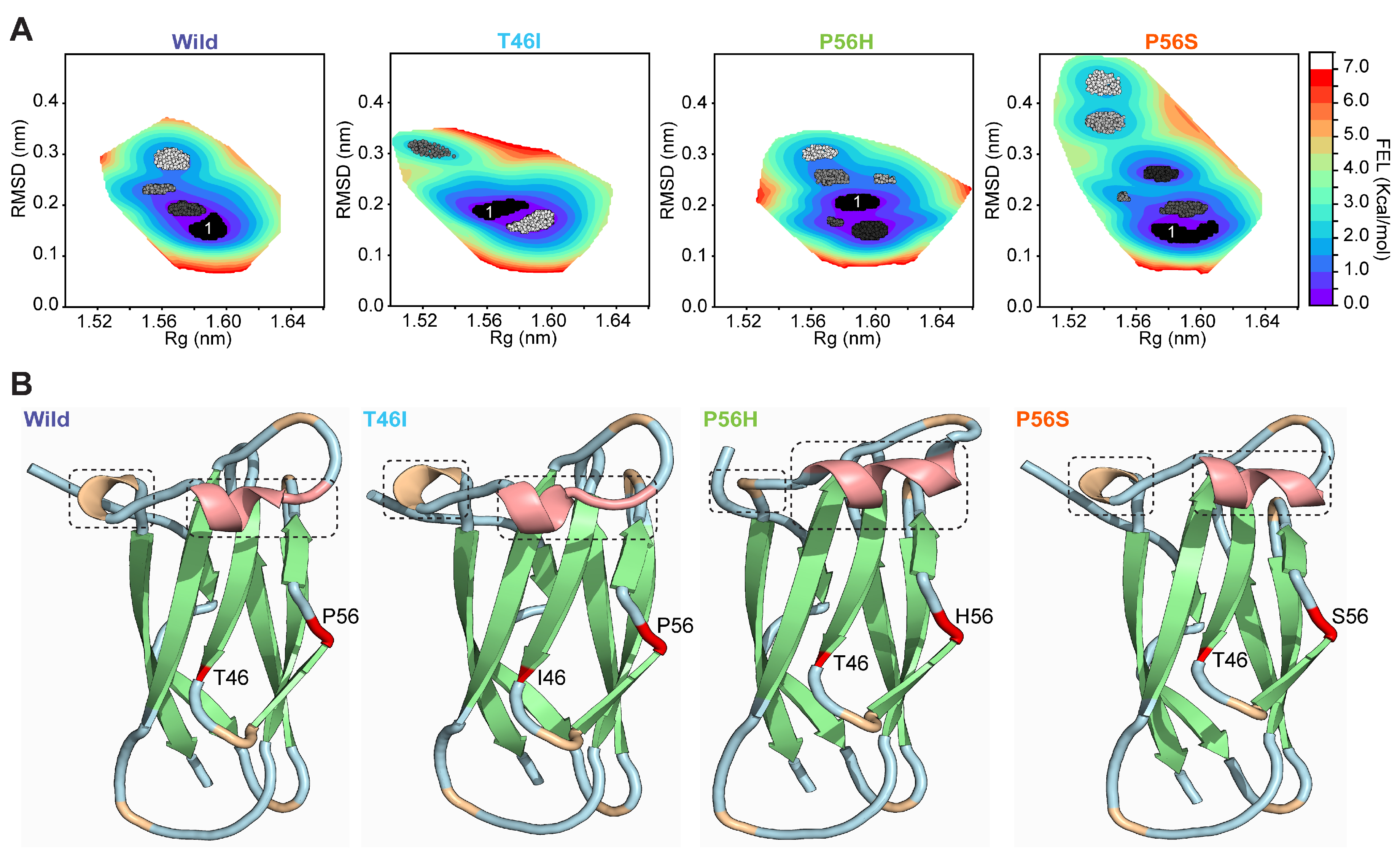
2.5. Pathogenic Variants Alter the Conformation of the Energy-Minima Basins of the VAPB MSP Domain
2.6. Pathogenic Variants Alter the Conformational Dynamics of the Energetically Favored State of the VAPB MSP Domain
2.7. Pathogenic Variants Alter the Organization of Secondary Structure in Energy-Minima Basins of the VAPB MSP Domain
3. Discussion
4. Materials and Methods
4.1. Preparation of the System for MD Simulation
4.2. Analysis of Protein Dynamics
4.3. Free Energy Landscape (FEL)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Costello, J.L.; Castro, I.G.; Hacker, C.; Schrader, T.A.; Metz, J.; Zeuschner, D.; Azadi, A.S.; Godinho, L.F.; Costina, V.; Findeisen, P.; et al. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol. 2017, 216, 331–342. [Google Scholar] [CrossRef]
- Kamemura, K.; Chihara, T. Multiple functions of the ER-resident VAP and its extracellular role in neural development and disease. J. Biochem. 2019, 165, 391–400. [Google Scholar] [CrossRef]
- Nishimura, Y.; Hayashi, M.; Inada, H.; Tanaka, T. Molecular cloning and characterization of mammalian homologues of vesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem. Biophys. Res. Commun. 1999, 254, 21–26. [Google Scholar] [CrossRef]
- Bullock, T.L.; Roberts, T.M.; Stewart, M. 2.5 A resolution crystal structure of the motile major sperm protein (MSP) of Ascaris suum. J. Mol. Biol. 1996, 263, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.E.; Brickner, J.H.; Reilein, A.R.; Fenn, T.D.; Walter, P.; Brunger, A.T. Structural basis of FFAT motif-mediated ER targeting. Structure 2005, 13, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Weir, M.L.; Klip, A.; Trimble, W.S. Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): A broadly expressed protein that binds to VAMP. Biochem. J. 1998, 333 Pt 2, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Kanekura, K.; Nishimoto, I.; Aiso, S.; Matsuoka, M. Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8). J. Biol. Chem. 2006, 281, 30223–30233. [Google Scholar] [CrossRef]
- Murphy, S.E.; Levine, T.P. VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta 2016, 1861, 952–961. [Google Scholar] [CrossRef]
- Kawano, M.; Kumagai, K.; Nishijima, M.; Hanada, K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 2006, 281, 30279–30288. [Google Scholar] [CrossRef]
- Ngo, M.; Ridgway, N.D. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol. Biol. Cell 2009, 20, 1388–1399. [Google Scholar] [CrossRef]
- De Vos, K.J.; Mórotz, G.M.; Stoica, R.; Tudor, E.L.; Lau, K.F.; Ackerley, S.; Warley, A.; Shaw, C.E.; Miller, C.C. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 2012, 21, 1299–1311. [Google Scholar] [CrossRef] [PubMed]
- Lindhout, F.W.; Cao, Y.; Kevenaar, J.T.; Bodzęta, A.; Stucchi, R.; Boumpoutsari, M.M.; Katrukha, E.A.; Altelaar, M.; MacGillavry, H.D.; Hoogenraad, C.C. VAP-SCRN1 interaction regulates dynamic endoplasmic reticulum remodeling and presynaptic function. Embo J. 2019, 38, e101345. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.G.; Liu, N.; Miao, G.; Chen, Y.; Zhao, H.; Zhang, H. The ER Contact Proteins VAPA/B Interact with Multiple Autophagy Proteins to Modulate Autophagosome Biogenesis. Curr. Biol. 2018, 28, 1234–1245.e4. [Google Scholar] [CrossRef] [PubMed]
- Gkogkas, C.; Middleton, S.; Kremer, A.M.; Wardrope, C.; Hannah, M.; Gillingwater, T.H.; Skehel, P. VAPB interacts with and modulates the activity of ATF6. Hum. Mol. Genet. 2008, 17, 1517–1526. [Google Scholar] [CrossRef]
- Furuita, K.; Jee, J.; Fukada, H.; Mishima, M.; Kojima, C. Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies. J. Biol. Chem. 2010, 285, 12961–12970. [Google Scholar] [CrossRef]
- Loewen, C.J.; Roy, A.; Levine, T.P. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003, 22, 2025–2035. [Google Scholar] [CrossRef]
- Shi, J.; Lua, S.; Tong, J.S.; Song, J. Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis. Biochemistry 2010, 49, 3887–3897. [Google Scholar] [CrossRef]
- Kors, S.; Costello, J.L.; Schrader, M. VAP Proteins—From Organelle Tethers to Pathogenic Host Interactors and Their Role in Neuronal Disease. Front. Cell Dev. Biol. 2022, 10, 895856. [Google Scholar] [CrossRef]
- Nishimura, A.L.; Mitne-Neto, M.; Silva, H.C.; Oliveira, J.R.; Vainzof, M.; Zatz, M. A novel locus for late onset amyotrophic lateral sclerosis/motor neurone disease variant at 20q13. J. Med. Genet. 2004, 41, 315–320. [Google Scholar] [CrossRef]
- Nishimura, A.L.; Mitne-Neto, M.; Silva, H.C.; Richieri-Costa, A.; Middleton, S.; Cascio, D.; Kok, F.; Oliveira, J.R.; Gillingwater, T.; Webb, J.; et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004, 75, 822–831. [Google Scholar] [CrossRef]
- Funke, A.D.; Esser, M.; Krüttgen, A.; Weis, J.; Mitne-Neto, M.; Lazar, M.; Nishimura, A.L.; Sperfeld, A.D.; Trillenberg, P.; Senderek, J.; et al. The p.P56S mutation in the VAPB gene is not due to a single founder: The first European case. Clin. Genet. 2010, 77, 302–303. [Google Scholar] [CrossRef] [PubMed]
- Millecamps, S.; Salachas, F.; Cazeneuve, C.; Gordon, P.; Bricka, B.; Camuzat, A.; Guillot-Noël, L.; Russaouen, O.; Bruneteau, G.; Pradat, P.F.; et al. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: Genotype-phenotype correlations. J. Med. Genet. 2010, 47, 554–560. [Google Scholar] [CrossRef]
- Di, L.; Chen, H.; Da, Y.; Wang, S.; Shen, X.M. Atypical familial amyotrophic lateral sclerosis with initial symptoms of pain or tremor in a Chinese family harboring VAPB-P56S mutation. J. Neurol. 2016, 263, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Guber, R.D.; Schindler, A.B.; Budron, M.S.; Chen, K.L.; Li, Y.; Fischbeck, K.H.; Grunseich, C. Nucleocytoplasmic transport defect in a North American patient with ALS8. Ann. Clin. Transl. Neurol. 2018, 5, 369–375. [Google Scholar] [CrossRef]
- Marques, V.D.; Barreira, A.A.; Davis, M.B.; Abou-Sleiman, P.M.; Silva, W.A., Jr.; Zago, M.A.; Sobreira, C.; Fazan, V.; Marques, W., Jr. Expanding the phenotypes of the Pro56Ser VAPB mutation: Proximal SMA with dysautonomia. Muscle Nerve 2006, 34, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Kosac, V.; Freitas, M.R.; Prado, F.M.; Nascimento, O.J.; Bittar, C. Familial adult spinal muscular atrophy associated with the VAPB gene: Report of 42 cases in Brazil. Arq. Neuropsiquiatr. 2013, 71, 788–790. [Google Scholar] [CrossRef]
- Chadi, G.; Maximino, J.R.; Jorge, F.M.H.; Borba, F.C.; Gilio, J.M.; Callegaro, D.; Lopes, C.G.; Santos, S.N.D.; Rebelo, G.N.S. Genetic analysis of patients with familial and sporadic amyotrophic lateral sclerosis in a Brazilian Research Center. Amyotroph. Lateral Scler. Front. Degener. 2017, 18, 249–255. [Google Scholar] [CrossRef]
- Sun, Y.M.; Dong, Y.; Wang, J.; Lu, J.H.; Chen, Y.; Wu, J.J. A novel mutation of VAPB in one Chinese familial amyotrophic lateral sclerosis pedigree and its clinical characteristics. J. Neurol. 2017, 264, 2387–2393. [Google Scholar] [CrossRef]
- de Alcântara, C.; Cruzeiro, M.M.; França, M.C., Jr.; Camargos, S.T.; de Souza, L.C. Amyotrophic lateral sclerosis type 8 is not a pure motor disease: Evidence from a neuropsychological and behavioural study. J. Neurol. 2019, 266, 1980–1987. [Google Scholar] [CrossRef]
- Trilico, M.L.C.; Lorenzoni, P.J.; Kay, C.S.K.; Ducci, R.D.P.; Fustes, O.J.H.; Werneck, L.C.; Scola, R.H. Characterization of the amyotrophic lateral sclerosis-linked P56S mutation of the VAPB gene in Southern Brazil. Amyotroph. Lateral Scler. Front. Degener. 2020, 21, 286–290. [Google Scholar] [CrossRef]
- Nunes Gonçalves, J.P.; Leoni, T.B.; Martins, M.P.; Peluzzo, T.M.; Dourado, M.E.T., Jr.; Saute, J.A.M.; Paranhos Miranda Covaleski, A.P.; Bulle de Oliveira, A.S.; Claudino, R.; Marques, W., Jr.; et al. Genetic epidemiology of familial ALS in Brazil. Neurobiol. Aging 2021, 102, 227.e1–227.e4. [Google Scholar] [CrossRef] [PubMed]
- Temp, A.G.M.; Dyrba, M.; Kasper, E.; Teipel, S.; Prudlo, J. Case Report: Cognitive Conversion in a Non-brazilian VAPB Mutation Carrier (ALS8). Front. Neurol. 2021, 12, 668772. [Google Scholar] [CrossRef]
- Leoni, T.B.; Rezende, T.J.R.; Peluzzo, T.M.; Martins, M.P.; Neto, A.R.C.; Gonzalez-Salazar, C.; Cruzeiro, M.M.; Camargos, S.T.; de Souza, L.C.; França, M.C., Jr. Structural brain and spinal cord damage in symptomatic and pre-symptomatic VAPB-related ALS. J. Neurol. Sci. 2022, 434, 120126. [Google Scholar] [CrossRef] [PubMed]
- Teuling, E.; Ahmed, S.; Haasdijk, E.; Demmers, J.; Steinmetz, M.O.; Akhmanova, A.; Jaarsma, D.; Hoogenraad, C.C. Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J. Neurosci. 2007, 27, 9801–9815. [Google Scholar] [CrossRef]
- Chai, A.; Withers, J.; Koh, Y.H.; Parry, K.; Bao, H.; Zhang, B.; Budnik, V.; Pennetta, G. hVAPB, the causative gene of a heterogeneous group of motor neuron diseases in humans, is functionally interchangeable with its Drosophila homologue DVAP-33A at the neuromuscular junction. Hum. Mol. Genet. 2008, 17, 266–280. [Google Scholar] [CrossRef] [PubMed]
- Ratnaparkhi, A.; Lawless, G.M.; Schweizer, F.E.; Golshani, P.; Jackson, G.R. A Drosophila model of ALS: Human ALS-associated mutation in VAP33A suggests a dominant negative mechanism. PLoS ONE 2008, 3, e2334. [Google Scholar] [CrossRef]
- Suzuki, H.; Kanekura, K.; Levine, T.P.; Kohno, K.; Olkkonen, V.M.; Aiso, S.; Matsuoka, M. ALS-linked P56S-VAPB, an aggregated loss-of-function mutant of VAPB, predisposes motor neurons to ER stress-related death by inducing aggregation of co-expressed wild-type VAPB. J. Neurochem. 2009, 108, 973–985. [Google Scholar] [CrossRef]
- Aliaga, L.; Lai, C.; Yu, J.; Chub, N.; Shim, H.; Sun, L.; Xie, C.; Yang, W.J.; Lin, X.; O’Donovan, M.J.; et al. Amyotrophic lateral sclerosis-related VAPB P56S mutation differentially affects the function and survival of corticospinal and spinal motor neurons. Hum. Mol. Genet. 2013, 22, 4293–4305. [Google Scholar] [CrossRef]
- Larroquette, F.; Seto, L.; Gaub, P.L.; Kamal, B.; Wallis, D.; Larivière, R.; Vallée, J.; Robitaille, R.; Tsuda, H. Vapb/Amyotrophic lateral sclerosis 8 knock-in mice display slowly progressive motor behavior defects accompanying ER stress and autophagic response. Hum. Mol. Genet. 2015, 24, 6515–6529. [Google Scholar] [CrossRef]
- Tokutake, Y.; Gushima, K.; Miyazaki, H.; Shimosato, T.; Yonekura, S. ALS-associated P56S-VAPB mutation restrains 3T3-L1 preadipocyte differentiation. Biochem. Biophys. Res. Commun. 2015, 460, 831–837. [Google Scholar] [CrossRef]
- Tripathi, P.; Guo, H.; Dreser, A.; Yamoah, A.; Sechi, A.; Jesse, C.M.; Katona, I.; Doukas, P.; Nikolin, S.; Ernst, S.; et al. Pathomechanisms of ALS8: Altered autophagy and defective RNA binding protein (RBP) homeostasis due to the VAPB P56S mutation. Cell Death Dis. 2021, 12, 466. [Google Scholar] [CrossRef]
- Chen, H.J.; Anagnostou, G.; Chai, A.; Withers, J.; Morris, A.; Adhikaree, J.; Pennetta, G.; de Belleroche, J.S. Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2010, 285, 40266–40281. [Google Scholar] [CrossRef] [PubMed]
- Kabashi, E.; El Oussini, H.; Bercier, V.; Gros-Louis, F.; Valdmanis, P.N.; McDearmid, J.; Mejier, I.A.; Dion, P.A.; Dupre, N.; Hollinger, D.; et al. Investigating the contribution of VAPB/ALS8 loss of function in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013, 22, 2350–2360. [Google Scholar] [CrossRef]
- van Blitterswijk, M.; van Es, M.A.; Koppers, M.; van Rheenen, W.; Medic, J.; Schelhaas, H.J.; van der Kooi, A.J.; de Visser, M.; Veldink, J.H.; van den Berg, L.H. VAPB and C9orf72 mutations in 1 familial amyotrophic lateral sclerosis patient. Neurobiol. Aging 2012, 33, 2950.e1–2950.e4. [Google Scholar] [CrossRef]
- Vinay Kumar, C.; Kumar, K.M.; Swetha, R.; Ramaiah, S.; Anbarasu, A. Protein aggregation due to nsSNP resulting in P56S VABP protein is associated with amyotrophic lateral sclerosis. J. Theor. Biol. 2014, 354, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Leal, S.S.; Ben Halevy, D.; Gomes, C.M.; Lev, S. Structural requirements for VAP-B oligomerization and their implication in amyotrophic lateral sclerosis-associated VAP-B(P56S) neurotoxicity. J. Biol. Chem. 2010, 285, 13839–13849. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Lim, L.; Wei, Y.; Gupta, G.; Song, J. Resolving the paradox for protein aggregation diseases: NMR structure and dynamics of the membrane-embedded P56S-MSP causing ALS imply a common mechanism for aggregation-prone proteins to attack membranes. F1000Research 2013, 2, 221. [Google Scholar] [CrossRef]
- Kirkitadze, M.D.; Condron, M.M.; Teplow, D.B. Identification and characterization of key kinetic intermediates in amyloid beta-protein fibrillogenesis. J. Mol. Biol. 2001, 312, 1103–1119. [Google Scholar] [CrossRef]
- Williamson, J.A.; Miranker, A.D. Direct detection of transient alpha-helical states in islet amyloid polypeptide. Protein Sci. 2007, 16, 110–117. [Google Scholar] [CrossRef]
- Ghosh, D.; Singh, P.K.; Sahay, S.; Jha, N.N.; Jacob, R.S.; Sen, S.; Kumar, A.; Riek, R.; Maji, S.K. Structure based aggregation studies reveal the presence of helix-rich intermediate during α-Synuclein aggregation. Sci. Rep. 2015, 5, 9228. [Google Scholar] [CrossRef]
- Dash, R.; Ali, M.C.; Dash, N.; Azad, M.A.K.; Hosen, S.M.Z.; Hannan, M.A.; Moon, I.S. Structural and Dynamic Characterizations Highlight the Deleterious Role of SULT1A1 R213H Polymorphism in Substrate Binding. Int. J. Mol. Sci. 2019, 20, 6256. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Choi, H.J.; Dash, R.; Sharif, S.R.; Oktaviani, D.F.; Seog, D.H.; Moon, I.S. N-Acetyl-D-Glucosamine Kinase Interacts with NudC and Lis1 in Dynein Motor Complex and Promotes Cell Migration. Int. J. Mol. Sci. 2020, 22, 129. [Google Scholar] [CrossRef] [PubMed]
- Hosen, S.M.Z.; Dash, R.; Junaid, M.; Mitra, S.; Absar, N. Identification and structural characterization of deleterious non-synonymous single nucleotide polymorphisms in the human SKP2 gene. Comput. Biol. Chem. 2019, 79, 127–136. [Google Scholar] [CrossRef]
- Arifuzzaman, M.; Mitra, S.; Das, R.; Hamza, A.; Absar, N.; Dash, R. In silico analysis of nonsynonymous single-nucleotide polymorphisms (nsSNPs) of the SMPX gene. Ann. Hum. Genet. 2020, 84, 54–71. [Google Scholar] [CrossRef]
- Fazil, M.H.; Kumar, S.; Rao, N.S.; Selvaraj, C.; Singh, S.K.; Pandey, H.P.; Singh, D.V. Comparative structural analysis of two proteins belonging to quorum sensing system in Vibrio cholerae. J. Biomol. Struct. Dyn. 2012, 30, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Maestro-Desmond Interoperability Tools. Maestro-Desmond Interoperability Tools, 2011 version; Schrödinger: New York, NY, USA, 2011.
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Shaw, D.E. A fast, scalable method for the parallel evaluation of distance-limited pairwise particle interactions. J. Comput. Chem. 2005, 26, 1318–1328. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the SC’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FA, USA, 11–17 November 2006; p. 43. [Google Scholar]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985, 31, 1695–1697. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Humphreys, D.D.; Friesner, R.A.; Berne, B.J. A Multiple-Time-Step Molecular Dynamics Algorithm for Macromolecules. J. Phys. Chem. 1994, 98, 6885–6892. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [PubMed]
- Salmas, R.E.; Yurtsever, M.; Durdagi, S. Investigation of Inhibition Mechanism of Chemokine Receptor CCR5 by Micro-second Molecular Dynamics Simulations. Sci. Rep. 2015, 5, 13180. [Google Scholar] [CrossRef]
- Shlens, J. A Tutorial on Principal Component Analysis. arXiv 2014, arXiv:1404.1100. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashar, M.A.; Dash, N.; Mitra, S.; Dash, R. Disclosing Pathogenic Variant Effects on the Structural Dynamics of the VAPB MSP Domain Causing Familial ALS. Int. J. Mol. Sci. 2025, 26, 6489. https://doi.org/10.3390/ijms26136489
Bashar MA, Dash N, Mitra S, Dash R. Disclosing Pathogenic Variant Effects on the Structural Dynamics of the VAPB MSP Domain Causing Familial ALS. International Journal of Molecular Sciences. 2025; 26(13):6489. https://doi.org/10.3390/ijms26136489
Chicago/Turabian StyleBashar, Md Abul, Nayan Dash, Sarmistha Mitra, and Raju Dash. 2025. "Disclosing Pathogenic Variant Effects on the Structural Dynamics of the VAPB MSP Domain Causing Familial ALS" International Journal of Molecular Sciences 26, no. 13: 6489. https://doi.org/10.3390/ijms26136489
APA StyleBashar, M. A., Dash, N., Mitra, S., & Dash, R. (2025). Disclosing Pathogenic Variant Effects on the Structural Dynamics of the VAPB MSP Domain Causing Familial ALS. International Journal of Molecular Sciences, 26(13), 6489. https://doi.org/10.3390/ijms26136489







