Genome-Wide Association Study and RNA-Seq Analysis Uncover Candidate Genes Controlling Growth Traits in Red Tilapia (Oreochromis spp.) Under Hyperosmotic Stress
Abstract
1. Introduction
2. Results
2.1. Statistics of Growth Traits
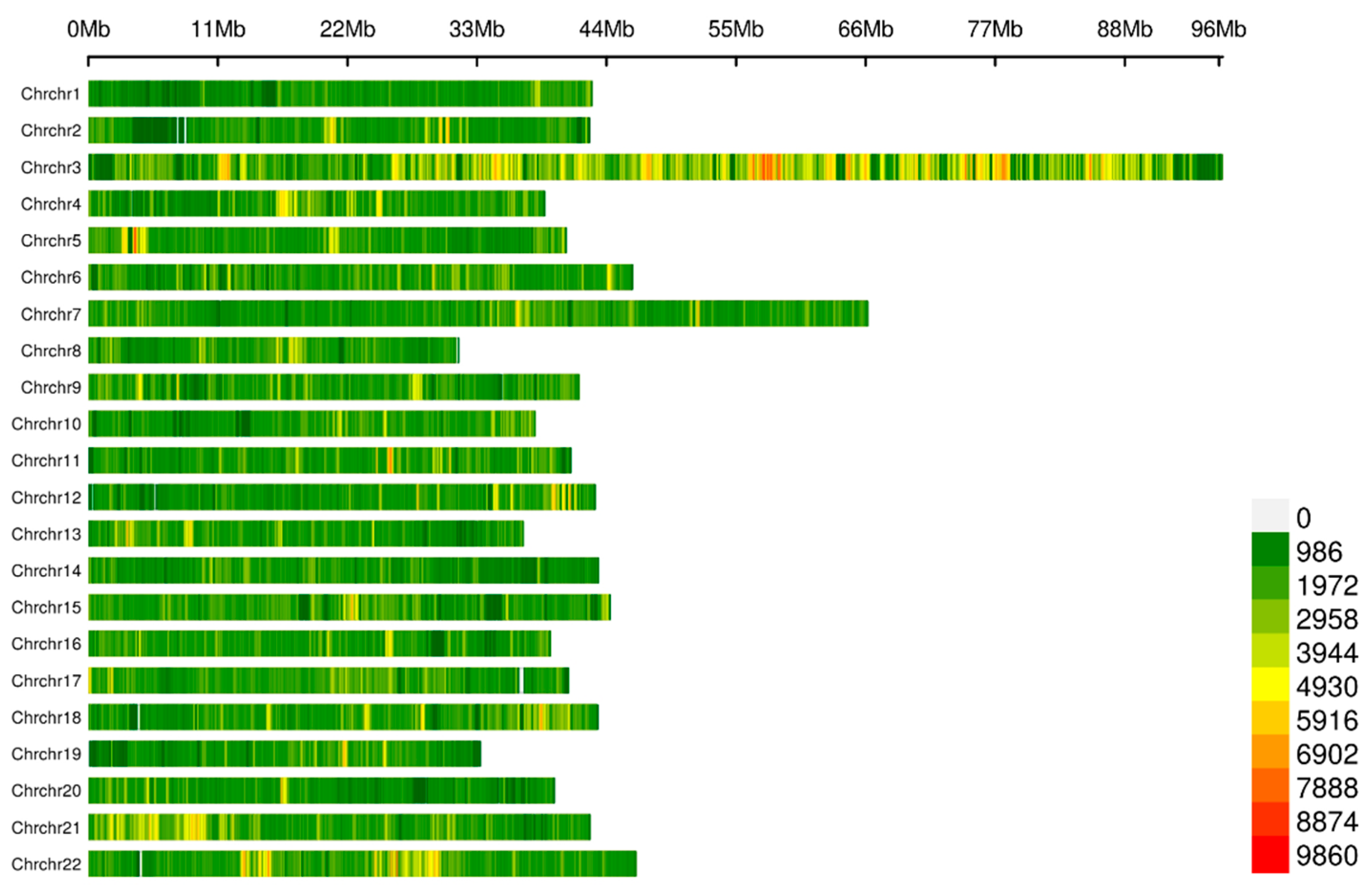
2.2. Quality Control for the Resequencing Data and Genotyping
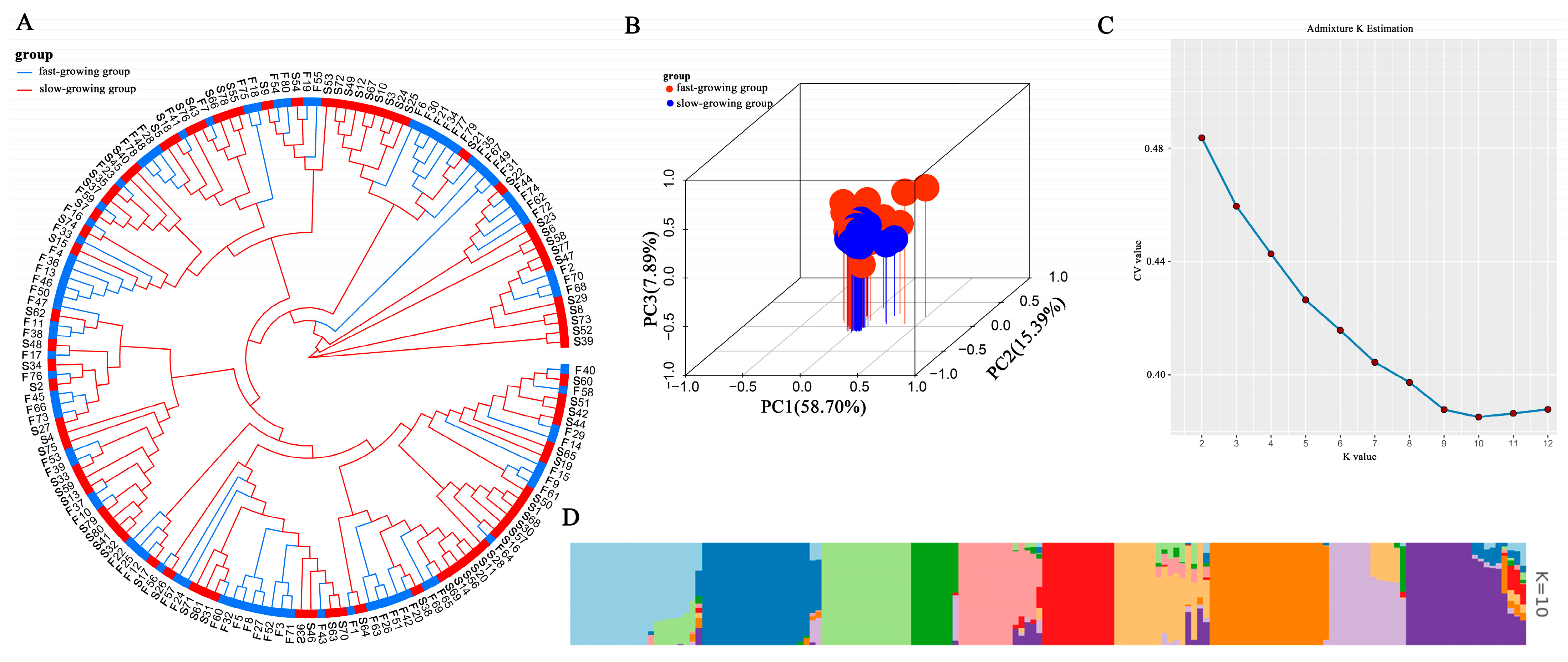
2.3. Analysis of Population Structure
2.4. GWAS for Growth Traits
2.5. Fine Mapping of Candidate Regions
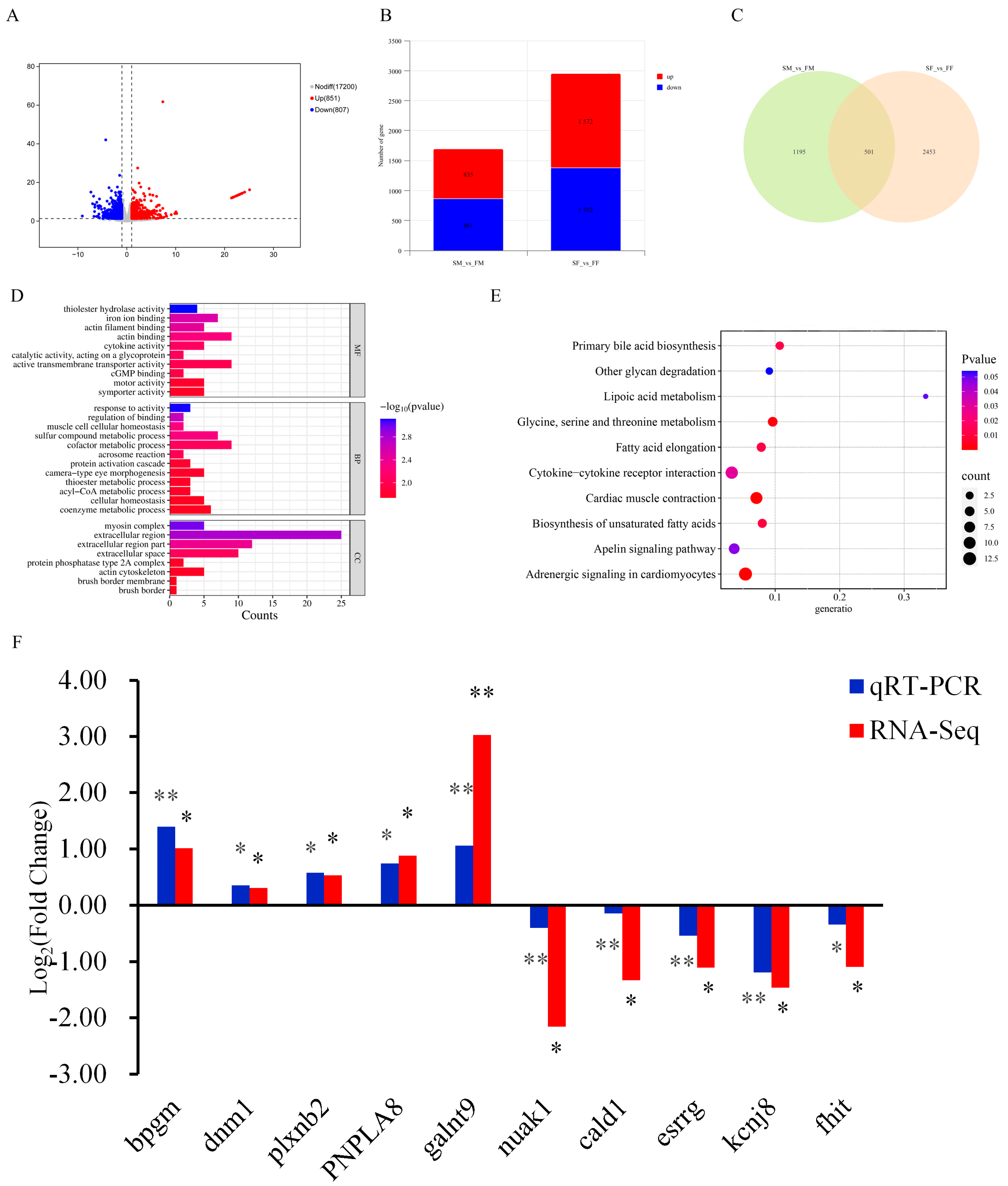
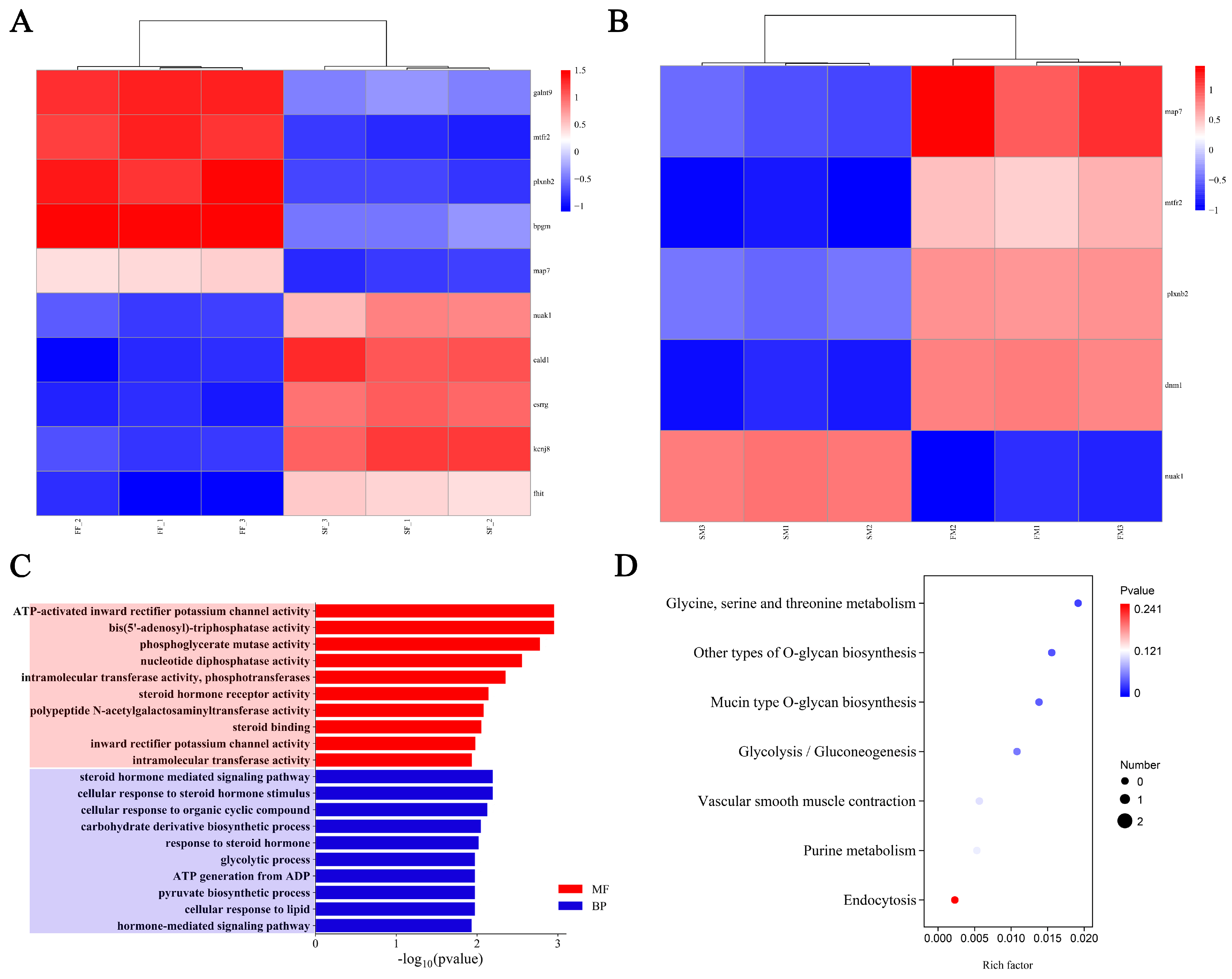
2.6. Transcriptome Analysis
2.7. Integration Analysis of GWAS and Transcriptome
3. Discussion
4. Materials and Methods
4.1. Fish Management
4.2. Growth Performance Analysis and Sampling
4.3. Library Construction and Sequencing
4.4. Sequence Data Filtering and Genotyping Based on Single-Nucleotide Polymorphisms
4.5. Population Structure Analysis and Linkage Disequilibrium
4.6. Genome-Wide Association Study
4.7. RNA Extraction and Transcriptome Sequencing Analysis
4.8. Validation of Candidate Genes by RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, B.; Fu, J.; Dong, Z.; Fang, M.; Zhu, W.; Wang, L. Maternal ancestry analyses of red tilapia strains based on D-loop sequences of seven tilapia populations. PeerJ 2019, 7, e7007. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.Z.; He, H.G. Effects of different salinity on juvenile growth of Oreochromis niloticus, Sarotherodon melanotheron and Israeli red tilapia. North. Chin. Fish. 2024, 43, 149–152. [Google Scholar]
- Chen, H.Q.; Bi, B.; Hu, Q. Effects of different salinity acclimation on tilapia muscle quality, serum biochemical profiles and Na+-K+-ATPase activity. J. Yunnan Agric. Univ. Nat. Sci. 2022, 37, 971–978. [Google Scholar]
- Head, W.D.; Zerbi, A.; Watanabe, W.O. Preliminary observations on the marketability of saltwater-cultured florida red tilapia in Puerto Rico. J. World Aquac. Soc. 1994, 25, 432–441. [Google Scholar] [CrossRef]
- Eknath, A.E.; Tayamen, M.M.; Palada-de Vera, M.S.; Danting, J.C.; Reyes, R.A.; Dionisio, E.E.; Capili, J.B.; Bolivar, H.L.; Abella, T.A.; Circa, A.V. Genetic improvement of farmed tilapias: The growth performance of eight strains of Oreochromis niloticus tested in different farm environments. In Genetics in Aquaculture; Elsevier: Amsterdam, The Netherlands, 1993; pp. 171–188. [Google Scholar]
- Jiang, B.; Huang, R.; Tao, Y.; Lu, S.; Hua, J.; Li, Y.; Dong, Y.; Xu, P.; Qiang, J. Multi-omics and biochemical analyses provide insights into hepatic glucolipid metabolism in red tilapia (Oreochromis spp.) under salinity stress. Aquaculture 2025, 599, 742203. [Google Scholar] [CrossRef]
- Brawand, D.; Wagner, C.E.; Li, Y.I.; Malinsky, M.; Keller, I.; Fan, S.; Simakov, O.; Ng, A.Y.; Lim, Z.W.; Bezault, E. The genomic substrate for adaptive radiation in African cichlid fish. Nature 2014, 513, 375–381. [Google Scholar] [CrossRef]
- Lee, B.Y.; Howe, A.E.; Conte, M.A.; D’Cotta, H.; Pepey, E.; Baroiller, J.F.; di Palma, F.; Carleton, K.L.; Kocher, T.D. An EST resource for tilapia based on 17 normalized libraries and assembly of 116,899 sequence tags. BMC Genom. 2010, 11, 278. [Google Scholar] [CrossRef]
- Zhu, F.; Sun, H.; Jiang, L.; Zhang, Q.; Liu, J. Genome-wide association study for growth-related traits in golden pompano (Trachinotus ovatus). Aquaculture 2023, 572, 739549. [Google Scholar] [CrossRef]
- Li, S.; Liu, H.; Bai, J.; Zhu, X. Transcriptome assembly and identification of genes and SNPs associated with growth traits in largemouth bass (Micropterus salmoides). Genetica 2017, 145, 175–187. [Google Scholar] [CrossRef]
- Liu, H.; Fu, B.; Pang, M.; Feng, X.; Wang, X.; Yu, X.; Tong, J. QTL fine mapping and identification of candidate genes for growth-related traits in bighead carp (Hypophthalmichehys nobilis). Aquaculture 2016, 465, 134–143. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, L.; Wu, X.; Li, B.; Huang, W.; Weng, Z.; Lin, Z.; Song, L.; Guo, Y.; Meng, Z. Identification of candidate growth-related SNPs and genes using GWAS in brown-marbled grouper (Epinephelus fuscoguttatus). Mar. Biotechnol. 2020, 22, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Hilsdorf, A.W.S.; Zhou, L.; Lin, Q.; Gao, Z.-X. Genetics and molecular breeding in aquaculture animals. Front. Genet. 2022, 13, 1071303. [Google Scholar] [CrossRef] [PubMed]
- Dufflocq, P.; Lhorente, J.P.; Bangera, R.; Neira, R.; Newman, S.; Yáñez, J.M. Correlated response of flesh color to selection for harvest weight in coho salmon (Oncorhynchus kisutch). Aquaculture 2017, 472, 38–43. [Google Scholar] [CrossRef]
- Taylor, J.; North, B.; Porter, M.; Bromage, N.; Migaud, H. Photoperiod can be used to enhance growth and improve feeding efficiency in farmed rainbow trout, Oncorhynchus mykiss. Aquaculture 2006, 256, 216–234. [Google Scholar] [CrossRef]
- Houston, R.D.; Bean, T.P.; Macqueen, D.J.; Gundappa, M.K.; Jin, Y.H.; Jenkins, T.L.; Selly, S.L.C.; Martin, S.A.; Stevens, J.R.; Santos, E.M. Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 2020, 21, 389–409. [Google Scholar] [CrossRef]
- Palaiokostas, C.; Houston, R.D. Genome-wide approaches to understanding and improving complex traits in aquaculture species. CABI Rev. 2018, 2017, 1–10. [Google Scholar] [CrossRef]
- Geng, X.; Sha, J.; Liu, S.; Bao, L.; Zhang, J.; Wang, R.; Yao, J.; Li, C.; Feng, J.; Sun, F. A genome-wide association study in catfish reveals the presence of functional hubs of related genes within QTLs for columnaris disease resistance. BMC Genom. 2015, 16, 196. [Google Scholar] [CrossRef]
- Gutierrez, A.P.; Yáñez, J.M.; Fukui, S.; Swift, B.; Davidson, W.S. Genome-wide association study (GWAS) for growth rate and age at sexual maturation in Atlantic salmon (Salmo salar). PLoS ONE 2015, 10, e0119730. [Google Scholar] [CrossRef]
- Udayantha, H.; Lee, S.; Liyanage, D.; Lim, C.; Jeong, T.; Omeka, W.; Yang, H.; Kim, G.; Kim, J.; Lee, J. Identification of candidate variants and genes associated with temperature tolerance in olive flounders by Genome-Wide Association Study (GWAS). Aquaculture 2023, 576, 739858. [Google Scholar] [CrossRef]
- Huang, P.; Guo, W.; Wang, Y.; Xiong, Y.; Ge, S.; Gong, G.; Lin, Q.; Xu, Z.; Gui, J.-F.; Mei, J. Genome-wide association study reveals the genetic basis of growth trait in yellow catfish with sexual size dimorphism. Genomics 2022, 114, 110380. [Google Scholar] [CrossRef]
- Oikonomou, S.; Kazlari, Z.; Papapetrou, M.; Papanna, K.; Papaharisis, L.; Manousaki, T.; Loukovitis, D.; Dimitroglou, A.; Kottaras, L.; Gourzioti, E. Genome Wide Association (GWAS) analysis and genomic heritability for parasite resistance and growth in European seabass. Aquac. Rep. 2022, 24, 101178. [Google Scholar] [CrossRef]
- Omeka, W.; Liyanage, D.; Lee, S.; Lim, C.; Yang, H.; Sandamalika, W.G.; Udayantha, H.; Kim, G.; Ganeshalingam, S.; Jeong, T. Genome-wide association study (GWAS) of growth traits in olive flounder (Paralichthys olivaceus). Aquaculture 2022, 555, 738257. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, H.C.; Wang, Y.Y.; Huang, H.Z. Genome-wide association study reveals growth-related SNPs and candidate genes in mandarin fish (Siniperca chuatsi). Aquaculture 2022, 550, 737879. [Google Scholar] [CrossRef]
- Wang, S.; Chen, L.; Xu, B. ELF3-induced miR-182 inhibits adipogenic differentiation in Graves’ orbitopathy by targeting thyrotropin receptor. Cent. Eur. J. Immunol. 2022, 47, 308–322. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wan, S.-M.; Zhang, S.-M.; Liu, J.-Q.; Sun, A.-L.; Wang, Y.; Zhu, Y.-F.; Gu, S.-X.; Gao, Z.-X. Identification of SNPs and candidate genes associated with growth using GWAS and transcriptome analysis in Coilia nasus. Aquaculture 2024, 586, 740777. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, J.; Cui, T.; Xia, W.; Luo, Q.; Fei, S.; Zhu, X.; Chen, K.; Zhao, J.; Ou, M. Genome-Wide Association Studies (GWAS) and Transcriptome Analysis Reveal Male Heterogametic Sex-Determining Regions and Candidate Genes in Northern Snakeheads (Channa argus). Int. J. Mol. Sci. 2024, 25, 10889. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, L.; Sun, F.; Wong, J.; Lee, M.; Yeo, S.; Wen, Y.; Yue, G.H. Salt tolerance candidate genes identified by QTL mapping, RNA-seq, and functional analysis in tilapia. Aquaculture 2025, 596, 741762. [Google Scholar] [CrossRef]
- Chen, C.H.; Li, B.J.; Gu, X.H.; Lin, H.R.; Xia, J.H. Marker-assisted selection of YY supermales from a genetically improved farmed tilapia-derived strain. Zool. Res. 2018, 40, 108. [Google Scholar]
- Lee, B.; Penman, D.; Kocher, T. Identification of a sex-determining region in Nile tilapia (Oreochromis niloticus) using bulked segregant analysis. Anim. Genet. 2003, 34, 379–383. [Google Scholar] [CrossRef]
- Ling, Y.; Qinglei, M.; Xian, L. Establishment of a female specific SCAR marker in blue tilapia Oreochromis aureus. Prog. Fish. Sci. 2011, 32, 34–40. [Google Scholar]
- Wang, J.; Yu, X.; Wu, S.; Jin, C.; Wang, M.; Ding, H.; Song, S.; Bao, Z.; Wang, B.; Hu, J. Identification of candidate SNPs and genes associated with resistance to nervous necrosis virus in leopard coral grouper (Plectropomus leopardus) using GWAS. Fish. Shellfish Immunol. 2024, 144, 109295. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, Y.; Wang, J.; Liu, C.; Gao, X.; Wu, Y.; Wang, J.; Wu, X.; Zhu, J.; Shen, W. Genome-wide association study identified candidate SNPs and genes associated with hypoxia tolerance in large yellow croaker (Larimichthys crocea). Aquaculture 2022, 560, 738472. [Google Scholar] [CrossRef]
- Li, Z.; Fang, M.; Tang, X.; Zhang, D.; Wang, Z. Disentangling genetic variation for endurance and resistance to visceral white-nodules disease in large yellow croaker (Larimichthys crocea) using genome information. Aquaculture 2023, 564, 739045. [Google Scholar] [CrossRef]
- Wang, Q.; Tian, F.; Pan, Y.; Buckler, E.S.; Zhang, Z. A super powerful method for genome wide association study. PLoS ONE 2014, 9, e107684. [Google Scholar] [CrossRef]
- Dai, X.; Zhang, W.; Zhuo, Z.; He, J.; Yin, Z. Neuroendocrine regulation of somatic growth in fishes. Sci. China Life Sci. 2015, 58, 137–147. [Google Scholar] [CrossRef]
- Wu, L.; Yang, Y.; Li, B.; Huang, W.; Wang, X.; Liu, X.; Meng, Z.; Xia, J. First genome-wide association analysis for growth traits in the largest coral reef-dwelling bony fishes, the Giant grouper (Epinephelus lanceolatus). Mar. Biotechnol. 2019, 21, 707–717. [Google Scholar] [CrossRef]
- Shi, Y.; Zhou, Z.; Liu, B.; Kong, S.; Chen, B.; Bai, H.; Li, L.; Pu, F.; Xu, P. Construction of a high-density genetic linkage map and QTL mapping for growth-related traits in Takifugu bimaculatus. Mar. Biotechnol. 2020, 22, 130–144. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Rastas, P.M.; Premachandra, H.; Knibb, W. First high-density linkage map and single nucleotide polymorphisms significantly associated with traits of economic importance in Yellowtail Kingfish Seriola lalandi. Front. Genet. 2018, 9, 127. [Google Scholar] [CrossRef]
- Yu, H.; You, X.; Li, J.; Zhang, X.; Zhang, S.; Jiang, S.; Lin, X.; Lin, H.-R.; Meng, Z.; Shi, Q. A genome-wide association study on growth traits in orange-spotted grouper (Epinephelus coioides) with RAD-seq genotyping. Sci. China Life Sci. 2018, 61, 934–946. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Yan, W.; Jenkins, C.M.; Han, X.; Mancuso, D.J.; Sims, H.F.; Yang, K.; Gross, R.W. The highly selective production of 2-arachidonoyl lysophosphatidylcholine catalyzed by purified calcium-independent phospholipase A2γ: Identification of a novel enzymatic mediator for the generation of a key branch point intermediate in eicosanoid signaling. J. Biol. Chem. 2005, 280, 26669–26679. [Google Scholar] [PubMed]
- Kinsey, G.R.; Blum, J.L.; Covington, M.D.; Cummings, B.S.; McHowat, J.; Schnellmann, R.G. Decreased iPLA2γ expression induces lipid peroxidation and cell death and sensitizes cells to oxidant-induced apoptosis. J. Lipid Res. 2008, 49, 1477–1487. [Google Scholar] [CrossRef]
- Saggar, J.K.; Chen, J.; Corey, P.; Thompson, L.U. The effect of secoisolariciresinol diglucoside and flaxseed oil, alone and in combination, on MCF-7 tumor growth and signaling pathways. Nutr. Cancer 2010, 62, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Heden, T.D.; Franklin, M.P.; Dailey, C.; Mashek, M.T.; Chen, C.; Mashek, D.G. Acot1 deficiency attenuates high-fat diet–induced fat mass gain by increasing energy expenditure. JCI Insight 2023, 8, e160987. [Google Scholar] [CrossRef] [PubMed]
- Pasini, E.; Baciu, C.; Angeli, M.; Arendt, B.; Pellegrina, D.; Reimand, J.; Patel, K.; Tomlinson, G.; Mazhab-Jafari, M.T.; Kotra, L.P. Acyl-CoA Thioesterase 1 Contributes to Transition of Steatosis to Metabolic-Associated Steatohepatitis. Int. J. Hepatol. 2024, 2024, 5560676. [Google Scholar] [CrossRef]
- Yu, W.; Goncalves, K.A.; Li, S.; Kishikawa, H.; Sun, G.; Yang, H.; Vanli, N.; Wu, Y.; Jiang, Y.; Hu, M.G. Plexin-B2 mediates physiologic and pathologic functions of angiogenin. Cell 2017, 171, 849–864.e25. [Google Scholar] [CrossRef]
- Zhang, M.; Pritchard, M.R.; Middleton, F.A.; Horton, J.A.; Damron, T.A. Microarray analysis of perichondral and reserve growth plate zones identifies differential gene expressions and signal pathways. Bone 2008, 43, 511–520. [Google Scholar] [CrossRef]
- Muirhead, H.; Watson, H. Glycolytic enzymes: From hexose to pyruvate. Curr. Opin. Struct. Biol. 1992, 2, 870–876. [Google Scholar] [CrossRef]
- Lametsch, R.; Kristensen, L.; Larsen, M.R.; Therkildsen, M.; Oksbjerg, N.; Ertbjerg, P. Changes in the muscle proteome after compensatory growth in pigs. J. Anim. Sci. 2006, 84, 918–924. [Google Scholar] [CrossRef]
- Tixier, V.; Bataillé, L.; Etard, C.; Jagla, T.; Weger, M.; DaPonte, J.P.; Strähle, U.; Dickmeis, T.; Jagla, K. Glycolysis supports embryonic muscle growth by promoting myoblast fusion. Proc. Natl. Acad. Sci. 2013, 110, 18982–18987. [Google Scholar] [CrossRef]
- Wang, N.; Tian, Y.; Zhang, J.; Li, Z.; Cheng, M.; Wu, Y. Involvement of glycolysis activation in flatfish sexual size dimorphism: Insights from transcriptomic analyses of Platichthys stellatus and Cynoglossus semilaevis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 39, 100832. [Google Scholar] [CrossRef]
- Tahir, M.N.; Adnan, A.; Strömberg, E.; Mischnick, P. Stability of lipase immobilized on O-pentynyl dextran. Bioprocess Biosyst. Eng. 2012, 35, 535–544. [Google Scholar] [CrossRef]
- Jourdain, I.; Gachet, Y.; Hyams, J.S. The dynamin related protein Dnm1 fragments mitochondria in a microtubule-dependent manner during the fission yeast cell cycle. Cell Motil. Cytoskelet. 2009, 66, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, W.O.; Ernst, D.H.; Chasar, M.P.; Wicklund, R.I.; Olla, B.L. The effects of temperature and salinity on growth and feed utilization of juvenile, sex-reversed male Florida red tilapia cultured in a recirculating system. Aquaculture 1993, 112, 309–320. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J. From FastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.11–11.10.33. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, s13742-015-0047-8. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Tanner, M.; Sharrard, M.; Rigby, A. Gene polymorphisms and the use of the bonferroni correction factor: When and when not to apply? Archives of Disease in Childhood. Arch. Dis. Child. 1997, 76, 385. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Love, M.I.; Soneson, C.; Patro, R. Swimming downstream: Statistical analysis of differential transcript usage following Salmon quantification. F1000Research 2018, 7, 952. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Wang, L.; Luo, M.; Fu, J.; Zhu, W.; Liu, W.; Dong, Z. Transcriptome analysis of skin color variation during and after overwintering of Malaysian red tilapia. Fish Physiol. Biochem. 2022, 48, 937. [Google Scholar]
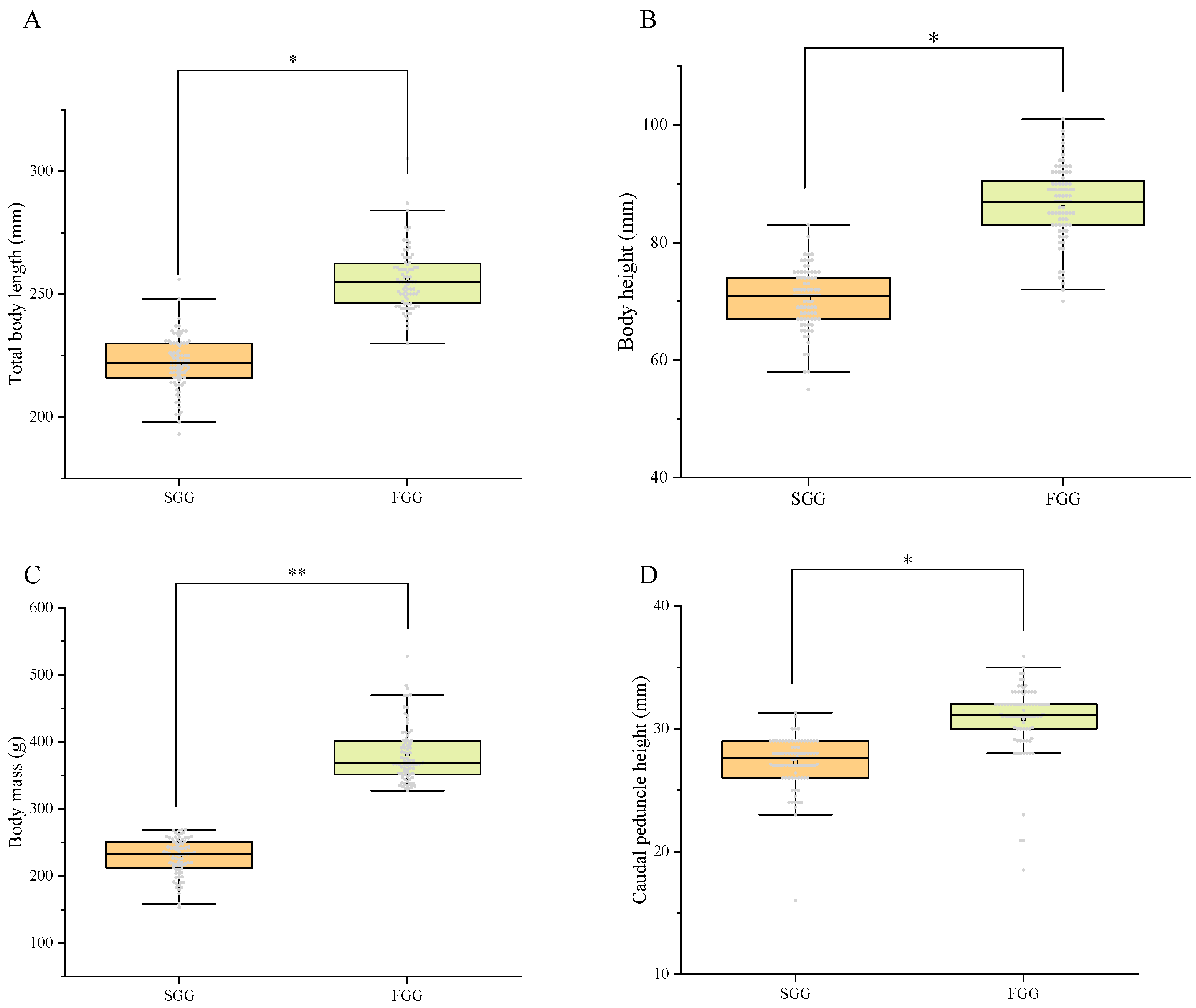


| Body Mass (g) | Total Body Length (mm) | Body Height (mm) | Caudal Peduncle Height (mm) | |
|---|---|---|---|---|
| Body mass | 1 | |||
| Total body length | 0.897 ** | 1 | ||
| Body height | 0.877 ** | 0.808 ** | 1 | |
| Caudal peduncle height | 0.634 ** | 0.643 ** | 0.616 ** | 1 |
| Trait | SNP ID | chr | Position | Allele | MAF | PVE (%) | −log(p-Value) | Location | Candidate Gene |
|---|---|---|---|---|---|---|---|---|---|
| total body length | chr7-48631309 | 7 | 48631309 | T/A | 0.1558 | 0.1333 | 8.38 | intergenic | pnpla8 |
| chr15-7606996 | 15 | 7606996 | A/C | 0.1688 | 0.0306 | 7.25 | intronic | nbas | |
| body height | chr2-35563795 | 2 | 35563795 | T/G | 0.3885 | 0.3873 | 7.31 | intergenic | tmsb |
| chr7-48631309 | 7 | 48631309 | T/A | 0.1558 | 0.1333 | 8.43 | intergenic | pnpla8 | |
| chr12-11404011 | 12 | 11404011 | T/C | 0.1469 | 0.0975 | 7.26 | intergenic | galnt9 | |
| chr12-11413800 | 12 | 11413800 | A/C | 0.1531 | 0.1102 | 7.48 | intergenic | galnt9 | |
| body mass | chr5-37611472 | 5 | 37611472 | T/C | 0.2000 | 0.0019 | 7.94 | intergenic | elf3 |
| chr7-47464467 | 7 | 47464467 | T/C | 0.4250 | 0.0036 | 7.57 | intronic | plxnb2 | |
| chr7-48631309 | 7 | 48631309 | T/A | 0.1558 | 0.1333 | 8.40 | intergenic | pnpla8 | |
| chr17-19727869 | 17 | 19727869 | A/C | 0.4594 | 0.0031 | 7.46 | upstream | acot1 | |
| caudal peduncle height | chr14-37611472 | 14 | 37611472 | A/G | 0.2000 | 1.3369 | 7.49 | intronic | lsamp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, B.; Tao, Y.; Tao, W.; Lu, S.; Badran, M.F.; Saleh, M.H.L.; Aboueleila, R.H.M.; Xu, P.; Qiang, J.; Liu, K. Genome-Wide Association Study and RNA-Seq Analysis Uncover Candidate Genes Controlling Growth Traits in Red Tilapia (Oreochromis spp.) Under Hyperosmotic Stress. Int. J. Mol. Sci. 2025, 26, 6492. https://doi.org/10.3390/ijms26136492
Jiang B, Tao Y, Tao W, Lu S, Badran MF, Saleh MHL, Aboueleila RHM, Xu P, Qiang J, Liu K. Genome-Wide Association Study and RNA-Seq Analysis Uncover Candidate Genes Controlling Growth Traits in Red Tilapia (Oreochromis spp.) Under Hyperosmotic Stress. International Journal of Molecular Sciences. 2025; 26(13):6492. https://doi.org/10.3390/ijms26136492
Chicago/Turabian StyleJiang, Bingjie, Yifan Tao, Wenjing Tao, Siqi Lu, Mohamed Fekri Badran, Moustafa Hassan Lotfy Saleh, Rahma Halim Mahmoud Aboueleila, Pao Xu, Jun Qiang, and Kai Liu. 2025. "Genome-Wide Association Study and RNA-Seq Analysis Uncover Candidate Genes Controlling Growth Traits in Red Tilapia (Oreochromis spp.) Under Hyperosmotic Stress" International Journal of Molecular Sciences 26, no. 13: 6492. https://doi.org/10.3390/ijms26136492
APA StyleJiang, B., Tao, Y., Tao, W., Lu, S., Badran, M. F., Saleh, M. H. L., Aboueleila, R. H. M., Xu, P., Qiang, J., & Liu, K. (2025). Genome-Wide Association Study and RNA-Seq Analysis Uncover Candidate Genes Controlling Growth Traits in Red Tilapia (Oreochromis spp.) Under Hyperosmotic Stress. International Journal of Molecular Sciences, 26(13), 6492. https://doi.org/10.3390/ijms26136492






