The Identification of Proteolytic Substrates of Calpain-5 with N-Terminomics
Abstract
1. Introduction
2. Results
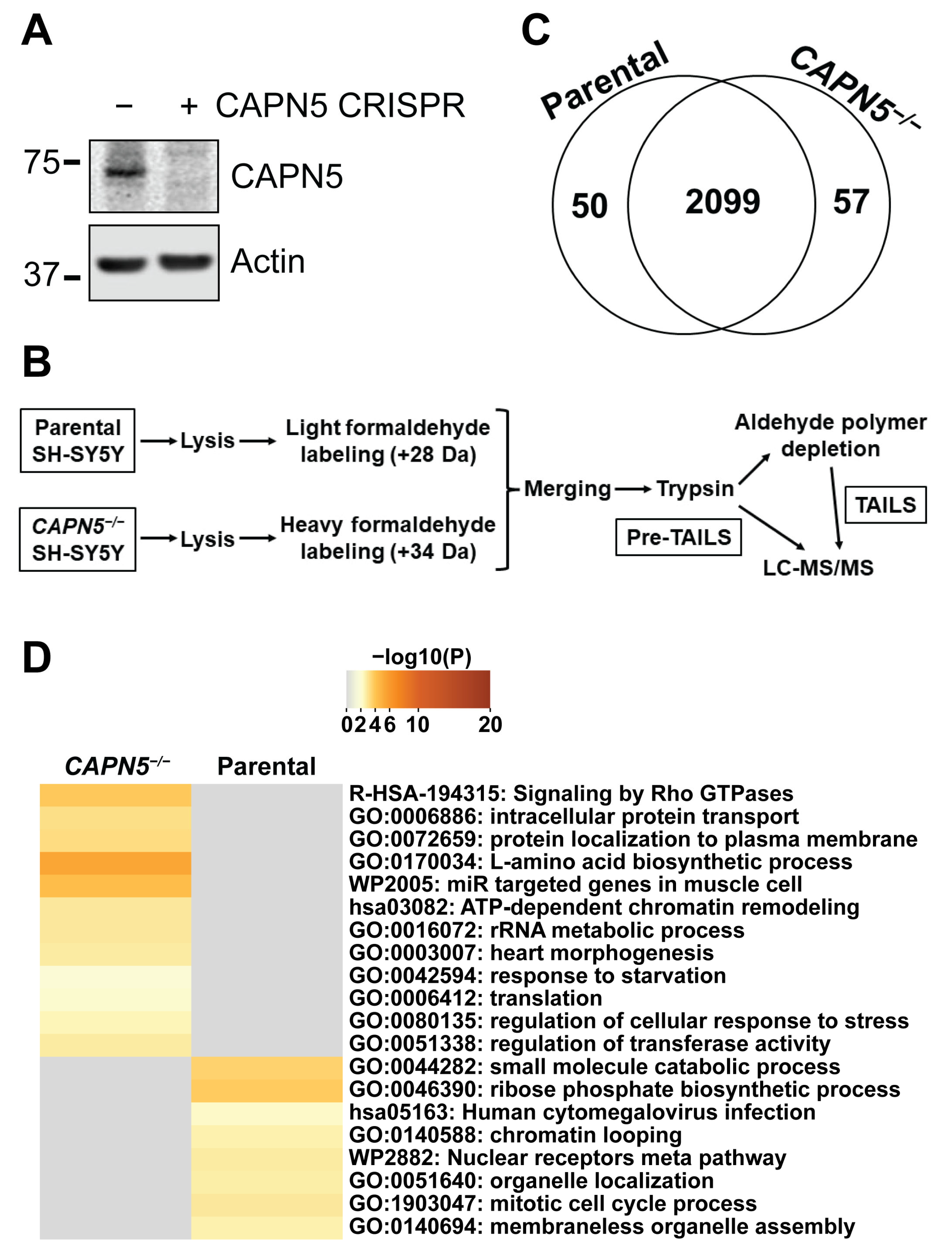
2.1. Inactivation of CAPN5 in the SH-SY5Y Cell Line
2.2. Comparison of the Total Proteomes of Parental and CAPN5−/− SH-SY5Y Cells
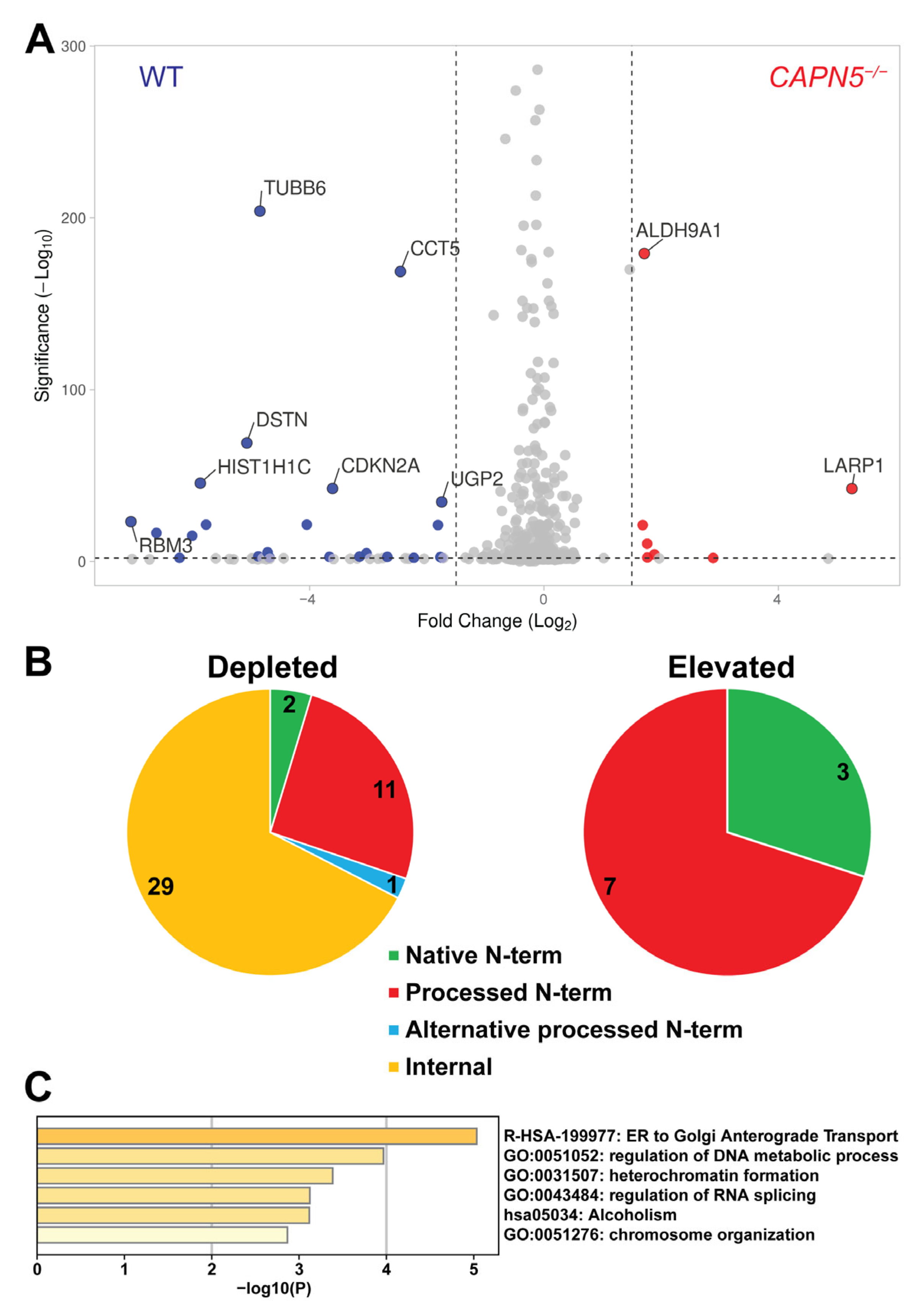
2.3. Identification of Candidate CAPN5 Substrates with N-Terminomics
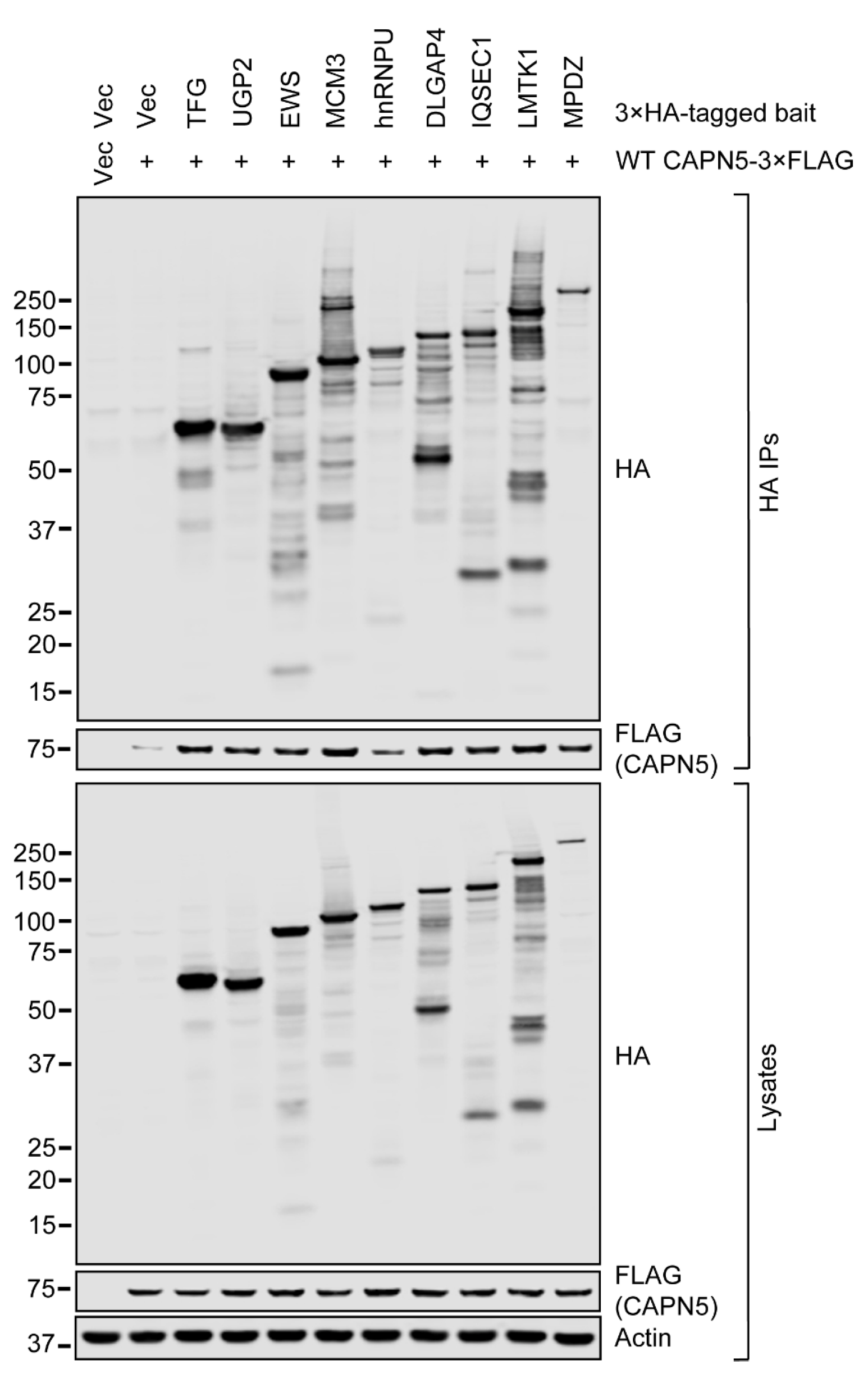
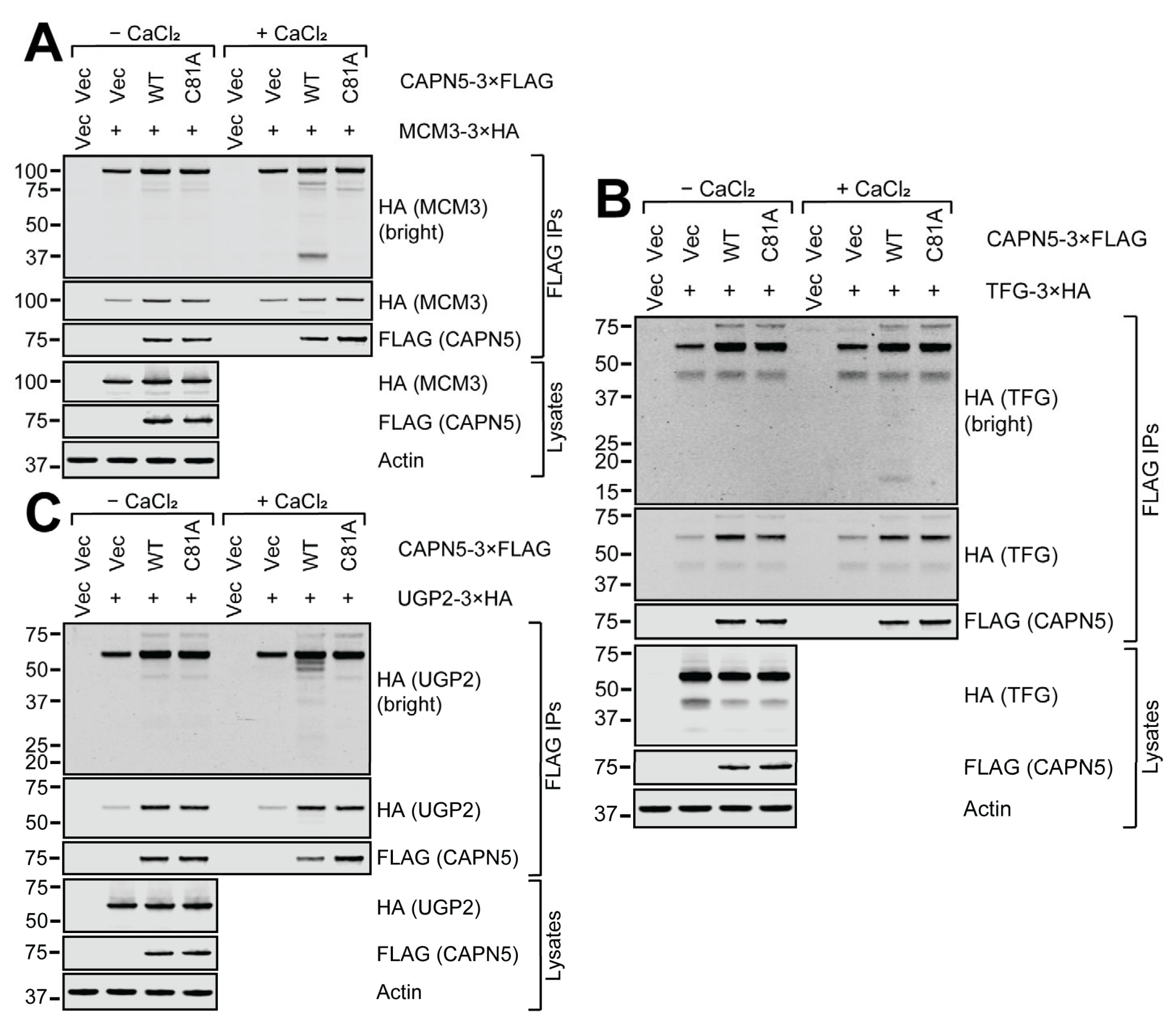
2.4. Confirmation of the Interaction Between CAPN5 and Its Potential Substrates
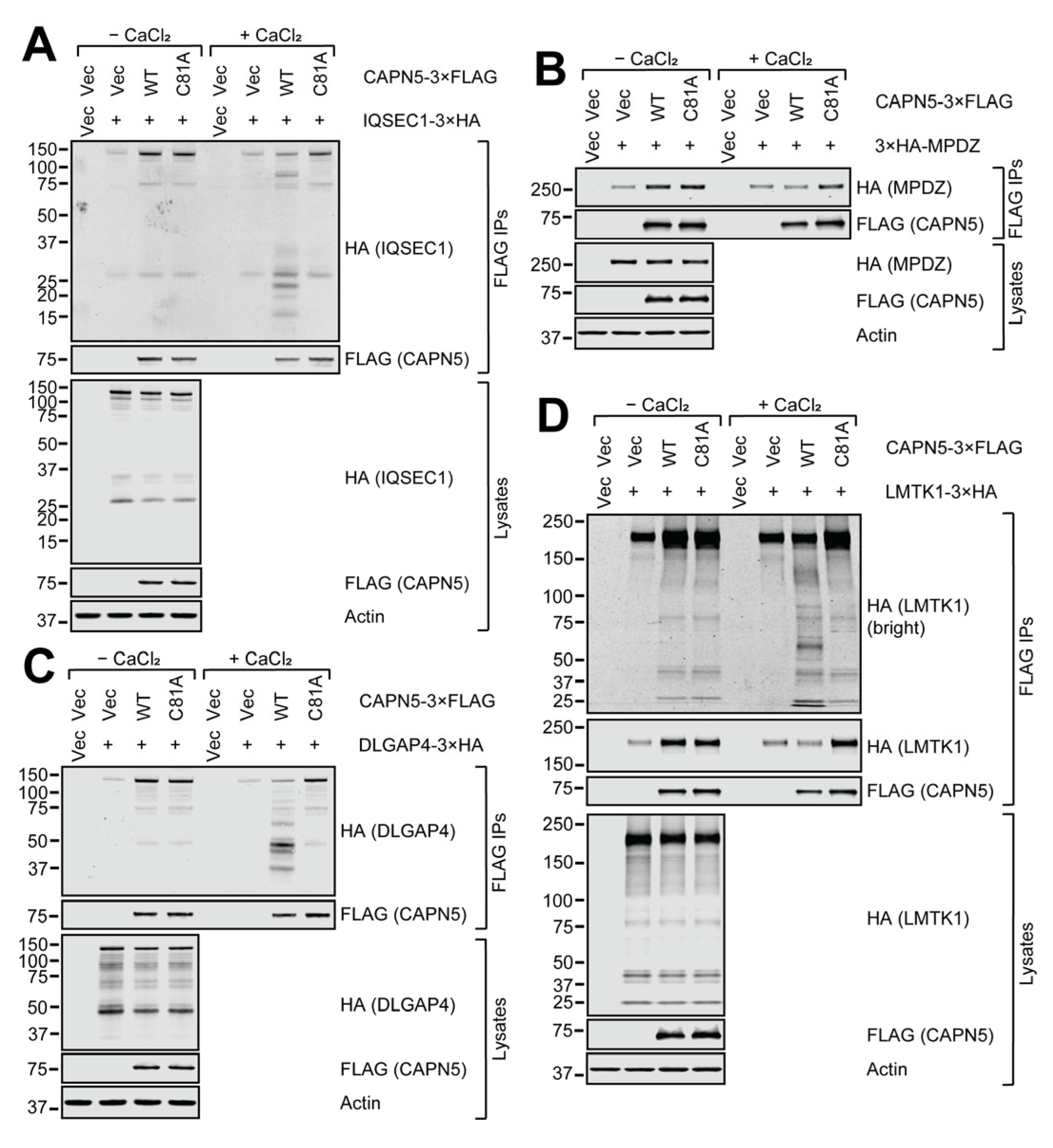
2.5. In Vitro CAPN5 Assays of the Substrate Candidates
2.6. Cellular Fragmentation of the Putative CAPN5 Substrates
3. Discussion
4. Materials and Methods
4.1. Cell Culture and the Generation of the CAPN5−/− SH-SY5Y Cell Line
4.2. Sample Preparation for Proteomics
4.3. N-Terminomics/Terminal Amine Isotopic Labeling of Substrates (TAILS) and Shotgun Proteomics
4.4. High Performance Liquid Chromatography (HPLC) and Mass Spectrometry
4.5. Proteomic Data and Bioinformatic Analysis
4.6. Plasmid Construction
4.7. Anti-HA Immunoprecipitation
4.8. In Vitro CAPN5 Assay
4.9. Denaturing Gel Electrophoresis and Immunoblotting
4.10. Databases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3×FLAG tag | Peptide with the sequence Asp-Tyr-Lys-Asp-His-Asp-Gly-Asp-Tyr-Lys-Asp-His-Asp-Ile-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys |
| AEBSF | 4-(2-Aminoethyl)-benzenesulfonylfluoride hydrochloride |
| CNS | Central nervous system |
| EDTA | Ethylenediaminetetraacetic acid |
| FBS | Fetal bovine serum |
| HA tag | Peptide with the sequence Tyr-Pro-Tyr-Asp-Val-Pro-Asp-Tyr-Ala |
| IP | Immunoprecipitation |
| LC-MS/MS | Liquid chromatography and tandem mass spectrometry |
| MW | Molecular weight |
| NIV | Neovascular inflammatory vitreoretinopathy |
| SDS-PAGE | Denaturing protein gel electrophoresis |
| SWATH | Sequential Window Acquisition of All Theoretical Mass Spectra |
| TAILS | Terminal amine isotopic labeling of substrates |
| WT | wild-type |
References
- Goll, D.E.; Thompson, V.F.; Li, H.; Wei, W.; Cong, J. The calpain system. Physiol. Rev. 2003, 83, 731–801. [Google Scholar] [CrossRef]
- Croall, D.E.; DeMartino, G.N. Calcium-activated neutral protease (calpain) system: Structure, function, and regulation. Physiol. Rev. 1991, 71, 813–847. [Google Scholar] [CrossRef]
- Ono, Y.; Sorimachi, H. Calpains: An elaborate proteolytic system. Biochim. Biophys. Acta 2012, 1824, 224–236. [Google Scholar] [CrossRef]
- Kapilan, A.; Bulluss, M.; Ziegler, A.R.; Dabaja, M.; Derakhshani, A.; Anowai, A.; Armstrong, V.; Campden, R.; Young, D.; Sun, Y.J.; et al. N-terminomics and proteomics analysis of Calpain-2 reveal key proteolytic processing of metabolic and cell adhesion proteins. Protein Sci. 2025, 34, e70144. [Google Scholar] [CrossRef]
- Dear, N.; Matena, K.; Vingron, M.; Boehm, T. A new subfamily of vertebrate calpains lacking a calmodulin-like domain: Implications for calpain regulation and evolution. Genomics 1997, 45, 175–184. [Google Scholar] [CrossRef]
- Bondada, V.; Gal, J.; Mashburn, C.; Rodgers, D.W.; Larochelle, K.E.; Croall, D.E.; Geddes, J.W. The C2 domain of calpain 5 contributes to enzyme activation and membrane localization. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119019. [Google Scholar] [CrossRef]
- Singh, R.; Brewer, M.K.; Mashburn, C.B.; Lou, D.; Bondada, V.; Graham, B.; Geddes, J.W. Calpain 5 is highly expressed in the central nervous system (CNS), carries dual nuclear localization signals, and is associated with nuclear promyelocytic leukemia protein bodies. J. Biol. Chem. 2014, 289, 19383–19394. [Google Scholar] [CrossRef]
- Schaefer, K.A.; Toral, M.A.; Velez, G.; Cox, A.J.; Baker, S.A.; Borcherding, N.C.; Colgan, D.F.; Bondada, V.; Mashburn, C.B.; Yu, C.G.; et al. Calpain-5 Expression in the Retina Localizes to Photoreceptor Synapses. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2509–2521. [Google Scholar] [CrossRef]
- Chukai, Y.; Iwamoto, T.; Itoh, K.; Tomita, H.; Ozaki, T. Characterization of mitochondrial calpain-5. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118989. [Google Scholar] [CrossRef]
- Gal, J.; Bondada, V.; Mashburn, C.B.; Rodgers, D.W.; Croall, D.E.; Geddes, J.W. S-acylation regulates the membrane association and activity of Calpain-5. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119298. [Google Scholar] [CrossRef]
- Geddes, J.W.; Bondada, V.; Croall, D.E.; Rodgers, D.W.; Gal, J. Impaired activity and membrane association of most calpain-5 mutants causal for neovascular inflammatory vitreoretinopathy. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166747. [Google Scholar] [CrossRef]
- Mahajan, V.B.; Skeie, J.M.; Bassuk, A.G.; Fingert, J.H.; Braun, T.A.; Daggett, H.T.; Folk, J.C.; Sheffield, V.C.; Stone, E.M. Calpain-5 mutations cause autoimmune uveitis, retinal neovascularization, and photoreceptor degeneration. PLoS Genet. 2012, 8, e1003001. [Google Scholar] [CrossRef]
- Wert, K.J.; Bassuk, A.G.; Wu, W.H.; Gakhar, L.; Coglan, D.; Mahajan, M.; Wu, S.; Yang, J.; Lin, C.S.; Tsang, S.H.; et al. CAPN5 mutation in hereditary uveitis: The R243L mutation increases calpain catalytic activity and triggers intraocular inflammation in a mouse model. Hum. Mol. Genet. 2015, 24, 4584–4598. [Google Scholar] [CrossRef]
- Velez, G.; Bassuk, A.G.; Schaefer, K.A.; Brooks, B.; Gakhar, L.; Mahajan, M.; Kahn, P.; Tsang, S.H.; Ferguson, P.J.; Mahajan, V.B. A novel de novo CAPN5 mutation in a patient with inflammatory vitreoretinopathy, hearing loss, and developmental delay. Cold Spring Harb. Mol. Case Stud. 2018, 4, a002519. [Google Scholar] [CrossRef]
- Velez, G.; Sun, Y.J.; Khan, S.; Yang, J.; Herrmann, J.; Chemudupati, T.; MacLaren, R.E.; Gakhar, L.; Wakatsuki, S.; Bassuk, A.G.; et al. Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants. Cell Rep. 2020, 30, 881–892.e5. [Google Scholar] [CrossRef]
- Tompa, P.; Buzder-Lantos, P.; Tantos, A.; Farkas, A.; Szilagyi, A.; Banoczi, Z.; Hudecz, F.; Friedrich, P. On the sequential determinants of calpain cleavage. J. Biol. Chem. 2004, 279, 20775–20785. [Google Scholar] [CrossRef]
- Sorimachi, H.; Mamitsuka, H.; Ono, Y. Understanding the substrate specificity of conventional calpains. Biol. Chem. 2012, 393, 853–871. [Google Scholar] [CrossRef]
- Shinkai-Ouchi, F.; Koyama, S.; Ono, Y.; Hata, S.; Ojima, K.; Shindo, M.; duVerle, D.; Ueno, M.; Kitamura, F.; Doi, N.; et al. Predictions of Cleavability of Calpain Proteolysis by Quantitative Structure-Activity Relationship Analysis Using Newly Determined Cleavage Sites and Catalytic Efficiencies of an Oligopeptide Array. Mol. Cell. Proteom. MCP 2016, 15, 1262–1280. [Google Scholar] [CrossRef]
- duVerle, D.A.; Mamitsuka, H. CalCleaveMKL: A Tool for Calpain Cleavage Prediction. Methods Mol. Biol. 2019, 1915, 121–147. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Zang, S.; Li, F.; Chen, Y.; Zhang, X.; Song, Z.; Peng, Q.; Gu, F. Photoreceptor Cell-Derived CAPN5 Regulates Retinal Pigment Epithelium Cell Proliferation Through Direct Regulation of SLIT2 Cleavage. Investig. Ophthalmol. Vis. Sci. 2018, 59, 1810–1821. [Google Scholar] [CrossRef]
- Chukai, Y.; Ito, G.; Konno, M.; Sakata, Y.; Ozaki, T. Mitochondrial calpain-5 truncates caspase-4 during endoplasmic reticulum stress. Biochem. Biophys. Res. Commun. 2022, 608, 156–162. [Google Scholar] [CrossRef]
- Biedler, J.L.; Roffler-Tarlov, S.; Schachner, M.; Freedman, L.S. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978, 38, 3751–3757. [Google Scholar]
- Kovalevich, J.; Langford, D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 2013, 1078, 9–21. [Google Scholar] [CrossRef]
- Xie, H.R.; Hu, L.S.; Li, G.Y. SH-SY5Y human neuroblastoma cell line: In vitro cell model of dopaminergic neurons in Parkinson’s disease. Chin. Med. J. 2010, 123, 1086–1092. [Google Scholar]
- Kleifeld, O.; Doucet, A.; auf dem Keller, U.; Prudova, A.; Schilling, O.; Kainthan, R.K.; Starr, A.E.; Foster, L.J.; Kizhakkedathu, J.N.; Overall, C.M. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 2010, 28, 281–288. [Google Scholar] [CrossRef]
- Kleifeld, O.; Doucet, A.; Prudova, A.; auf dem Keller, U.; Gioia, M.; Kizhakkedathu, J.N.; Overall, C.M. Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat. Protoc. 2011, 6, 1578–1611. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Spitzer, M.; Wildenhain, J.; Rappsilber, J.; Tyers, M. BoxPlotR: A web tool for generation of box plots. Nat. Methods 2014, 11, 121–122. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Arnesen, T.; Van Damme, P.; Polevoda, B.; Helsens, K.; Evjenth, R.; Colaert, N.; Varhaug, J.E.; Vandekerckhove, J.; Lillehaug, J.R.; Sherman, F.; et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA 2009, 106, 8157–8162. [Google Scholar] [CrossRef]
- Duggleby, R.G.; Chao, Y.C.; Huang, J.G.; Peng, H.L.; Chang, H.Y. Sequence differences between human muscle and liver cDNAs for UDPglucose pyrophosphorylase and kinetic properties of the recombinant enzymes expressed in Escherichia coli. Eur. J. Biochem. 1996, 235, 173–179. [Google Scholar] [CrossRef]
- Perenthaler, E.; Nikoncuk, A.; Yousefi, S.; Berdowski, W.M.; Alsagob, M.; Capo, I.; van der Linde, H.C.; van den Berg, P.; Jacobs, E.H.; Putar, D.; et al. Loss of UGP2 in brain leads to a severe epileptic encephalopathy, emphasizing that bi-allelic isoform-specific start-loss mutations of essential genes can cause genetic diseases. Acta Neuropathol. 2020, 139, 415–442. [Google Scholar] [CrossRef]
- Yin, X.; Zeng, D.; Liao, Y.; Tang, C.; Li, Y. The Function of H2A Histone Variants and Their Roles in Diseases. Biomolecules 2024, 14, 993. [Google Scholar] [CrossRef]
- Wei, H.; Zhang, Y.; Jia, Y.; Chen, X.; Niu, T.; Chatterjee, A.; He, P.; Hou, G. Heat shock protein 90: Biological functions, diseases, and therapeutic targets. MedComm 2024, 5, e470. [Google Scholar] [CrossRef]
- Gonzalez, L.; Nebreda, A.R. RINGO/Speedy proteins, a family of non-canonical activators of CDK1 and CDK2. Semin. Cell Dev. Biol. 2020, 107, 21–27. [Google Scholar] [CrossRef]
- Chauhan, S.; Zheng, X.; Tan, Y.Y.; Tay, B.H.; Lim, S.; Venkatesh, B.; Kaldis, P. Evolution of the Cdk-activator Speedy/RINGO in vertebrates. Cell Mol. Life Sci. 2012, 69, 3835–3850. [Google Scholar] [CrossRef]
- McKenna, E.D.; Sarbanes, S.L.; Cummings, S.W.; Roll-Mecak, A. The Tubulin Code, from Molecules to Health and Disease. Annu. Rev. Cell Dev. Biol. 2023, 39, 331–361. [Google Scholar] [CrossRef]
- Rygiel, A.M.; Beer, S.; Simon, P.; Wertheim-Tysarowska, K.; Oracz, G.; Kucharzik, T.; Tysarowski, A.; Niepokoj, K.; Kierkus, J.; Jurek, M.; et al. Gene conversion between cationic trypsinogen (PRSS1) and the pseudogene trypsinogen 6 (PRSS3P2) in patients with chronic pancreatitis. Hum. Mutat. 2015, 36, 350–356. [Google Scholar] [CrossRef]
- Chen, J.M.; Ferec, C. Genes, cloned cDNAs, and proteins of human trypsinogens and pancreatitis-associated cationic trypsinogen mutations. Pancreas 2000, 21, 57–62. [Google Scholar] [CrossRef]
- Zha, C.; Huang, A.; Kailasam, S.; Young, D.; Dufour, A.; Sossin, W.S. Identifying putative substrates of Calpain-15 in neurodevelopment. PLoS ONE 2025, 20, e0319489. [Google Scholar] [CrossRef]
- Agbani, E.O.; Young, D.; Chen, S.A.; Smith, S.; Lee, A.; Poole, A.W.; Dufour, A.; Poon, M.C. Membrane procoagulation and N-terminomics/TAILS profiling in Montreal platelet syndrome kindred with VWF p.V1316M mutation. Commun. Med. 2023, 3, 125. [Google Scholar] [CrossRef]
- Ameen, S.S.; Griem-Krey, N.; Dufour, A.; Hossain, M.I.; Hoque, A.; Sturgeon, S.; Nandurkar, H.; Draxler, D.F.; Medcalf, R.L.; Kamaruddin, M.A.; et al. N-Terminomic Changes in Neurons During Excitotoxicity Reveal Proteolytic Events Associated with Synaptic Dysfunctions and Potential Targets for Neuroprotection. Mol. Cell. Proteom. MCP 2023, 22, 100543. [Google Scholar] [CrossRef]
- Das, N.; de Almeida, L.G.N.; Derakhshani, A.; Young, D.; Mehdinejadiani, K.; Salo, P.; Rezansoff, A.; Jay, G.D.; Sommerhoff, C.P.; Schmidt, T.A.; et al. Tryptase beta regulation of joint lubrication and inflammation via proteoglycan-4 in osteoarthritis. Nat. Commun. 2023, 14, 1910. [Google Scholar] [CrossRef]
- Piatkov, K.I.; Oh, J.H.; Liu, Y.; Varshavsky, A. Calpain-generated natural protein fragments as short-lived substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 2014, 111, E817–E826. [Google Scholar] [CrossRef]
- Al-Dosari, M.S.; Al-Owain, M.; Tulbah, M.; Kurdi, W.; Adly, N.; Al-Hemidan, A.; Masoodi, T.A.; Albash, B.; Alkuraya, F.S. Mutation in MPDZ causes severe congenital hydrocephalus. J. Med. Genet. 2013, 50, 54–58. [Google Scholar] [CrossRef]
- Ali, M.; Hocking, P.M.; McKibbin, M.; Finnegan, S.; Shires, M.; Poulter, J.A.; Prescott, K.; Booth, A.; Raashid, Y.; Jafri, H.; et al. Mpdz null allele in an avian model of retinal degeneration and mutations in human leber congenital amaurosis and retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2011, 52, 7432–7440. [Google Scholar] [CrossRef]
- Ansar, M.; Chung, H.L.; Al-Otaibi, A.; Elagabani, M.N.; Ravenscroft, T.A.; Paracha, S.A.; Scholz, R.; Abdel Magid, T.; Sarwar, M.T.; Shah, S.F.; et al. Bi-allelic Variants in IQSEC1 Cause Intellectual Disability, Developmental Delay, and Short Stature. Am. J. Hum. Genet. 2019, 105, 907–920. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Dal Cin, P.; Nafa, K.; Teot, L.A.; Surti, U.; Fletcher, C.D.; Ladanyi, M. EWSR1-CREB1 is the predominant gene fusion in angiomatoid fibrous histiocytoma. Genes. Chromosom. Cancer 2007, 46, 1051–1060. [Google Scholar] [CrossRef]
- Ayhan, O.; Balkan, M.; Guven, A.; Hazan, R.; Atar, M.; Tok, A.; Tolun, A. Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J. Med. Genet. 2014, 51, 239–244. [Google Scholar] [CrossRef]
- Beetz, C.; Johnson, A.; Schuh, A.L.; Thakur, S.; Varga, R.E.; Fothergill, T.; Hertel, N.; Bomba-Warczak, E.; Thiele, H.; Nurnberg, G.; et al. Inhibition of TFG function causes hereditary axon degeneration by impairing endoplasmic reticulum structure. Proc. Natl. Acad. Sci. USA 2013, 110, 5091–5096. [Google Scholar] [CrossRef]
- Bielack, S.S.; Paulussen, M.; Kohler, G. A patient with two Ewing’s sarcomas with distinct EWS fusion transcripts. N. Engl. J. Med. 2004, 350, 1364–1365. [Google Scholar] [CrossRef]
- Carvill, G.L.; Heavin, S.B.; Yendle, S.C.; McMahon, J.M.; O’Roak, B.J.; Cook, J.; Khan, A.; Dorschner, M.O.; Weaver, M.; Calvert, S.; et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 2013, 45, 825–830. [Google Scholar] [CrossRef]
- Colombo, R.; Pontoglio, A.; Bini, M. Two Novel TEX15 Mutations in a Family with Nonobstructive Azoospermia. Gynecol. Obstet. Investig. 2017, 82, 283–286. [Google Scholar] [CrossRef]
- Delattre, O.; Zucman, J.; Plougastel, B.; Desmaze, C.; Melot, T.; Peter, M.; Kovar, H.; Joubert, I.; de Jong, P.; Rouleau, G.; et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992, 359, 162–165. [Google Scholar] [CrossRef]
- Fazeli, W.; Herkenrath, P.; Stiller, B.; Neugebauer, A.; Fricke, J.; Lang-Roth, R.; Nurnberg, G.; Thoenes, M.; Becker, J.; Altmuller, J.; et al. A TUBB6 mutation is associated with autosomal dominant non-progressive congenital facial palsy, bilateral ptosis and velopharyngeal dysfunction. Hum. Mol. Genet. 2017, 26, 4055–4066. [Google Scholar] [CrossRef]
- Hallor, K.H.; Mertens, F.; Jin, Y.; Meis-Kindblom, J.M.; Kindblom, L.G.; Behrendtz, M.; Kalen, A.; Mandahl, N.; Panagopoulos, I. Fusion of the EWSR1 and ATF1 genes without expression of the MITF-M transcript in angiomatoid fibrous histiocytoma. Genes Chromosom. Cancer 2005, 44, 97–102. [Google Scholar] [CrossRef]
- Hamdan, F.F.; Srour, M.; Capo-Chichi, J.M.; Daoud, H.; Nassif, C.; Patry, L.; Massicotte, C.; Ambalavanan, A.; Spiegelman, D.; Diallo, O.; et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014, 10, e1004772. [Google Scholar] [CrossRef]
- Harlalka, G.V.; McEntagart, M.E.; Gupta, N.; Skrzypiec, A.E.; Mucha, M.W.; Chioza, B.A.; Simpson, M.A.; Sreekantan-Nair, A.; Pereira, A.; Gunther, S.; et al. Novel Genetic, Clinical, and Pathomechanistic Insights into TFG-Associated Hereditary Spastic Paraplegia. Hum. Mutat. 2016, 37, 1157–1161. [Google Scholar] [CrossRef]
- Ishiura, H.; Sako, W.; Yoshida, M.; Kawarai, T.; Tanabe, O.; Goto, J.; Takahashi, Y.; Date, H.; Mitsui, J.; Ahsan, B.; et al. The TRK-fused gene is mutated in hereditary motor and sensory neuropathy with proximal dominant involvement. Am. J. Hum. Genet. 2012, 91, 320–329. [Google Scholar] [CrossRef]
- Jeon, I.S.; Davis, J.N.; Braun, B.S.; Sublett, J.E.; Roussel, M.F.; Denny, C.T.; Shapiro, D.N. A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene 1995, 10, 1229–1234. [Google Scholar]
- Kanadome, T.; Shibata, H.; Kuwata, K.; Takahara, T.; Maki, M. The calcium-binding protein ALG-2 promotes endoplasmic reticulum exit site localization and polymerization of Trk-fused gene (TFG) protein. FEBS J. 2017, 284, 56–76. [Google Scholar] [CrossRef]
- Knierim, E.; Gill, E.; Seifert, F.; Morales-Gonzalez, S.; Unudurthi, S.D.; Hund, T.J.; Stenzel, W.; Schuelke, M. A recessive mutation in beta-IV-spectrin (SPTBN4) associates with congenital myopathy, neuropathy, and central deafness. Hum. Genet. 2017, 136, 903–910. [Google Scholar] [CrossRef]
- Luscan, R.; Mechaussier, S.; Paul, A.; Tian, G.; Gerard, X.; Defoort-Dellhemmes, S.; Loundon, N.; Audo, I.; Bonnin, S.; LeGargasson, J.F.; et al. Mutations in TUBB4B Cause a Distinctive Sensorineural Disease. Am. J. Hum. Genet. 2017, 101, 1006–1012. [Google Scholar] [CrossRef]
- Maddirevula, S.; Alzahrani, F.; Al-Owain, M.; Al Muhaizea, M.A.; Kayyali, H.R.; AlHashem, A.; Rahbeeni, Z.; Al-Otaibi, M.; Alzaidan, H.I.; Balobaid, A.; et al. Autozygome and high throughput confirmation of disease genes candidacy. Genet. Med. 2019, 21, 736–742. [Google Scholar] [CrossRef]
- Okutman, O.; Muller, J.; Baert, Y.; Serdarogullari, M.; Gultomruk, M.; Piton, A.; Rombaut, C.; Benkhalifa, M.; Teletin, M.; Skory, V.; et al. Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum. Mol. Genet. 2015, 24, 5581–5588. [Google Scholar] [CrossRef]
- Peter, M.; Couturier, J.; Pacquement, H.; Michon, J.; Thomas, G.; Magdelenat, H.; Delattre, O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 1997, 14, 1159–1164. [Google Scholar] [CrossRef]
- Shaheen, R.; Sebai, M.A.; Patel, N.; Ewida, N.; Kurdi, W.; Altweijri, I.; Sogaty, S.; Almardawi, E.; Seidahmed, M.Z.; Alnemri, A.; et al. The genetic landscape of familial congenital hydrocephalus. Ann. Neurol. 2017, 81, 890–897. [Google Scholar] [CrossRef]
- Tessadori, F.; Duran, K.; Knapp, K.; Fellner, M.; Deciphering Developmental Disorders Study; Smithson, S.; Beleza Meireles, A.; Elting, M.W.; Waisfisz, Q.; O’Donnell-Luria, A.; et al. Recurrent de novo missense variants across multiple histone H4 genes underlie a neurodevelopmental syndrome. Am. J. Hum. Genet. 2022, 109, 750–758. [Google Scholar] [CrossRef]
- Tessadori, F.; Giltay, J.C.; Hurst, J.A.; Massink, M.P.; Duran, K.; Vos, H.R.; van Es, R.M.; Deciphering Developmental Disorders Study; Scott, R.H.; van Gassen, K.L.I.; et al. Germline mutations affecting the histone H4 core cause a developmental syndrome by altering DNA damage response and cell cycle control. Nat. Genet. 2017, 49, 1642–1646. [Google Scholar] [CrossRef]
- Tessadori, F.; Rehman, A.U.; Giltay, J.C.; Xia, F.; Streff, H.; Duran, K.; Bakkers, J.; Lalani, S.R.; van Haaften, G. A de novo variant in the human HIST1H4J gene causes a syndrome analogous to the HIST1H4C-associated neurodevelopmental disorder. Eur. J. Hum. Genet. 2020, 28, 674–678. [Google Scholar] [CrossRef]
- Wang, X.; Jin, H.R.; Cui, Y.Q.; Chen, J.; Sha, Y.W.; Gao, Z.L. Case study of a patient with cryptozoospermia associated with a recessive TEX15 nonsense mutation. Asian J. Androl. 2018, 20, 101–102. [Google Scholar] [CrossRef]
- Potz, B.A.; Abid, M.R.; Sellke, F.W. Role of Calpain in Pathogenesis of Human Disease Processes. J. Nat. Sci. 2016, 2, e218. [Google Scholar]
- Vosler, P.S.; Brennan, C.S.; Chen, J. Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol. Neurobiol. 2008, 38, 78–100. [Google Scholar] [CrossRef]
- Storr, S.J.; Carragher, N.O.; Frame, M.C.; Parr, T.; Martin, S.G. The calpain system and cancer. Nat. Rev. Cancer 2011, 11, 364–374. [Google Scholar] [CrossRef]
- Stone, E.M.; Kimura, A.E.; Folk, J.C.; Bennett, S.R.; Nichols, B.E.; Streb, L.M.; Sheffield, V.C. Genetic linkage of autosomal dominant neovascular inflammatory vitreoretinopathy to chromosome 11q13. Hum. Mol. Genet. 1992, 1, 685–689. [Google Scholar] [CrossRef]
- Bennett, S.R.; Folk, J.C.; Kimura, A.E.; Russell, S.R.; Stone, E.M.; Raphtis, E.M. Autosomal dominant neovascular inflammatory vitreoretinopathy. Ophthalmology 1990, 97, 1125–1135, discussion 1135–1136. [Google Scholar] [CrossRef]
- Bassuk, A.G.; Yeh, S.; Wu, S.; Martin, D.F.; Tsang, S.H.; Gakhar, L.; Mahajan, V.B. Structural modeling of a novel CAPN5 mutation that causes uveitis and neovascular retinal detachment. PLoS ONE 2015, 10, e0122352. [Google Scholar] [CrossRef]
- Randazzo, N.M.; Shanks, M.E.; Clouston, P.; MacLaren, R.E. Two Novel CAPN5 Variants Associated with Mild and Severe Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy Phenotypes. Ocul. Immunol. Inflamm. 2019, 27, 693–698. [Google Scholar] [CrossRef]
- Sakagami, H.; Katsumata, O.; Hara, Y.; Tamaki, H.; Watanabe, M.; Harvey, R.J.; Fukaya, M. Distinct synaptic localization patterns of brefeldin A-resistant guanine nucleotide exchange factors BRAG2 and BRAG3 in the mouse retina. J. Comp. Neurol. 2013, 521, 860–876. [Google Scholar] [CrossRef]
- Becamel, C.; Figge, A.; Poliak, S.; Dumuis, A.; Peles, E.; Bockaert, J.; Lubbert, H.; Ullmer, C. Interaction of serotonin 5-hydroxytryptamine type 2C receptors with PDZ10 of the multi-PDZ domain protein MUPP1. J. Biol. Chem. 2001, 276, 12974–12982. [Google Scholar] [CrossRef]
- Krapivinsky, G.; Medina, I.; Krapivinsky, L.; Gapon, S.; Clapham, D.E. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron 2004, 43, 563–574. [Google Scholar] [CrossRef]
- Wang, T.; Bai, Y.; Zheng, X.; Liu, X.; Xing, S.; Wang, L.; Wang, H.; Feng, G.; Li, C. Sapap4 deficiency leads to postsynaptic defects and abnormal behaviors relevant to hyperkinetic neuropsychiatric disorder in mice. Cereb. Cortex 2023, 33, 1104–1118. [Google Scholar] [CrossRef]
- Tonami, K.; Kurihara, Y.; Aburatani, H.; Uchijima, Y.; Asano, T.; Kurihara, H. Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Mol. Cell. Biol. 2007, 27, 2548–2561. [Google Scholar] [CrossRef]
- Anderson, B.M.; de Almeida, L.G.N.; Sekhon, H.; Young, D.; Dufour, A.; Edgington-Mitchell, L.E. N-Terminomics/TAILS Profiling of Macrophages after Chemical Inhibition of Legumain. Biochemistry 2020, 59, 329–340. [Google Scholar] [CrossRef]
- Ziegler, A.R.; Dufour, A.; Scott, N.E.; Edgington-Mitchell, L.E. Ion Mobility-Based Enrichment-Free N-Terminomics Analysis Reveals Novel Legumain Substrates in Murine Spleen. Mol. Cell. Proteom. MCP 2024, 23, 100714. [Google Scholar] [CrossRef]
- Wang, L.; Main, K.; Wang, H.; Julien, O.; Dufour, A. Biochemical Tools for Tracking Proteolysis. J. Proteome Res. 2021, 20, 5264–5279. [Google Scholar] [CrossRef]
- Mainoli, B.; Hirota, S.; Edgington-Mitchell, L.E.; Lu, C.; Dufour, A. Proteomics and Imaging in Crohn’s Disease: TAILS of Unlikely Allies. Trends Pharmacol. Sci. 2020, 41, 74–84. [Google Scholar] [CrossRef]
- Jagdeo, J.M.; Dufour, A.; Klein, T.; Solis, N.; Kleifeld, O.; Kizhakkedathu, J.; Luo, H.; Overall, C.M.; Jan, E. N-Terminomics TAILS Identifies Host Cell Substrates of Poliovirus and Coxsackievirus B3 3C Proteinases That Modulate Virus Infection. J. Virol. 2018, 92, e02211-17. [Google Scholar] [CrossRef]
- Mallia-Milanes, B.; Dufour, A.; Philp, C.; Solis, N.; Klein, T.; Fischer, M.; Bolton, C.E.; Shapiro, S.; Overall, C.M.; Johnson, S.R. TAILS proteomics reveals dynamic changes in airway proteolysis controlling protease activity and innate immunity during COPD exacerbations. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 315, L1003–L1014. [Google Scholar] [CrossRef]
- Bhardwaj, S.; Bulluss, M.; D’Aubeterre, A.; Derakhshani, A.; Penner, R.; Mahajan, M.; Mahajan, V.B.; Dufour, A. Integrating the analysis of human biopsies using post-translational modifications proteomics. Protein Sci. 2024, 33, e4979. [Google Scholar] [CrossRef]
- Anowai, A.; Chopra, S.; Mainoli, B.; Young, D.; Dufour, A. N-Terminomics/TAILS of Tissue and Liquid Biopsies. Methods Mol. Biol. 2022, 2456, 85–94. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Perez-Riverol, Y.; Sharma, V.; Carver, J.J.; Mendoza, L.; Kundu, D.J.; Wang, S.; Bandla, C.; Kamatchinathan, S.; et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023, 51, D1539–D1548. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S.; et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef]
- Wang, H.Y.; Li, Y.; Xue, T.; Cheng, N.; Du, H.N. Construction of a series of pCS2+ backbone-based Gateway vectors for overexpressing various tagged proteins in vertebrates. Acta Biochim. Biophys. Sin. 2016, 48, 1128–1134. [Google Scholar] [CrossRef]
- Couthouis, J.; Hart, M.P.; Erion, R.; King, O.D.; Diaz, Z.; Nakaya, T.; Ibrahim, F.; Kim, H.J.; Mojsilovic-Petrovic, J.; Panossian, S.; et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012, 21, 2899–2911. [Google Scholar] [CrossRef]
- Cormier, C.Y.; Mohr, S.E.; Zuo, D.; Hu, Y.; Rolfs, A.; Kramer, J.; Taycher, E.; Kelley, F.; Fiacco, M.; Turnbull, G.; et al. Protein Structure Initiative Material Repository: An open shared public resource of structural genomics plasmids for the biological community. Nucleic Acids Res. 2010, 38, D743–D749. [Google Scholar] [CrossRef]
- Cormier, C.Y.; Park, J.G.; Fiacco, M.; Steel, J.; Hunter, P.; Kramer, J.; Singla, R.; LaBaer, J. PSI:Biology-materials repository: A biologist’s resource for protein expression plasmids. J. Struct. Funct. Genom. 2011, 12, 55–62. [Google Scholar] [CrossRef][Green Version]
- Seiler, C.Y.; Park, J.G.; Sharma, A.; Hunter, P.; Surapaneni, P.; Sedillo, C.; Field, J.; Algar, R.; Price, A.; Steel, J.; et al. DNASU plasmid and PSI:Biology-Materials repositories: Resources to accelerate biological research. Nucleic Acids Res. 2014, 42, D1253–D1260. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]


| UniProt AC | Gene Name | Protein Name | Cleavage Site |
|---|---|---|---|
| RNA binding proteins | |||
| Q13838 | DDX39B | Spliceosome RNA helicase DDX39B | 355R V356 |
| Q01844 | EWSR1 | RNA-binding protein EWS | 614R R615 |
| Q00839 | HNRNPU | Heterogeneous nuclear ribonucleoprotein U | 733R G734 |
| Q9BPZ3 | PAIP2 | Polyadenylate-binding protein-interacting protein 2 | 47I E48 |
| P98179 | RBM3 | RNA-binding protein 3 | 101R S102 |
| Synaptic proteins | |||
| Q9Y2H0 | DLGAP4 | Disks large-associated protein 4 | 944K A945 |
| Q6DN90 | IQSEC1 | IQ motif and SEC7 domain-containing protein 1 | 158R S159 |
| O75970 | MPDZ | Multiple PDZ domain protein | 1138L Q1139 |
| Cytoskeletal proteins | |||
| Q9H254 | SPTBN4 | Spectrin beta chain, non-erythrocytic 4 | 855R V856 |
| P68371 | TUBB4B | Tubulin beta-4B chain * | 62R A63 |
| Q9BUF5 | TUBB6 | Tubulin beta-6 chain | 62R A63 |
| Regulators of chromosomal processes | |||
| Q8TB52 | FBXO30 | F-box only protein 30 | 588G V589 |
| P0C0S5 | H2AZ1 | Histone H2A.Z * | 23R A24 |
| P62805 | H4C12 (+13) | Histone H4 | 46R I47, 96R T97 |
| Q6ZMQ8 | AATK | Serine/threonine-protein kinase LMTK1 | 403A T404 |
| P25205 | MCM3 | DNA replication licensing factor MCM3 | 680K S681 |
| O75528 | TADA3 | Transcriptional adapter 3 | 385R M386 |
| Q9BXT5 | TEX15 | Testis-expressed protein 15 | 1653R K1654 |
| Q7Z2Z1 | TICRR | Treslin | 186K Q187 |
| Q9H091 | ZMYND15 | Zinc finger MYND domain-containing protein 15 | 114E G115 |
| Protein processing | |||
| Q9H3G5 | CPVL | Probable serine carboxypeptidase CPVL | 457R A458 |
| P07478 | PRSS2 | Trypsin-2 * | 72R L73 |
| Miscellaneous functions | |||
| P27482 | CALML3 | Calmodulin-like protein 3 | 112R L113 |
| P08238 | HSP90AB1 | Heat shock protein HSP 90-beta * | 378R G379 |
| Q86WN2 | IFNE | Interferon epsilon | 160Y S161 |
| O15018 | PDZD2 | PDZ domain-containing protein 2 | 2574S V2575 |
| A6NHP3 | SPDYE2B | Speedy protein E2B * | 110R V111 |
| Q92734 | TFG | Protein TFG | 383R N384 |
| Q16851 | UGP2 | UTP-glucose-1-phosphate uridylyltransferase | 12M S13 |
| Gene | Disease | Abbreviation, MIM Code | References |
|---|---|---|---|
| MPDZ | Hydrocephalus, congenital, 2, with or without brain or eye anomalies | HYC2, MIM:615219 | [45,67] |
| Leber congenital amaurosis, Retinitis pigmentosa | LCA, RP | [46] | |
| H4 | Tessadori-van Haaften neurodevelopmental syndrome 1, 2, 3 and 4 | TEVANED1–4 MIM:619758, 619759, 619950 and 619951 | [68,69,70] |
| IQSEC1 | Intellectual developmental disorder with short stature and behavioral abnormalities | IDDSSBA, MIM:618687 | [47] |
| SPTBN4 | Neurodevelopmental disorder with hypotonia, neuropathy and deafness | NEDHND, MIM:617519 | [62] |
| HNRNPU | Developmental and epileptic encephalopathy 54 | DEE54, MIM:617391 | [52,57] |
| UGP2 | Developmental and epileptic encephalopathy 83 | DEE83, MIM:618744 | [32] |
| TFG | Neuropathy, hereditary motor and sensory, Okinawa type | HMSNO, MIM:604484 | [59,61] |
| Spastic paraplegia 57, autosomal recessive | SPG57, MIM:615658 | [50,58,61,64] | |
| TUBB4B | Leber congenital amaurosis with early-onset deafness | LCAEOD, MIM:617879 | [63] |
| TUBB6 | Facial palsy, congenital, with ptosis and velopharyngeal dysfunction | FPVEPD, MIM:617732 | [55] |
| EWS | Ewing sarcoma | ES, MIM:612219 | [51,54,60,66] |
| Angiomatoid fibrous histiocytoma | AFH, MIM:612160 | [48,56] | |
| ZMYND15 | Spermatogenic failure 14 | SPGF14, MIM:615842 | [49] |
| TEX15 | Spermatogenic failure 25 | SPGF25, MIM:617960 | [53,65,71] |
| Gene | Entry Clone |
|---|---|
| AATK | HsCD00821095 |
| DLGAP4 | HsCD00829035 |
| IQSEC1 | HsCD00878932 |
| MCM3 | HsCD00041579 |
| TFG | HsCD00041139 |
| UGP2 | HsCD00513800 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gal, J.; Dufour, A.; Young, D.; Yang, E.S.; Geddes, J.W. The Identification of Proteolytic Substrates of Calpain-5 with N-Terminomics. Int. J. Mol. Sci. 2025, 26, 6459. https://doi.org/10.3390/ijms26136459
Gal J, Dufour A, Young D, Yang ES, Geddes JW. The Identification of Proteolytic Substrates of Calpain-5 with N-Terminomics. International Journal of Molecular Sciences. 2025; 26(13):6459. https://doi.org/10.3390/ijms26136459
Chicago/Turabian StyleGal, Jozsef, Antoine Dufour, Daniel Young, Eddy S. Yang, and James W. Geddes. 2025. "The Identification of Proteolytic Substrates of Calpain-5 with N-Terminomics" International Journal of Molecular Sciences 26, no. 13: 6459. https://doi.org/10.3390/ijms26136459
APA StyleGal, J., Dufour, A., Young, D., Yang, E. S., & Geddes, J. W. (2025). The Identification of Proteolytic Substrates of Calpain-5 with N-Terminomics. International Journal of Molecular Sciences, 26(13), 6459. https://doi.org/10.3390/ijms26136459





