Taxonomy, Phylogeny, Genomes, and Repeatomes in the Subgenera Salvia, Sclarea, and Glutinaria (Salvia, Lamiaceae)
Abstract
1. Introduction
2. Taxonomy and Phylogeny of the Genus Salvia
3. Genomes and Repeatomes in the Subgenera Salvia, Sclarea, and Glutinaria
3.1. Karyological Studies of the Salvia Species
| Sections and Species | Chromosome Number | Source | DNA Content, pg/1C |
|---|---|---|---|
| Sect. Plethiosphace | |||
| S. nemorosa | 2n = 2x = 14 | [70,72,83] | 0.56–0.6 [19] 0.55 [81] |
| S. tesquicola (S. nemorosa subsp. tesquicola) | 2n = 2x = 14 | [67] | |
| S. algeriensis | x = 7, 9 2n = 4x = 36 2n = 38 2n = 6x = 42 | [61] | |
| S. amplexicaulis | 2n = 2x = 20 | [83] | 0.74 [19] |
| S. austriaca | 2n = 2x = 18 | [72,83] | |
| S. deserta | 2n = 2x = 14, 16 | [67] | 0.56 [19] |
| S. nutans | 2n = 2x = 22 | [67] | 0.50 [19] |
| S. pratensis | 2n = 16, 18, 32 | [67] | 0.56 [19] 0.46 [81] |
| S. transsylvanica | 2n = 2x = 16 | [61] | |
| S. verbenaca | 2n = 14, 16, 42, 54, 56, 60, 62, 64 | [67] | 0.48–0.49 [19] |
| S. dumetorum | 2n = 2x = 14 | [72] | 0.60 [19] |
| S. jurisicii | 2n = 2x = 22 | [61,67] | |
| S. grandifolia | 2n = 4x = 40 | [88] | |
| Sect. Aethiopis | |||
| S. aethiopis | 2n = 2x = 22 | [67] | 1.50 [19] |
| S. sclarea | 2n = 2x = 22 | [67] | 0.66–0.69 [19] 0.58 [81] |
| S. verbascifolia | 2n = 2x = 16, 18, 22 | [67,72] | |
| S. karabachensis | 2n = 2x = 20 | [67] | |
| S. argentea | 2n = 2x = 20, 22 | [59,72,83] | |
| S. frigida | 2n = 2x = 20 | [59] | |
| S. fominii | 2n = 16 | [67] | |
| S. palestina | 2n = 2x = 20 | [59,67] | |
| S. poculata | 2n = 2x = 20 | [59] | |
| S. limbata | 2n = 2x = 22 | [59] | 0.86 [80] |
| S. desoleana | 2n = 4x = 44 | [67] | 0.78 [19] |
| Sect. Horminum | |||
| S. viridis | 2n = 2x = 16 | [70,72,83] | 0.53 [19] 0.43 [81] |
3.2. Integration of the Repeatomic and Cytogenomic Data
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Drew, B.T.; González-Gallegos, J.G.; Xiang, C.L.; Kriebel, R.; Drummond, C.P.; Walked, J.B.; Sytsma, K.J. Salvia united: The greatest good for the greatest number. Taxon 2017, 66, 133–145. [Google Scholar] [CrossRef]
- Kriebel, R.; Drew, B.T.; Drummond, C.P.; González-Gallegos, J.G.; Celep, F.; Mahdjoub, M.M.; Rose, J.P.; Xiang, C.L.; Hu, G.X.; Walker, J.B.; et al. Tracking temporal shifts in area, biomes, and pollinators in the radiation of Salvia (sages) across continents: Leveraging anchored hybrid enrichment and targeted sequence data. Am. J. Bot. 2019, 106, 573–597. [Google Scholar] [CrossRef] [PubMed]
- Baikova, E.V. Genus Salvia: Morphiology, Evolution, Prospects of Cultivation; Nauka: Novosibirsk, Russia, 2006; p. 248. [Google Scholar]
- Wu, Y.B.; Ni, Z.Y.; Shi, Q.W.; Dong, M.; Kiyota, H.; Gu, Y.C.; Cong, B. Constituents from Salvia species and their biological activities. Chem. Rev. 2012, 112, 5967–6026. [Google Scholar] [CrossRef]
- Zhumaliyeva, G.; Zhussupova, A.; Zhusupova, G.E.; Błońska-Sikora, E.; Cerreto, A.; Omirbekova, N.; Zhunusbayeva, Z.; Gemejiyeva, N.; Ramazanova, M.; Wrzosek, M.; et al. Natural Compounds of Salvia L. Genus and Molecular Mechanism of Their Biological Activity. Biomedicines 2023, 11, 3151. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, A.; Esmaeilizadeh, M. Pharmacological properties of Salvia officinalis and its components. J. Tradit. Complement. Med. 2017, 7, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Hafez Ghoran, S.; Taktaz, F.; Mozafari, A.A.; Tunçtürk, M.; Sekeroglu, N.; Kijjoa, A. Uncommon Terpenoids from Salvia Species: Chemistry, Biosynthesis and Biological Activities. Molecules 2022, 27, 1128. [Google Scholar] [CrossRef]
- Sharifi-Rad, M.; Ozcelik, B.; Altı, G.; Daşkaya-Dikmen, C.; Martorell, M.; Ramírez-Alarcón, K.; Alarcón-Zapata, P.; Morais-Braga, M.F.B.; Carneiro, J.N.P.; Borges Leal, A.L.A.; et al. Salvia spp. plants-from farm to food applications and phytopharmacotherapy. Trends Food Sci. Technol. 2018, 80, 242–263. [Google Scholar] [CrossRef]
- Will, M.; Claßen-Bockhoff, R. Time to split Salvia s.l. (Lamiaceae)—New insights from Old World Salvia phylogeny. Mol. Phylogenet. Evol. 2017, 109, 33–58. [Google Scholar] [CrossRef]
- Bentham, G. Labiatae. In Prodromus Systematis Universalis Regni Vegetabilis; Candolle, A.P., Ed.; Treuttel et Würtz: Paris, France, 1848; Volume 12, pp. 212–226. [Google Scholar]
- Briquet, J. Labiatae. In Die Naturlichen Pflanzenfamilien; Engler, H.G.A., Prantl, K.A.E., Eds.; Wilhelm Engelmann: Leipzig, Germany, 1897; Volume 4, pp. 182–286. [Google Scholar]
- Hedge, I.C. A revision of Salvia in Africa, including Madagascar and the Canary Islands. Notes R. Bot. Gard. Edinb. 1974, 33, 1–121. [Google Scholar]
- Pobedimova, E.G. Salvia L. In Flora USSR; Schischkin, B.K., Ed.; Akad. Scient.; URSS: Moscow, Leningrad, 1954; Volume 21, pp. 244–363. [Google Scholar]
- Walker, J.B.; Sytsma, K.J.; Treutlein, J.; Wink, M. Salvia (Lamiaceae) Is Not Monophyletic: Implications for the Systematics, Radiation, and Ecological Specializations of Salvia and Tribe Mentheae. Am. J. Bot. 2004, 91, 1115–1125. [Google Scholar] [CrossRef]
- Will, M.; Schmalz, N.; Classen-Bockhoff, R. Towards a new classification of Salvia s.l.: (Re) establishing the genus Pleudia Raf. Turk. J. Bot. 2015, 39, 693–707. [Google Scholar] [CrossRef]
- Hu, G.X.; Takano, A.; Drew, B.T.; Liu, E.D.; Soltis, D.E.; Soltis, P.S.; Peng, H.; Xiang, C.L. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann. Bot. 2018, 122, 649–668. [Google Scholar] [CrossRef]
- Celep, F.; Kahraman, A.; Guerin, G.R.; Karabacak, E.; Akaydın, G.; Doğan, M. Nutlet micromorphology and its taxonomic and phylogenetic significance in Salvia (Lamiaceae). Plant Biosyst. 2022, 156, 271–283. [Google Scholar] [CrossRef]
- Ranjbar, M.; Pakatchi, A.; Babataheri, Z.J. Chromosome number evolution, biogeography and phylogenetic relationships in Salvia (Lamiaceae). Webbia J. Plant Taxon Geogr. 2015, 70, 293–312. [Google Scholar] [CrossRef]
- Maynard, R.C.I.; Ruter, J.M. DNA Content estimation in the genus Salvia. J. Amer. Soc. Hort. Sci. 2022, 147, 123–134. [Google Scholar] [CrossRef]
- Liu, B.; Chen, C.; Li, X.; Chen, R.; Song, W. Physical mapping of 45S rDNA to metaphase chromosomes in 30 taxonomically diverse plant species. J. Hortic. Sci. Biotechnol. 2005, 80, 287–290. [Google Scholar] [CrossRef]
- Song, Z.; Lin, C.; Xing, P.; Fen, Y.; Jin, H.; Zhou, C.; Gu, Y.Q.; Wang, J.; Li, X. A high-quality reference genome sequence of Salvia miltiorrhiza provides insights into tanshinone synthesis in its red rhizomes. Plant Genome 2020, 13, e20041. [Google Scholar] [CrossRef]
- Muravenko, O.V.; Yurkevich, O.Y.; Kalnyuk, J.V.; Samatadze, T.E.; Zoshchuk, S.A.; Amosova, A.V. Integration of Repeatomic and Cytogenetic Data on Satellite DNA for the Genome Analysis in the Genus Salvia (Lamiaceae). Plants 2022, 11, 2244. [Google Scholar] [CrossRef]
- Ali, M.; Li, P.; She, G.; Chen, D.; Wan, X.; Zhao, J. Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis). Sci. Rep. 2017, 7, 16074. [Google Scholar] [CrossRef]
- Jia, K.H.; Liu, H.; Zhang, R.G.; Xu, J.; Zhou, S.S.; Jiao, S.Q.; Yan, X.M.; Tian, X.C.; Shi, T.L.; Luo, H.; et al. Chromosome-scale assembly and evolution of the tetraploid Salvia splendens (Lamiaceae) Genome. Hortic. Res. 2021, 8, 177. [Google Scholar] [CrossRef]
- Han, D.; Li, W.; Hou, Z.; Lin, C.; Xie, Y.; Zhou, X.; Gao, Y.; Huang, J.; Lai, J.; Wang, L.; et al. The Chromosome-Scale Assembly of the Salvia Rosmarinus Genome Provides Insight into Carnosic Acid Biosynthesis. Plant J. 2023, 113, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Ceylan, F.; Uncu, A.O.; Soyturk Patat, A.; Uncu, A.T. Whole-genome resequencing identifies exonic single-nucleotide variations in terpenoid biosynthesis genes of the medicinal and aromatic plant common sage (Salvia officinalis L.). Genet. Resour. Crop. Evol. 2024, 71, 4171–4181. [Google Scholar] [CrossRef]
- Li, C.Y.; Yang, L.; Liu, Y.; Xu, Z.G.; Gao, J.; Huang, Y.B.; Xu, J.J.; Fan, H.; Kong, Y.; Wei, Y.K.; et al. The sage genome provides insight into the evolutionary dynamics of diterpene biosynthesis gene cluster in plants. Cell Rep. 2022, 40, 111236. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kang, Y.; Kim, C. Chromosome-level genome assembly of Salvia sclarea. Sci. Data 2025, 12, 14. [Google Scholar] [CrossRef]
- Royal Botanic Gardens, Kew. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:30000096-2 (accessed on 30 June 2025).
- WFO Plant List. Available online: https://wfoplantlist.org/taxon/wfo-4000033888-2025-06 (accessed on 30 June 2025).
- Claßen-Bockhoff, R.; Speck, T. Diversity and evolution in Salvia—presentation of a new research project. Vitex 2000, 1, 3–4. [Google Scholar] [CrossRef]
- Claβen-Bockhoff, R.; Wester, P.; Tweraser, E. The Staminal Lever Mechanism in Salvia L. (Lamiaceae)—A Review. Plant Biol. 2003, 5, 33–41. [Google Scholar] [CrossRef]
- Speck, T.; Rowe, N.P.; Civeyrel, L.; Claßen-Bockhoff, R.; Neinhuis, C.; Spatz, C. The potential of plant biomechanics in functional biology and systematics. In Deep Morphology: Towards a Renaissance of Morphology in Plant Systematics; Stuessy, T., Hörandl, F., Mayer, V., Eds.; Koeltz: Königstein, Germany, 2003; pp. 241–271. [Google Scholar]
- Claßen-Bockhoff, R.; Speck, T.; Tweraser, E.; Wester, P.; Thimm, S.; Reith, M. The staminal lever mechanism in Salvia L. (Lamiaceae): A key innovation for adaptive radiation? Organisms Divers. Evol. 2004, 4, 189–205. [Google Scholar] [CrossRef]
- Zhang, B.; Claßen-Bockhoff, R.; Zhang, Z.Q.; Sun, S.; Luo, Y.J.; Li, Q.J. Functional implications of the staminal lever mechanism in Salvia cyclostegia (Lamiaceae). Ann. Bot. 2011, 107, 621–628. [Google Scholar] [CrossRef]
- Drew, B.T.; Sytsma, K.J. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am. J. Bot. 2012, 99, 933–953. [Google Scholar] [CrossRef]
- Claßen-Bockhoff, R. Stamen construction, development and evolution in Salvia s.l. Nat. Volatiles Essent. Oils 2017, 4, 28–48. [Google Scholar]
- Bentham, G. Labiatae. In Genera Plantarum; Bentham, G., Hooker, J.D., Eds.; Reeve and Co.: London, UK, 1876; Volume 2, pp. 1160–1196. [Google Scholar]
- Walker, J.B.; Sytsma, K.J. Staminal evolution in the genus Salvia (Lamiaceae): Molecular phylogenetic evidence for multiple origins of the staminal lever. Ann. Bot. 2007, 100, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Karaca, M.; Ince, A.G. Molecular markers in Salvia L.: Past, present and future. In Salvia biotechnology; Georgiev, V., Pavlov, A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 291–398. [Google Scholar] [CrossRef]
- Jug-Dujaković, M.; Ninčević, T.; Liber, Z.; Grdiša, M.; Šatović, Z. Salvia officinalis survived in situ Pleistocene glaciation in “refugia within refugia” as inferred from AFLP markers. Plant Syst. Evol. 2020, 306, 2. [Google Scholar] [CrossRef]
- Javan, Z.S.; Rahmani, F.; Heidari, R. Assessment of genetic variation of genus Salvia by RAPD and ISSR markers. Aust. J. Crop. Sci. 2012, 6, 1068–1073. [Google Scholar]
- Sunar, S.; Korkmaz, M.; Siğmaz, B.; Ağar, G. Determination of the Genetic Relationships among Salvia Species by RAPD and ISSR Analyses. Turk. J. Pharm. Sci. 2020, 17, 480–485. [Google Scholar] [CrossRef]
- Zheng, R.; Zhao, S.; Khayyatnezhad, M.; Shah, S.A. Comparative Study and Genetic Diversity in Salvia (Lamiaceae) Using RAPD Molecular Markers. Caryologia 2021, 74, 45–56. [Google Scholar] [CrossRef]
- Radosavljević, I.; Satovic, Z.; Jakse, J.; Javornik, B.; Greguraš, D.; Jug-Dujaković, M.; Liber, Z. Development of New Microsatellite Markers for Salvia officinalis L. and Its Potential Use in Conservation-Genetic Studies of Narrow Endemic Salvia brachyodon Vandas. Int. J. Mol. Sci. 2012, 13, 12082–12093. [Google Scholar] [CrossRef] [PubMed]
- Ince, A.G.; Karaca, M. E-Microsatellite Markers for Some Naturally Occurring Salvia Species in the Mediterranean Region. Turk. J. Biol. 2015, 39, 69–77. [Google Scholar] [CrossRef]
- Moein, F.; Jamzad, Z.; Rahiminejad, M.; Landis, J.B.; Mirtadzadini, M.; Soltis, D.E.; Soltis, P.S. Towards a global perspective for Salvia L.: Phylogeny, diversification and floral evolution. J. Evol. Biol. 2023, 36, 589–604. [Google Scholar] [CrossRef]
- Rose, J.P.; Kriebel, R.; Kahan, L.; DiNicola, A.; González-Gallegos, J.G.; Celep, F.; Lemmon, E.M.; Lemmon, A.R.; Sytsma, K.J.; Drew, B.T. Sage insights into the phylogeny of Salvia: Dealing with sources of discordance within and across genomes. Front Plant Sci. 2021, 12, 767478. [Google Scholar] [CrossRef]
- Yu, D.; Pei, Y.; Cui, N.; Zhao, G.; Hou, M.; Chen, Y.; Chen, J.; Li, X. Comparative and phylogenetic analysis of complete chloroplast genome sequences of Salvia regarding its worldwide distribution. Sci. Rep. 2023, 13, 14268. [Google Scholar] [CrossRef]
- Gao, C.; Wu, C.; Zhang, Q.; Zhao, X.; Wu, M.; Chen, R.; Zhao, Y.; Li, Z. Characterization of chloroplast genomes from two Salvia medicinal plants and gene transfer among their mitochondrial and chloroplast genomes. Front. Genet. 2020, 11, 574962. [Google Scholar] [CrossRef]
- Zhao, F.; Drew, B.T.; Chen, Y.P.; Hu, G.X.; Li, B.; Xiang, C.L. The chloroplast genome of Salvia: Genomic characterization and phylogenetic analysis. Int. J. Plant Sci. 2020, 181, 812–830. [Google Scholar] [CrossRef]
- Du, Q.; Yang, H.; Zeng, J.; Chen, Z.; Zhou, J.; Sun, S.; Wang, B.; Liu, C. Comparative Genomics and Phylogenetic Analysis of the Chloroplast Genomes in Three Medicinal Salvia Species for Bioexploration. Int. J. Mol. Sci. 2022, 23, 12080. [Google Scholar] [CrossRef]
- Yang, H.; Chen, H.; Ni, Y.; Li, J.; Cai, Y.; Wang, J.; Liu, C. Mitochondrial genome sequence of Salvia officinalis (Lamiales: Lamiaceae) suggests diverse genome structures in cogeneric species and finds the stop gain of genes through RNA editing events. Int. J. Mol. Sci. 2023, 24, 5372. [Google Scholar] [CrossRef]
- Winkler, H. Verbreitung und Ursache der Parthenogenese im Pflanzen-und Tierreich; Fischer: Jena, Germany, 1920; p. 165. [Google Scholar]
- Zelenin, A.V.; Rodionov, A.V.; Bolsheva, N.L.; Badaeva, E.D.; Muravenko, O.V. Genome: Origins and evolution of the term. Mol. Biol. 2016, 50, 542–550. [Google Scholar] [CrossRef]
- Epling, C.; Lewis, H.; Raven, P.H. Chromosomes of Salvia: Section Audibertia. Aliso 1962, 5, 217–221. [Google Scholar] [CrossRef]
- Haque, M.S. Chromosome numbers in the genus Salvia Linn. Proc. Indian Acad. Sci. 1981, 47, 419–426. [Google Scholar]
- Ward, D.E. Chromosome counts from New Mexico and Mexico [List of plant species, cytogeography]. Phytologia 1984, 56, 55–60. [Google Scholar]
- Martin, E.; Cetin, O.; Kahraman, A.; Celep, F.; Dogan, M. A cytomorphological study in some taxa of the genus Salvia L. (Lamiaceae). Caryologia 2011, 64, 272–287. [Google Scholar] [CrossRef]
- Martin, E.; Altınordu, F.; Celep, F.; Kahraman, A.; Doğan, M. Karyomorphological studies in seven taxa of the genus Salvia (Lamiaceae) in Turkey. Caryologia 2015, 68, 13–18. [Google Scholar] [CrossRef]
- Martin, E.; Celep, F.; Eroğlu, H.E. Comparative chromosomal features and new karyological data in Salvia: B-chromosomes, polyploidy, dysploidy and symmetric karyotypes. Braz. J. Bot. 2022, 45, 625–634. [Google Scholar] [CrossRef]
- Celep, F.; Raders, E.; Drew, B. Two new hybrid species of Salvia (S.× karamanensis and S.× doganii) from Turkey: Evidence from molecular and morphological studies. Turk. J. Bot. 2020, 44, 647–660. [Google Scholar] [CrossRef]
- Ramakrishnan, R.; Banadka, A.; Sudheer, W.N.; Bajrang, C.P.; Al-Khayri, J.M.; Nagella, P. Advances in Breeding Strategies of Common Sage (Salvia officinalis L.). In Biodiversity and Genetic Improvement of Herbs and Spices; Al-Khayri, J.M., Jain, S.M., Penna, S., Eds.; Advances in Plant Breeding Strategies; Springer: Cham, Switzerland, 2025; Volume 8, pp. 345–372. [Google Scholar] [CrossRef]
- Copetta, A.; Ruffoni, B. Biodiversity and Breeding in Salvia officinalis L. In Breeding of Ornamental Crops: Annuals and Cut Flowers; Al-Khayri, J.M., Jain, S.M., Wani, M.A., Eds.; Advances in Plant Breeding Strategies; Springer: Cham, Switzerland, 2025; Volume 6, pp. 263–296. [Google Scholar] [CrossRef]
- Tychonievich, J.; Warner, R. Interspecific crossability of selected Salvia species and po-tential use for crop improvement. J. Am. Soc. Hortic. Sci. 2011, 136, 41–47. [Google Scholar] [CrossRef]
- Morrison, E.G.; Drew, B.T. Potential hybridization among two species of California Salvia. Trans. Nebr. Acad. Sci. 2023, 43, 25–33. [Google Scholar] [CrossRef]
- Missouri Botanical Garden. Index to Plant Chromosome Numbers (IPCN); Missouri Botanical Garden: St. Louis, MO, USA; Available online: http://legacy.tropicos.org/Project/IPCN (accessed on 17 April 2025).
- Houben, A. B Chromosomes—A matter of chromosome drive. Front Plant Sci. 2017, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.S.; Ghoshal, K.K. Karyotypes and chromosome morphology in the genus Salvia Linn. Cytologia 1980, 45, 627–640. [Google Scholar] [CrossRef]
- Afzal-Rafii, Z. Étude cytotaxonomique et phylogénétique de quelques Salvia de la région méditerranéenne: Groupe du Salvia officinalis L. Bull. Soc. Bot. Fr. 1976, 123, 515–527. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, L.; Zhao, H.; Yang, R.; Ding, C.; Zhou, Y.; Wan, D. Chromosome numbers of some species of Salvia (Lamiaceae) from the Sichuan Province, China. Nord. J. Bot. 2009, 27, 287–291. [Google Scholar] [CrossRef]
- Patudin, A.V. Chromosome numbers in some species of Salvia L. (Lamiaceae). Bot. Zhurn. 1975, 60, 529–534. [Google Scholar]
- Alberto, C.M.; Sanso, A.M.; Xifreda, C.C. Chromosomal studies in species of Salvia (Lamiaceae) from Argentina. Bot. J. Linn. Soc. 2003, 141, 483–490. [Google Scholar] [CrossRef]
- Rodionov, A.V. Tandem duplications, eupolyploidy and secondary diploidization—genetic mechanisms of plant speciation and progressive evolution. Turczaninowia 2022, 25, 87–121. [Google Scholar] [CrossRef]
- Yang, Z.; Xun, G.; Pan, Y. Cytological study of six Salvia species (Lamiaceae) from the Hengduanshan Mountains region of China. Caryologia 2004, 57, 360–366. [Google Scholar] [CrossRef]
- Zhao, H.X.; Zhang, L.; Fan, X.; Yang, R.W.; Ding, C.B.; Zhou, Y.H. Studies on chromosome numbers of Salvia miltiorrhiza, S. flava and S. evansiana. China J. Chin. Mater. Med. 2006, 31, 1847–1849. [Google Scholar]
- Wang, M.; Zhang, L.; Ding, C.B.; Yang, R.W.; Zhou, Y.H.; Yang, Z.J.; Yin, Z.Q. Meiotic observations of eight taxa in the genus Salvia. Caryologia 2009, 62, 334–340. [Google Scholar]
- Hu, G.X.; Xiang, C.L.; Liu, E.D.; Dong, H.J.; Funamoto, T. Karyotypic study of eighteen taxa of Salvia (Lamiaceae) from China. Caryologia 2016, 69, 50–57. [Google Scholar] [CrossRef]
- Murin, A. Karyotaxonomy of some medicinal and aromatic plants. Thaiszia 1997, 7, 75–88. [Google Scholar]
- Hesamzadeh Hejazi, S.M.; Ziaei-Nasab, M. Investigating Inter-and Intra-specific Karyotypic Diversity of Some Species of Salvia Genus in Iran. Iran. J. Sci. 2024, 48, 45–60. [Google Scholar] [CrossRef]
- Leitch, I.J.; Johnston, E.; Pellicer, J.; Hidalgo, O.; Bennett, M.D. Plant DNA C-values Database (Release 7.1, April 2019). Royal Botanic Gardens, Kew. Available online: https://cvalues.science.kew.org/ (accessed on 17 April 2025).
- Zhen-Qiao, S.; Wang, J.H.; Xie, Y.L. Karyological studies of Salvia miltiorrhiza in China. Caryologia 2010, 63, 269–277. [Google Scholar] [CrossRef]
- Markova, M.L.; Ivanova, P.S. Karyological study of the genus Salvia L. in Bulgaria. Filologija 1982, 20, 3–19. (In Bulgarian) [Google Scholar]
- Aryavand, A. In IOPB chromosome number reports LVII. Taxon 1977, 26, 443–452. [Google Scholar]
- Sheidai, M.; Alijanpoo, B. Karyotype analysis in some Salvia species (Lamiaceae) of Iran. Cytologia 2011, 76, 425–429. [Google Scholar] [CrossRef]
- Özdemir, C.; Şenel, G. The morphological, anatomical and karyological properties of Salvia sclarea L. Turk. J. Bot. 1999, 23, 7–18. [Google Scholar]
- Kharazian, N. Karyotypic study of some Salvia (Lamiaceae) species from Iran. J. Appl. Biol. Sci. 2011, 5, 21–25. [Google Scholar]
- Hu, G.X.; Liu, E.D.; Wu, Z.K.; Sytsma, K.J.; Drew, B.T.; Xiang, C. Integrating DNA sequences with morphological analysis clarifies phylogenetic position of Salvia grandifolia (Lamiaceae): An enigmatic species endemic to southwestern China. Int. J. Plant Sci. 2020, 181, 787–799. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J. Nuclear genome size: Are we getting closer? Cytometry Part A 2010, 77, 635–642. [Google Scholar] [CrossRef]
- Özkan, U.; Benlioğlu, B.; Özgen, Y. Karyological Studies on Mediterranean Sage (Salvia aethiopis L.). J. Appl. Biol. Sci. 2017, 11, 33–34. [Google Scholar]
- Kursat, M.; Emre, I.; Gedik, O.; Kiran, Y. Karyological reports for some Salvia taxa from Turkey. Caryologia 2018, 71, 120–127. [Google Scholar] [CrossRef]
- Liu, Q.; Li, X.; Zhou, X.; Li, M.; Zhang, F.; Schwarzacher, T.; Heslop-Harrison, J.S. The repetitive DNA landscape in Avena (Poaceae): Chromosome and genome evolution defined by major repeat classes in whole-genome sequence reads. BMC Plant Biol. 2019, 19, 226. [Google Scholar] [CrossRef]
- Zagorski, D.; Hartmann, M.; Bertrand, Y.J.; Paštová, L.; Slavíková, R.; Josefiová, J.; Fehrer, J. Characterization and dynamics of repeatomes in closely related species of Hieracium (Asteraceae) and their synthetic and apomictic hybrids. Front. Plant Sci. 2020, 11, 591053. [Google Scholar] [CrossRef]
- Dogan, M.; Pouch, M.; Mandáková, T.; Hloušková, P.; Guo, X.; Winter, P.; Chumová, Z.; Van Niekerk, A.; Mummenhoff, K.; Al-Shehbaz, I.; et al. Evolution of tandem repeats is mirroring post-polyploid cladogenesis in Heliophila (Brassicaceae). Front. Plant Sci. 2021, 11, 607893. [Google Scholar] [CrossRef]
- Maumus, F.; Quesneville, H. Deep investigation of Arabidopsis thaliana junk DNA reveals a continuum between repetitive elements and genomic dark matter. PLoS ONE 2014, 9, e94101. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S. Comparative genome organization in plants: From sequence and markers to chromatin and chromosomes. Plant Cell 2000, 12, 617–635. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, Z.; Li, Y.; Hu, H.; Wang, Z.; Du, X. Which factors contribute most to genome size variation within angiosperms? Ecol. Evol. 2021, 11, 2660–2668. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Satish, L.; Sharma, A.; Vinod, K.K.; Emamverdian, A.; Zhou, M.; Wei, Q. Transposable elements in plants: Recent advancements, tools and prospects. Plant Mol. Biol. Rep. 2022, 40, 628–645. [Google Scholar] [CrossRef]
- Neumann, P.; Navrátilová, A.; Koblížková, A.; Kejnovský, E.; Hřibová, E.; Hobza, R.; Widmer, A.; Doležel, J.; Macas, J. Plant centromeric retrotransposons: A structural and cytogenetic perspective. Mob. DNA 2011, 2, 4. [Google Scholar] [CrossRef]
- Wells, J.N.; Feschotte, C. A field guide to eukaryotic transposable elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Papolu, P.K.; Ramakrishnan, M.; Mullasseri, S.; Kalendar, R.; Wei, Q.; Zou, L.; Ahmad, Z.; Vinod, K.K.; Yang, P.; Zhou, M. Retrotransposons: How the continuous evolutionary front shapes plant genomes for response to heat stress. Front. Plant Sci. 2022, 13, 1064847. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.H.; Mokhtar, M.M.; El Allali, A. Transposable elements: Multifunctional players in the plant genome. Front. Plant Sci. 2024, 14, 1330127. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O. LTR retrotransposons and flowering plant genome size: Emergence of the increase/decrease model. Cytogenet. Genome Res. 2005, 110, 91–107. [Google Scholar] [CrossRef]
- Galindo-González, L.; Mhiri, C.; Deyholos, M.K.; Grandbastien, M.-A. LTR-retrotransposons in plants: Engines of evolution. Gene 2017, 626, 14–25. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Ribosomal RNA genes in plants: Variability in copy number and in intergenic spacer. Plant Mol. Biol. 1987, 9, 509–520. [Google Scholar] [CrossRef]
- Garcia, S.; Kovařík, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Amosova, A.V.; Gnutikov, A.A.; Rodionov, A.V.; Loskutov, I.G.; Nosov, N.N.; Yurkevich, O.Y.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Genome Variability in artificial allopolyploid hybrids of Avena sativa L. and Avena macrostachya Balansa ex Coss. et Durieu based on marker sequences of satellite DNA and the ITS1–5.8S rDNA region. Int. J. Mol. Sci. 2024, 25, 5534. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrovic, N.; Mravinac, B. Satellite DNA evolution. In Repetitive DNA; Garrido-Ramos, M.A., Ed.; Karger: Granada, Spain, 2012; pp. 126–152. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA in Plants: More than Just Rubbish. Cytogenet. Genome Res. 2015, 146, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Amosova, A.V.; Ghukasyan, L.; Yurkevich, O.Y.; Bolsheva, N.L.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Cytogenomics of Deschampsia P. Beauv. (Poaceae) Species Based on Sequence Analyses and FISH Mapping of CON/COM Satellite DNA Families. Plants 2021, 10, 1105. [Google Scholar] [CrossRef]
- Macas, J.; Koblížková, A.; Navrátilová, A.; Neumann, P. Hypervariable 3′ UTR region of plant LTR-retrotransposons as a source of novel satellite repeats. Gene 2009, 448, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Becher, H.; Powell, R.F.; Brown, M.R.; Metherell, C.; Pellicer, J.; Leitch, I.J.; Twyford, A.D. The nature of intraspecific and interspecific genome size variation in taxonomically complex eyebrights. Ann. Bot. 2021, 128, 639–651. [Google Scholar] [CrossRef]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef]
- Muravenko, O.V.; Kalnyuk, J.V.; Yurkevich, O.Y.; Korotkikh, I.N.; Nevkrytaya, N.V.; Grunina, E.N.; Shmarayeva, A.N.; Popov, K.; Samatadze, T.E. Intraspecific variability of Salvia officinalis (Lamiaceae) according to DNA repeats chromosome patterns. Probl. Bot. South. Sib. Mong. 2023, 22, 207–211. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Kotseruba, V.V.; Fisenko, A.V.; Chikida, N.N.; Belousova, M.K.; Zhurbenko, P.M.; Surzhikov, S.A.; Dragovich, A.Y. Intraspecific divergence of diploid grass Aegilops comosa is associated with structural chromosome changes. Comp. Cytogenet. 2023, 17, 75–112. [Google Scholar] [CrossRef] [PubMed]
- Vondrak, T.; Ávila Robledillo, L.; Novák, P.; Koblížková, A.; Neumann, P.; Macas, J. Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant J. 2020, 101, 484–500. [Google Scholar] [CrossRef] [PubMed]
- Orooji, F.; Mirzaghaderi, G.; Kuo, Y.T.; Fuchs, J. Variation in the number and position of rDNA loci contributes to the diversification and speciation in Nigella (Ranunculaceae). Front. Plant Sci. 2022, 13, 917310. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.; Hufnagel, B.; Soriano, A.; Péret, B. The highly repeat-diverse (peri)centromeres of white lupin (Lupinus albus L.). Front. Plant Sci. 2022, 13, 862079. [Google Scholar] [CrossRef]
- Yurkevich, O.Y.; Samatadze, T.E.; Zoshchuk, S.A.; Semenov, A.R.; Morozov, A.I.; Selyutina, I.Y.; Amosova, A.V.; Muravenko, O.V. Repeatome Analysis and Satellite DNA Chromosome Patterns in Hedysarum Species. Int. J. Mol. Sci. 2024, 25, 12340. [Google Scholar] [CrossRef]

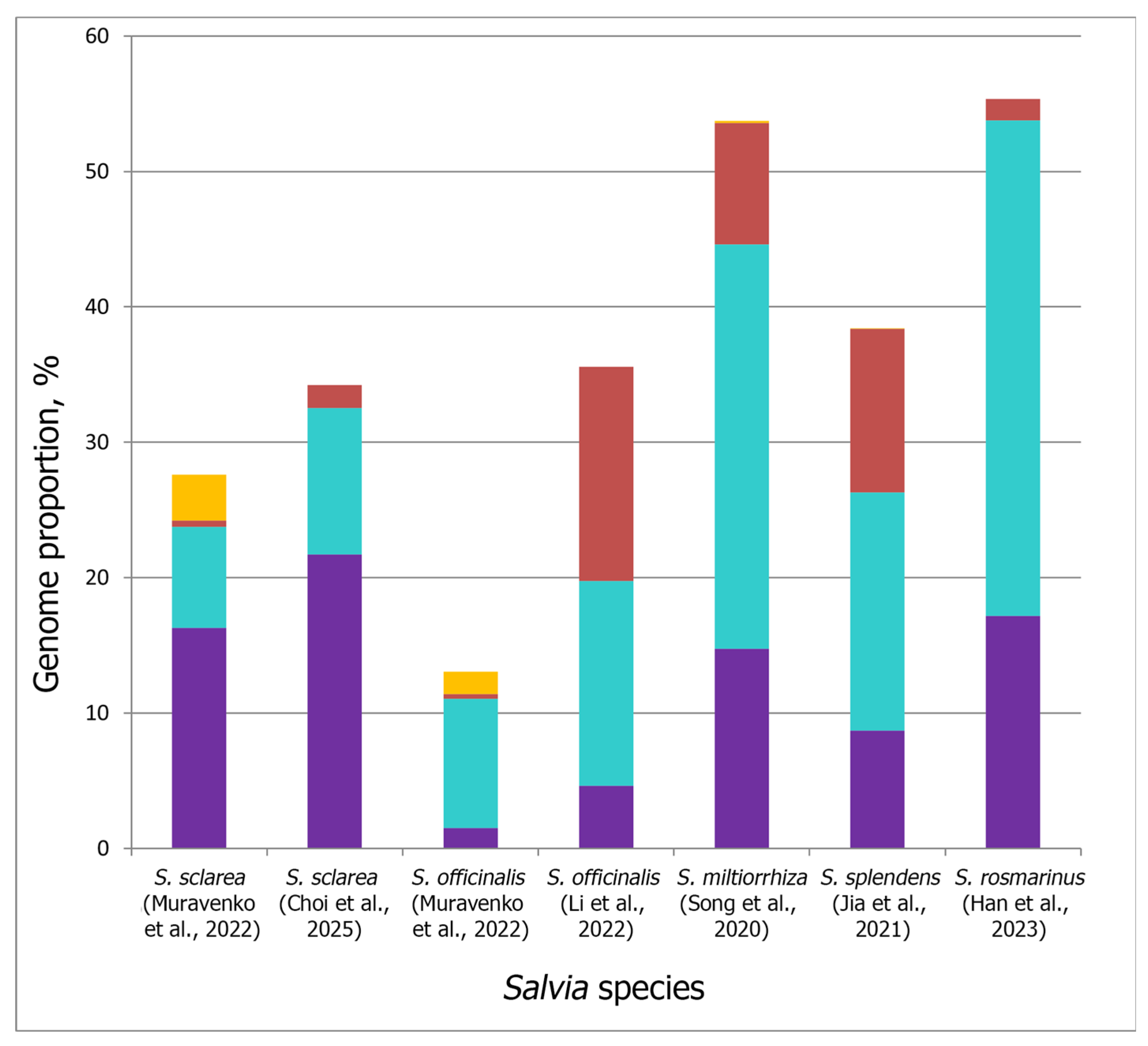
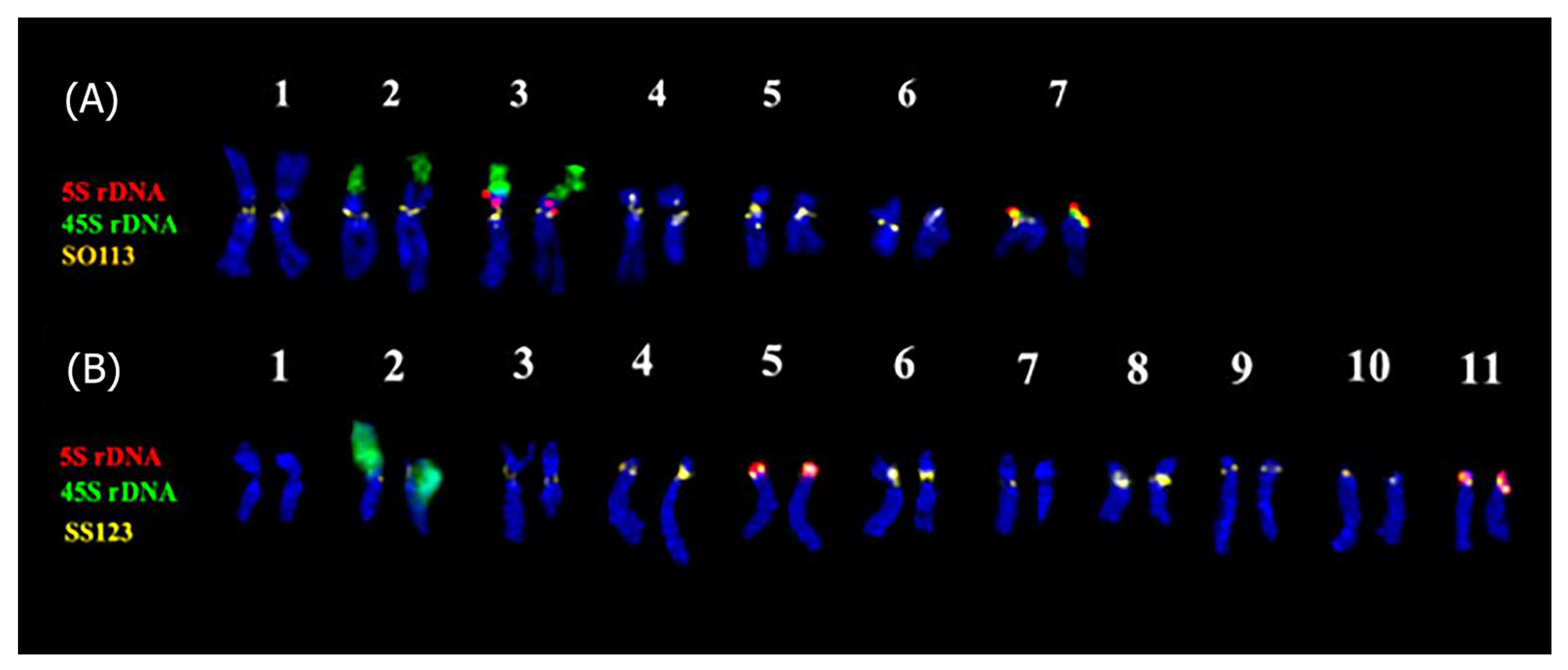
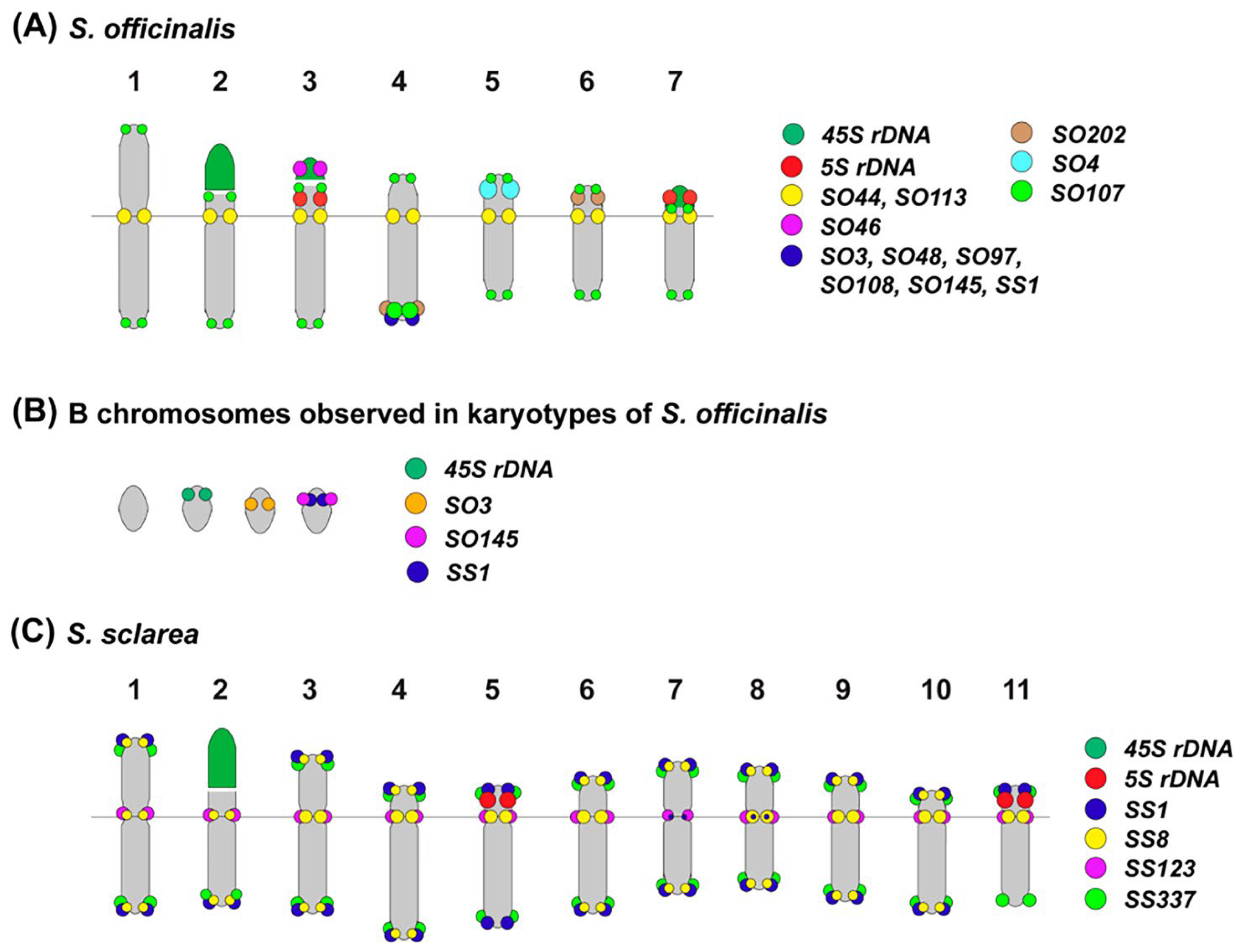
| Subgenus (Subgeneric Classification of Salvia [2] | Estimated Number of Species | Clade Number According to Will and Claßen-Bockhoff [9] | Staminal Type According to Walker and Sytsma [39] |
|---|---|---|---|
| Salvia L. | 70 | Clade I | A |
| Sclarea (Moench) Benth. | 120 | Clade I | B |
| “Heterosphace” | 43 | Clade I | A |
| Rosmarinus (L.) | 3 | C | |
| Perovskia (Kar.) | 8 | D | |
| Calosphace (Benth.) Epling | 550 | Clade II | E, F, G |
| Audibertia (Benth.) | 19 | Clade II | H, I |
| Meriandra (Benth.) | 2 | J | |
| Dorystaechas (Boiss. & Heldr. ex Benth.) | 1 | K | |
| Zhumeria (Rech. f. & Wendelbo) | 31 | Clade III (in part) | L, M |
| Glutinaria (Raf.) | 100 | Clade IV | N |
| Sections and Species | Chromosome Number | Sources | DNA Content, pg/1C |
|---|---|---|---|
| Sect. Sonchifoliae | No data | ||
| Sect. Notiosphace | |||
| S. plebeia | 2n = 2x = 16 + 0–2B, 32 | [67] | |
| Sect. Substoloniferae | |||
| S. trijuga | 2n = 2x = 16 | [75,78] | 0.42 [19] |
| Sect. Glutinaria | |||
| S. glutinosa | 2n = 2x = 16 | [61,80] | 0.83 [80] 1.07 [81] |
| S. nubicola | 2n = 2x = 16 | [67] | 1.11 [19] |
| S. nipponica | 2n = 2x = 16 | [67] | 1.21 [19] |
| Sect. Annuae | |||
| S. roborowskii | 2n = 2x = 16 | [75,78] | |
| Sect. Eurysphace | |||
| S. campanulata | n = 8 | [67] | |
| 2n = 4x = 32 | [78] | ||
| S. przewalskii | 2n = 2x = 16 | [72] | |
| 2n = 4x = 32 | [71,75,78] | ||
| S. evansiana | 2n = 2x = 16 | [78] | |
| 2n = 4x = 32 | [71,76] | ||
| Sect. Drymosphace | |||
| S. miltiorrhiza | 2n = 2x = 16 | [71,75,76,82] | 0.7 [19] 0.6 [21] 2.05 [81] |
| Sect. Sobiso | |||
| S. pygmaea | 2n = 2x = 16 | [67] | |
| Sections and Species | Chromosome Number | Sources | DNA Content, pg/1C |
|---|---|---|---|
| Sect. Salvia | |||
| S. officinalis | 2n = 2x = 14 | [59,72,79] | 0.59 [19] 0.49 [81] |
| 2n = 2x = 14 + 1–2B | [70] | ||
| S. fruticosa | 2n = 2x = 14 | [59] | 0.58 [19] 0.84 [81] |
| S. grandiflora | 2n = 2x = 14, 16 | [72,83] | |
| S. scabiosifolia | 2n = 2x = 14 | [83] | 0.77 [19] |
| S. aucheri subsp. canescens | 2n = 2x = 18 | [59] | |
| S. ringens | 2n = 2x = 12, 16 | [67] | 0.61 [81] |
| Sect. Holochilus | |||
| S. bucharica | 2n = 4x = 32 | [19] | 1.22 [19] |
| Sect. Hymenosphace | |||
| S. pomifera | 2n = 2x = 14 | [59,70] | |
| S. hydrangea | 2n = 2x = 14 | [59,70] | |
| S. multicaulis | 2n = 4x = 28 | [59] | 1.13 [81] |
| 2n = 16, 32, 30 | [70,84] | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalnyuk, J.V.; Yurkevich, O.Y.; Badaeva, E.D.; Semenov, A.R.; Zoshchuk, S.A.; Amosova, A.V.; Muravenko, O.V. Taxonomy, Phylogeny, Genomes, and Repeatomes in the Subgenera Salvia, Sclarea, and Glutinaria (Salvia, Lamiaceae). Int. J. Mol. Sci. 2025, 26, 6436. https://doi.org/10.3390/ijms26136436
Kalnyuk JV, Yurkevich OY, Badaeva ED, Semenov AR, Zoshchuk SA, Amosova AV, Muravenko OV. Taxonomy, Phylogeny, Genomes, and Repeatomes in the Subgenera Salvia, Sclarea, and Glutinaria (Salvia, Lamiaceae). International Journal of Molecular Sciences. 2025; 26(13):6436. https://doi.org/10.3390/ijms26136436
Chicago/Turabian StyleKalnyuk, Julia V., Olga Yu. Yurkevich, Ekaterina D. Badaeva, Alexey R. Semenov, Svyatoslav A. Zoshchuk, Alexandra V. Amosova, and Olga V. Muravenko. 2025. "Taxonomy, Phylogeny, Genomes, and Repeatomes in the Subgenera Salvia, Sclarea, and Glutinaria (Salvia, Lamiaceae)" International Journal of Molecular Sciences 26, no. 13: 6436. https://doi.org/10.3390/ijms26136436
APA StyleKalnyuk, J. V., Yurkevich, O. Y., Badaeva, E. D., Semenov, A. R., Zoshchuk, S. A., Amosova, A. V., & Muravenko, O. V. (2025). Taxonomy, Phylogeny, Genomes, and Repeatomes in the Subgenera Salvia, Sclarea, and Glutinaria (Salvia, Lamiaceae). International Journal of Molecular Sciences, 26(13), 6436. https://doi.org/10.3390/ijms26136436








