Short-Term Probiotic Colonization Alters Molecular Dynamics of 3D Oral Biofilms
Abstract
1. Introduction
2. Results
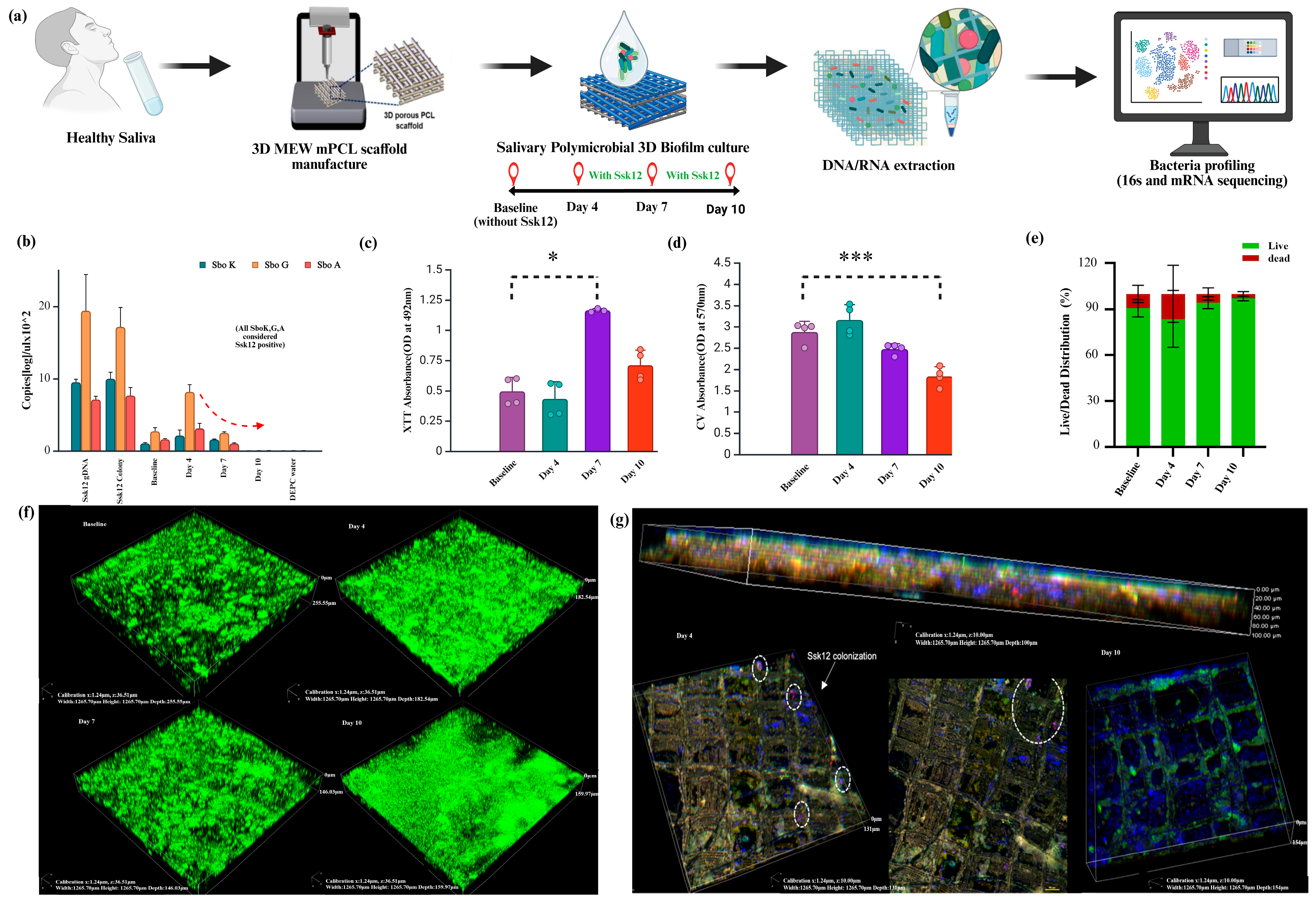
2.1. Transient Colonization of the Probiotic Bacteria into Oral Biofilm
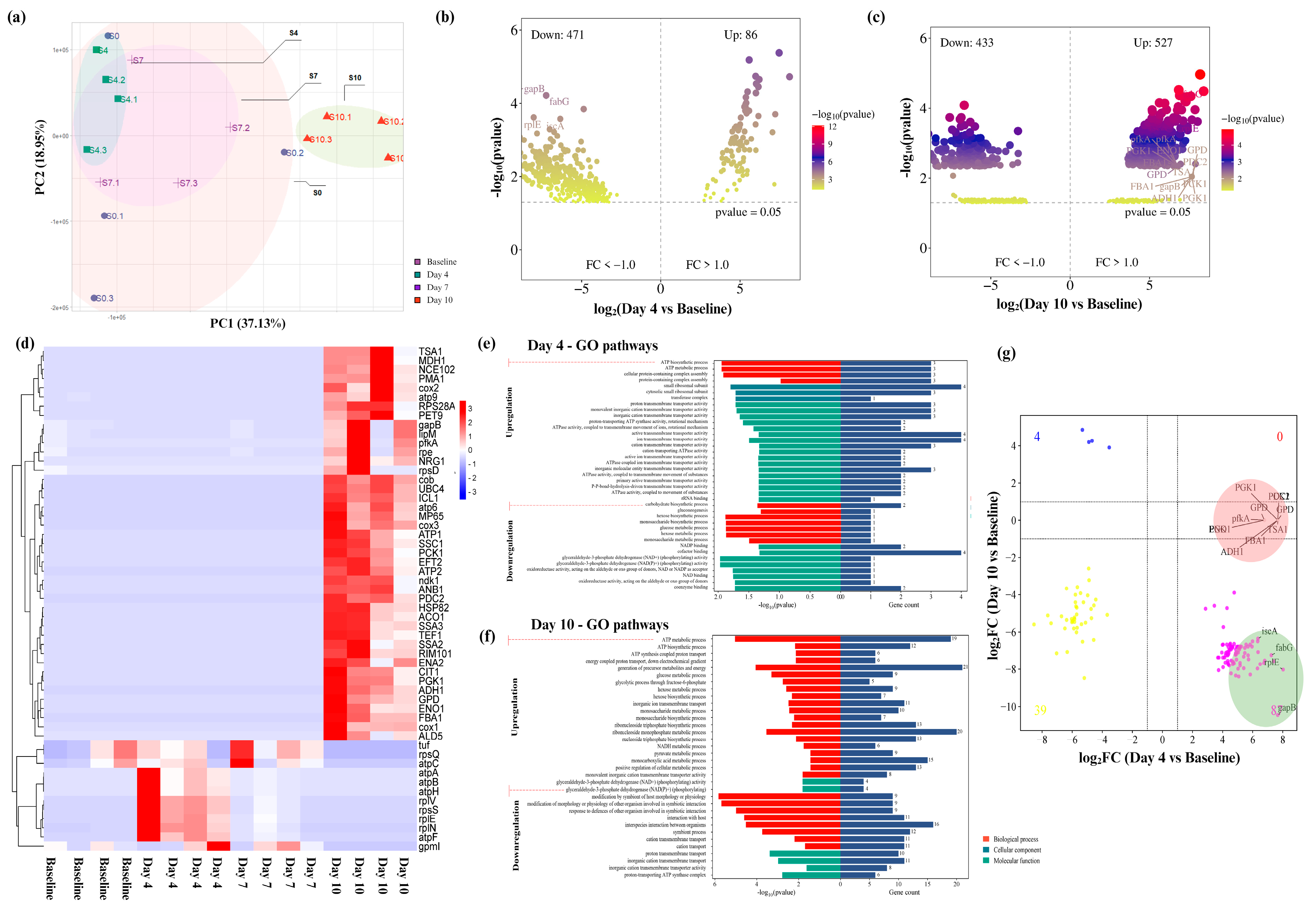
2.2. Longitudinal Microbiota Composition Changes with SsK12 Probiotic Colonization
2.3. Functional Role of Ssk12 Colonization in Oral Biofilm Modulation with a Stronger Link to Metabolic Redox Regulation
3. Discussion
4. Materials and Methods
4.1. Salivary Sample Collection and Establishment of Ex Vivo Community
4.2. Manufacture of Melt Electrowriting MEW PCL Scaffolds
4.3. Salivary Biofilm Culture and Ssk12 Inoculation
4.4. Characterization of Salivary Biofilm Biomass
4.5. Validation of Colonization Ssk12 in Salivary Biofilm Biomass Using Real-Time Quantitative PCR (RT-PCR) and smFISH (Single Molecule FISH)
4.6. Microbial Profile Using 16srRNA Sequencing mRNA Sequencing
4.7. Identification of Differentially Abundant Genes and GO Functional Pathway Enrichment Analyses
4.8. Identification of Top Hub Genes and Antiparallel Gene Expression Between Peak and Decline of Probiotic Ssk12 Colonization
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Rajendhran, J.; Gunasekaran, P. Human microbiomics. Indian J. Microbiol. 2010, 50, 109–112. [Google Scholar] [CrossRef]
- Deandra, F.A.; Ketherin, K.; Rachmasari, R.; Sulijaya, B.; Takahashi, N. Probiotics and metabolites regulate the oral and gut microbiome composition as host modulation agents in periodontitis: A narrative review. Heliyon 2023, 9, e13475. [Google Scholar] [CrossRef]
- Luo, S.C.; Wei, S.M.; Luo, X.T.; Yang, Q.Q.; Wong, K.H.; Cheung, P.C.K.; Zhang, B.B. How probiotics, prebiotics, synbiotics, and postbiotics prevent dental caries: An oral microbiota perspective. NPJ Biofilms Microbiomes 2024, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, V.H.; Bandara, H.M.; Ishikawa, K.H.; Mayer, M.P.; Samaranayake, L.P. The role of probiotic bacteria in managing periodontal disease: A systematic review. Expert Rev. Anti Infect. Ther. 2016, 14, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Nibali, L.; Koidou, V.P.; Hamborg, T.; Donos, N. Empirical or microbiologically guided systemic antimicrobials as adjuncts to non-surgical periodontal therapy? A systematic review. J. Clin. Periodontol. 2019, 46, 999–1012. [Google Scholar] [CrossRef]
- Routier, A.; Blaizot, A.; Agossa, K.; Dubar, M. What do we know about the mechanisms of action of probiotics on factors involved in the pathogenesis of periodontitis? A scoping review of in vitro studies. Arch. Oral Biol. 2021, 129, 105196. [Google Scholar] [CrossRef]
- Lundtorp-Olsen, C.; Markvart, M.; Twetman, S.; Belstrøm, D. Effect of Probiotic Supplements on the Oral Microbiota—A Narrative Review. Pathogens 2024, 13, 419. [Google Scholar] [CrossRef]
- Dassi, E.; Ballarini, A.; Covello, G.; Quattrone, A.; Jousson, O.; De Sanctis, V.; Bertorelli, R.; Denti, M.A.; Segata, N. Enhanced microbial diversity in the saliva microbiome induced by short-term probiotic intake revealed by 16S rRNA sequencing on the IonTorrent PGM platform. J. Biotechnol. 2014, 190, 30–39. [Google Scholar] [CrossRef]
- Dassi, E.; Ferretti, P.; Covello, G.; Bertorelli, R.; Denti, M.A.; De Sanctis, V.; Tett, A.; Segata, N. The short-term impact of probiotic consumption on the oral cavity microbiome. Sci. Rep. 2018, 8, 10476. [Google Scholar] [CrossRef]
- Burton, J.P.; Chilcott, C.N.; Moore, C.J.; Speiser, G.; Tagg, J.R. A preliminary study of the effect of probiotic Streptococcus salivarius K12 on oral malodour parameters. J. Appl. Microbiol. 2006, 100, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, K.; Chen, T.; Paster, B.J. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017, 23, 276–286. [Google Scholar] [CrossRef] [PubMed]
- López-López, A.; Camelo-Castillo, A.; Ferrer, M.D.; Simon-Soro, Á.; Mira, A. Health-Associated Niche Inhabitants as Oral Probiotics: The Case of Streptococcus dentisani. Front. Microbiol. 2017, 8, 379. [Google Scholar] [CrossRef]
- Burton, J.P.; Cowley, S.; Simon, R.R.; McKinney, J.; Wescombe, P.A.; Tagg, J.R. Evaluation of safety and human tolerance of the oral probiotic Streptococcus salivarius K12: A randomized, placebo-controlled, double-blind study. Food Chem. Toxicol. 2011, 49, 2356–2364. [Google Scholar] [CrossRef]
- Cosseau, C.; Devine, D.A.; Dullaghan, E.; Gardy, J.L.; Chikatamarla, A.; Gellatly, S.; Yu, L.L.; Pistolic, J.; Falsafi, R.; Tagg, J.; et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect. Immun. 2008, 76, 4163–4175. [Google Scholar] [CrossRef]
- Tagg, J.R.; Harold, L.K.; Jain, R.; Hale, J.D.F. Beneficial modulation of human health in the oral cavity and beyond using bacteriocin-like inhibitory substance-producing streptococcal probiotics. Front. Microbiol. 2023, 14, 1161155. [Google Scholar] [CrossRef] [PubMed]
- Jansen, P.M.; Abdelbary, M.M.H.; Conrads, G. A concerted probiotic activity to inhibit periodontitis-associated bacteria. PLoS ONE 2021, 16, e0248308. [Google Scholar] [CrossRef]
- MacDonald, K.W.; Chanyi, R.M.; Macklaim, J.M.; Cadieux, P.A.; Reid, G.; Burton, J.P. Streptococcus salivarius inhibits immune activation by periodontal disease pathogens. BMC Oral Health 2021, 21, 245. [Google Scholar] [CrossRef]
- Moman, R.; O’Neill, C.A.; Ledder, R.G.; Cheesapcharoen, T.; McBain, A.J. Mitigation of the Toxic Effects of Periodontal Pathogens by Candidate Probiotics in Oral Keratinocytes, and in an Invertebrate Model. Front. Microbiol. 2020, 11, 999. [Google Scholar] [CrossRef]
- Poorni, S.; Nivedhitha, M.S.; Srinivasan, M.; Balasubramaniam, A. Effect of Probiotic Streptococcus salivarius K12 and M18 Lozenges on the Cariogram Parameters of Patients With High Caries Risk: A Randomised Control Trial. Cureus 2022, 14, e23282. [Google Scholar] [CrossRef]
- Do, H.; Li, Z.R.; Tripathi, P.K.; Mitra, S.; Guerra, S.; Dash, A.; Weerasekera, D.; Makthal, N.; Shams, S.; Aggarwal, S.; et al. Engineered probiotic overcomes pathogen defences using signal interference and antibiotic production to treat infection in mice. Nat. Microbiol. 2024, 9, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Babina, K.; Salikhova, D.; Polyakova, M.; Svitich, O.; Samoylikov, R.; Ahmad El-Abed, S.; Zaytsev, A.; Novozhilova, N. The Effect of Oral Probiotics (Streptococcus salivarius k12) on the Salivary Level of Secretory Immunoglobulin A, Salivation Rate, and Oral Biofilm: A Pilot Randomized Clinical Trial. Nutrients 2022, 14, 1124. [Google Scholar] [CrossRef]
- Burton, J.P.; Wescombe, P.A.; Moore, C.J.; Chilcott, C.N.; Tagg, J.R. Safety assessment of the oral cavity probiotic Streptococcus salivarius K12. Appl. Environ. Microbiol. 2006, 72, 3050–3053. [Google Scholar] [CrossRef] [PubMed]
- Horz, H.P.; Meinelt, A.; Houben, B.; Conrads, G. Distribution and persistence of probiotic Streptococcus salivarius K12 in the human oral cavity as determined by real-time quantitative polymerase chain reaction. Oral Microbiol. Immunol. 2007, 22, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Power, D.A.; Burton, J.P.; Chilcott, C.N.; Dawes, P.J.; Tagg, J.R. Preliminary investigations of the colonisation of upper respiratory tract tissues of infants using a paediatric formulation of the oral probiotic Streptococcus salivarius K12. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 1261–1263. [Google Scholar] [CrossRef]
- Comelli, E.M.; Guggenheim, B.; Stingele, F.; Neeser, J.R. Selection of dairy bacterial strains as probiotics for oral health. Eur. J. Oral Sci. 2002, 110, 218–224. [Google Scholar] [CrossRef]
- Haukioja, A.; Yli-Knuuttila, H.; Loimaranta, V.; Kari, K.; Ouwehand, A.C.; Meurman, J.H.; Tenovuo, J. Oral adhesion and survival of probiotic and other lactobacilli and bifidobacteria in vitro. Oral Microbiol. Immunol. 2006, 21, 326–332. [Google Scholar] [CrossRef]
- Groll, J.; Boland, T.; Blunk, T.; Burdick, J.A.; Cho, D.W.; Dalton, P.D.; Derby, B.; Forgacs, G.; Li, Q.; Mironov, V.A.; et al. Biofabrication: Reappraising the definition of an evolving field. Biofabrication 2016, 8, 013001. [Google Scholar] [CrossRef]
- Han, P.; Vaquette, C.; Abdal-Hay, A.; Ivanovski, S. The Mechanosensing and Global DNA Methylation of Human Osteoblasts on MEW Fibers. Nanomaterials 2021, 11, 2943. [Google Scholar] [CrossRef]
- Ivanovski, S.; Breik, O.; Carluccio, D.; Alayan, J.; Staples, R.; Vaquette, C. 3D printing for bone regeneration: Challenges and opportunities for achieving predictability. Periodontol. 2000 2023, 93, 358–384. [Google Scholar] [CrossRef]
- Ramachandra, S.S.; Abdal-Hay, A.; Han, P.; Lee, R.S.B.; Ivanovski, S. Fabrication and characterization of a 3D polymicrobial microcosm biofilm model using melt electrowritten scaffolds. Biomater. Adv. 2023, 145, 213251. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Udawatte, N.S.; Liaw, A.; Staples, R.; Salomon, C.; Seneviratne, C.J.; Ivanovski, S.; Han, P. Microbial DNA Profiles of Bacterial Extracellular Vesicles from 3D Salivary Polymicrobial Biofilms—A Pilot Study. Adv. Healthc. Mater. 2025, 14, e2403300. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra, S.S.; Wright, P.; Han, P.; Abdal-Hay, A.; Lee, R.S.B.; Ivanovski, S. Evaluating models and assessment techniques for understanding oral biofilm complexity. Microbiologyopen 2023, 12, e1377. [Google Scholar] [CrossRef]
- Aruni, A.W.; Dou, Y.; Mishra, A.; Fletcher, H.M. The biofilm community—Rebels with a cause. Curr. Oral Health Rep. 2015, 2, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Páez, A.; Belda-Ferre, P.; Simón-Soro, A.; Mira, A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genom. 2014, 15, 311. [Google Scholar] [CrossRef]
- Jakubovics, N.S.; Gill, S.R.; Iobst, S.E.; Vickerman, M.M.; Kolenbrander, P.E. Regulation of gene expression in a mixed-genus community: Stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J. Bacteriol. 2008, 190, 3646–3657. [Google Scholar] [CrossRef]
- Kolenbrander, P.E. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 2000, 54, 413–437. [Google Scholar] [CrossRef]
- Gazzaniga, F.; Stebbins, R.; Chang, S.Z.; McPeek, M.A.; Brenner, C. Microbial NAD metabolism: Lessons from comparative genomics. Microbiol. Mol. Biol. Rev. 2009, 73, 529–541. [Google Scholar] [CrossRef]
- Zhu, L.; Li, H.; Yang, X.; Xue, L.; Li, X.; Du, J. Effects of Streptococcus salivarius K12 on Experimental Periodontitis and Oral Microbiota in Mice. J. Biosci. Med. 2019, 7, 95–111. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services. Probiotics. Fact Sheet for Health Professional. 3 November 2023. Available online: https://ods.od.nih.gov/factsheets/Probiotics-HealthProfessional/ (accessed on 3 July 2024).
- Van Holm, W.; Carvalho, R.; Delanghe, L.; Eilers, T.; Zayed, N.; Mermans, F.; Bernaerts, K.; Boon, N.; Claes, I.; Lebeer, S.; et al. Antimicrobial potential of known and novel probiotics on in vitro periodontitis biofilms. NPJ Biofilms Microbiomes 2023, 9, 3. [Google Scholar] [CrossRef]
- Ling, Z.N.; Jiang, Y.F.; Ru, J.N.; Lu, J.H.; Ding, B.; Wu, J. Amino acid metabolism in health and disease. Signal Transduct. Target. Ther. 2023, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Lamont, R.J.; Koo, H. Oral polymicrobial communities: Assembly, function, and impact on diseases. Cell Host Microbe 2023, 31, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Abdulkareem, A.A.; Al-Taweel, F.B.; Al-Sharqi, A.J.; Gul, S.S.; Sha, A.; Chapple, I.L. Current concepts in the pathogenesis of periodontitis: From symbiosis to dysbiosis. J. Oral Microbiol. 2023, 15, 2197779. [Google Scholar] [CrossRef]
- Cheng, Y.; Song, Z.; Chen, J.; Tang, Z.; Wang, B. Molecular basis, potential biomarkers, and future prospects of OSCC and PD-1/PD-L1 related immunotherapy methods. Heliyon 2024, 10, e25895. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Luo, J.; Liu, H.; Li, Y.; Liu, J.; Zhu, L.; Wang, J.; Zeng, H. Dual recombinase polymerase amplification system combined with lateral flow immunoassay for simultaneous detection of Staphylococcus aureus and Vibrio parahaemolyticus. J. Pharm. Biomed. Anal. 2025, 255, 116621. [Google Scholar] [CrossRef]
- Xiang, E.; Gómez-Cerezo, M.N.; Ali, Y.; Ramachandra, S.S.; Yang, N.; Dargusch, M.; Moran, C.S.; Ivanovski, S.; Abdal-Hay, A. Surface Modification of Pure Zinc by Acid Etching: Accelerating the Corrosion Rate and Enhancing Biocompatibility and Antibacterial Characteristics. ACS Appl. Mater. Interfaces 2022, 14, 22554–22569. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.V.; Herst, P.M.; Tan, A.S. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. Biotechnol. Annu. Rev. 2005, 11, 127–152. [Google Scholar] [CrossRef]
- Reid, P.; Heng, N.C.K.; Hale, J.D.; Krishnan, D.; Crane, J.; Tagg, J.R.; Milne, T.J. A TaqMan™-based quantitative PCR screening assay for the probiotic Streptococcus salivarius K12 based on the specific detection of its megaplasmid-associated salivaricin B locus. J. Microbiol. Methods 2020, 170, 105837. [Google Scholar] [CrossRef]
- Han, K.; Liu, S.; Cheng, Y.-S. HuluFISH non-denaturing in situ detection of genomic DNA opened by CRISPR-Cas9 Nickase and Exonuclease. bioRxiv 2021. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Lu, Y.; Zhou, G.; Ewald, J.; Pang, Z.; Shiri, T.; Xia, J. MicrobiomeAnalyst 2.0: Comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res. 2023, 51, W310–W318. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; KA, L.C. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, P.; Bhide, M. Development of a bioinformatics platform for analysis of quantitative transcriptomics and proteomics data: The OMnalysis. PeerJ 2021, 9, e12415. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Jorth, P.; Turner, K.H.; Gumus, P.; Nizam, N.; Buduneli, N.; Whiteley, M. Metatranscriptomics of the human oral microbiome during health and disease. mBio 2014, 5, e01012-14. [Google Scholar] [CrossRef] [PubMed]
- Ewing, B.; Hillier, L.; Wendl, M.C.; Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998, 8, 175–185. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]
- Zapala, M.A.; Schork, N.J. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc. Natl. Acad. Sci. USA 2006, 103, 19430–19435. [Google Scholar] [CrossRef] [PubMed]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, J.; White, K.P. RNA-seq differential expression studies: More sequence or more replication? Bioinformatics 2014, 30, 301–304. [Google Scholar] [CrossRef]
- Yao, Y.; Rao, S.; Habimana, O. Active microbiome structure and functional analyses of freshwater benthic biofilm samples influenced by RNA extraction methods. Front. Microbiol. 2021, 12, 588025. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Udawatte, N.S.; Liu, C.; Staples, R.; Han, P.; Kumar, P.S.; Arumugam, T.V.; Ivanovski, S.; Seneviratne, C.J. Short-Term Probiotic Colonization Alters Molecular Dynamics of 3D Oral Biofilms. Int. J. Mol. Sci. 2025, 26, 6403. https://doi.org/10.3390/ijms26136403
Udawatte NS, Liu C, Staples R, Han P, Kumar PS, Arumugam TV, Ivanovski S, Seneviratne CJ. Short-Term Probiotic Colonization Alters Molecular Dynamics of 3D Oral Biofilms. International Journal of Molecular Sciences. 2025; 26(13):6403. https://doi.org/10.3390/ijms26136403
Chicago/Turabian StyleUdawatte, Nadeeka S., Chun Liu, Reuben Staples, Pingping Han, Purnima S. Kumar, Thiruma V. Arumugam, Sašo Ivanovski, and Chaminda J. Seneviratne. 2025. "Short-Term Probiotic Colonization Alters Molecular Dynamics of 3D Oral Biofilms" International Journal of Molecular Sciences 26, no. 13: 6403. https://doi.org/10.3390/ijms26136403
APA StyleUdawatte, N. S., Liu, C., Staples, R., Han, P., Kumar, P. S., Arumugam, T. V., Ivanovski, S., & Seneviratne, C. J. (2025). Short-Term Probiotic Colonization Alters Molecular Dynamics of 3D Oral Biofilms. International Journal of Molecular Sciences, 26(13), 6403. https://doi.org/10.3390/ijms26136403








