Genome-Wide Identification and Functional Analysis of the PEBP Gene Family in Begonia semperflorens ‘Super Olympia’ Reveal Its Potential Role in Regulating Flowering
Abstract
1. Introduction
2. Results
2.1. Identification of PEBP-like Genes in B. semperflorens ‘Super Olympia’
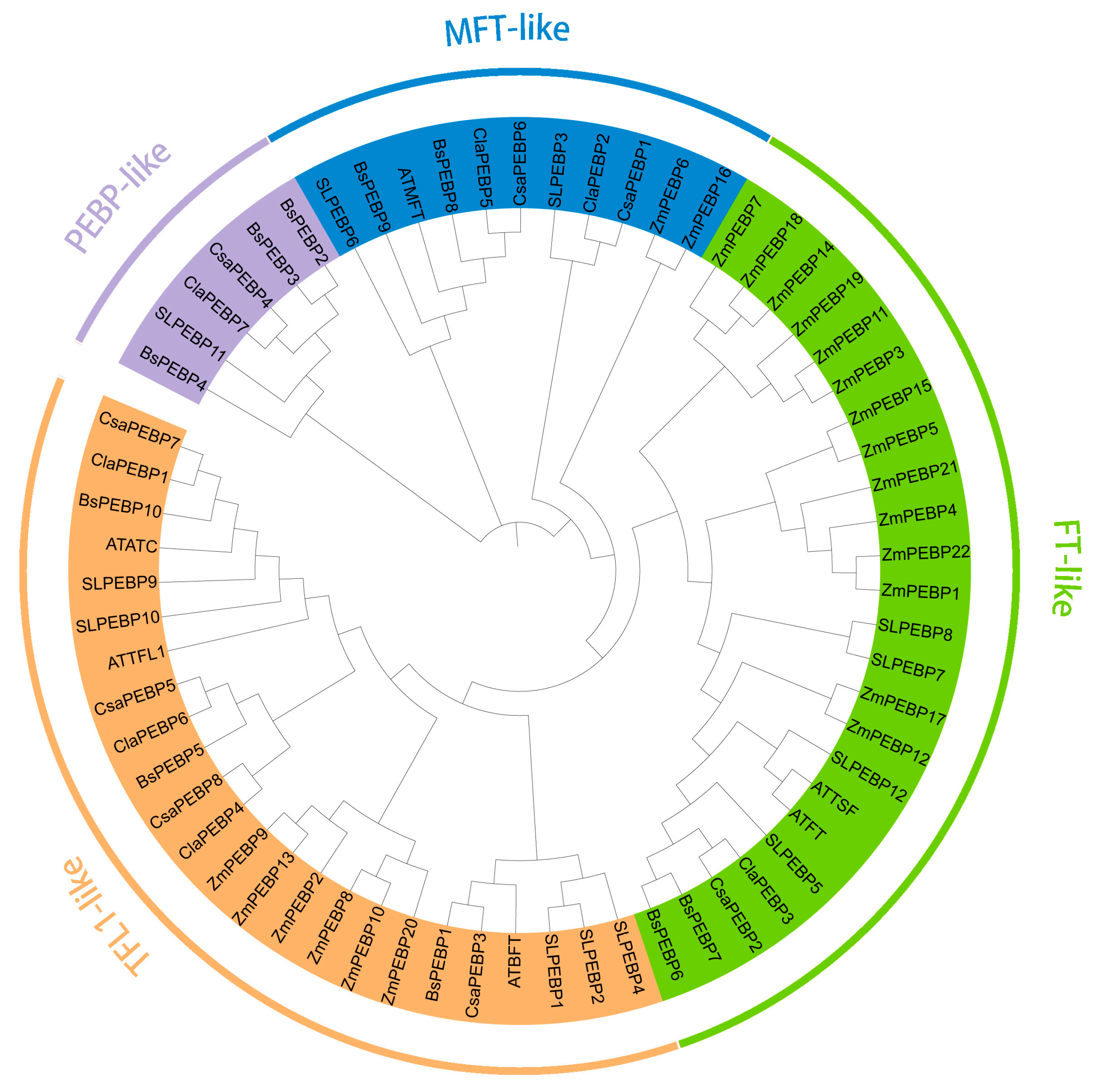
2.2. Sequence Alignment and Phylogenetic Relationship Analysis
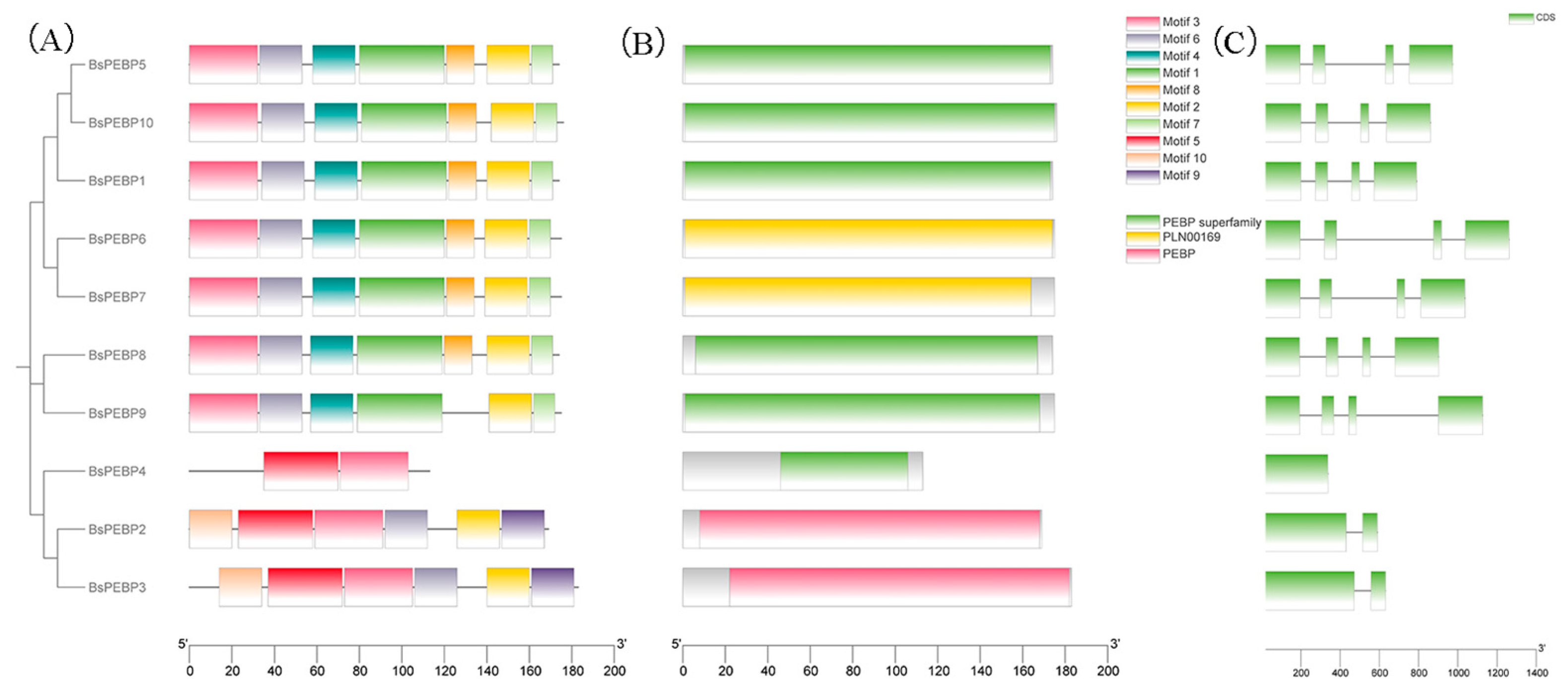
2.3. Gene Structure, Conserved Domains, and Motif Analysis
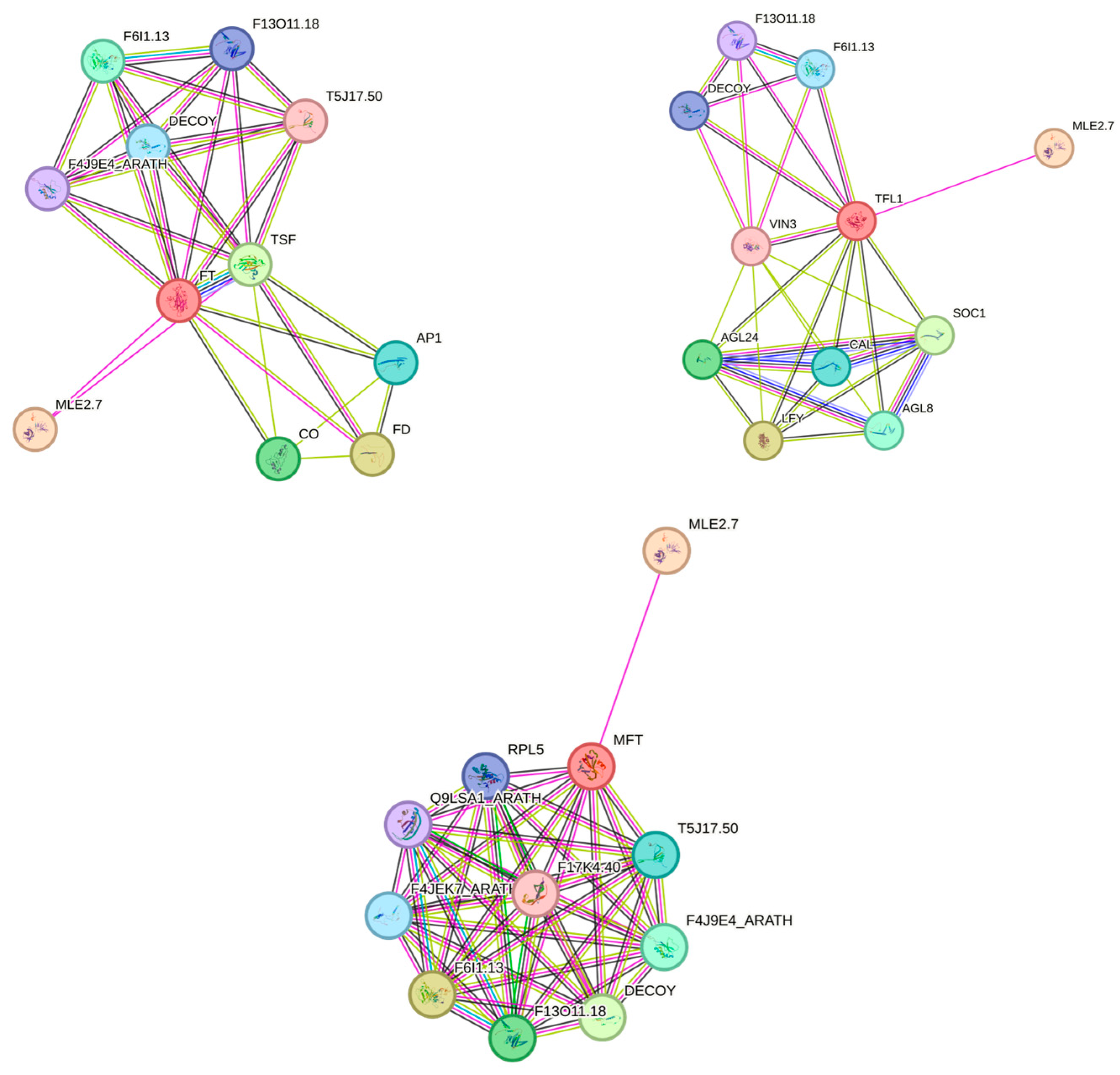
2.4. Prediction of the Protein-Protein Interaction (PPI) Network
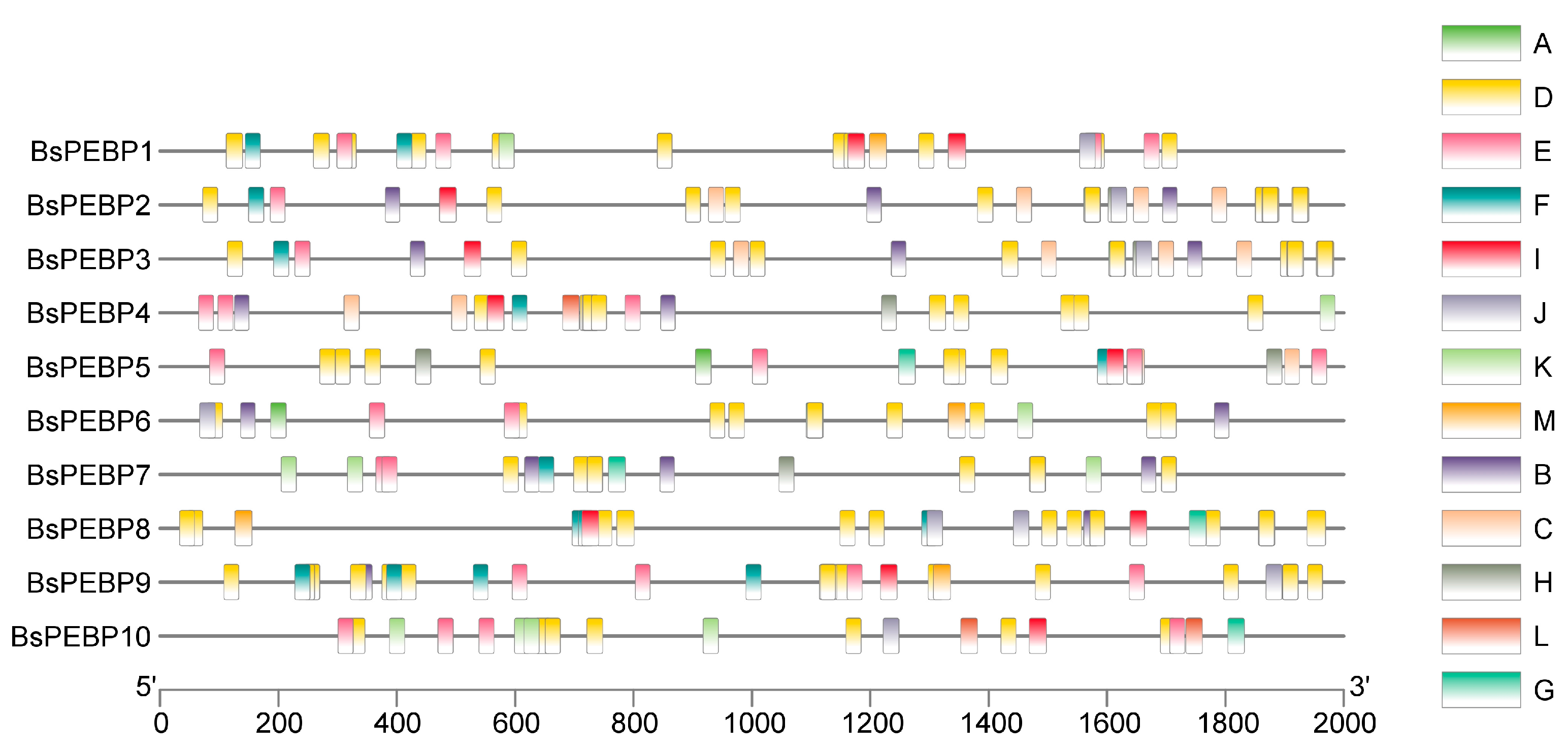
2.5. Analysis of Promoter Cis-Acting Elements
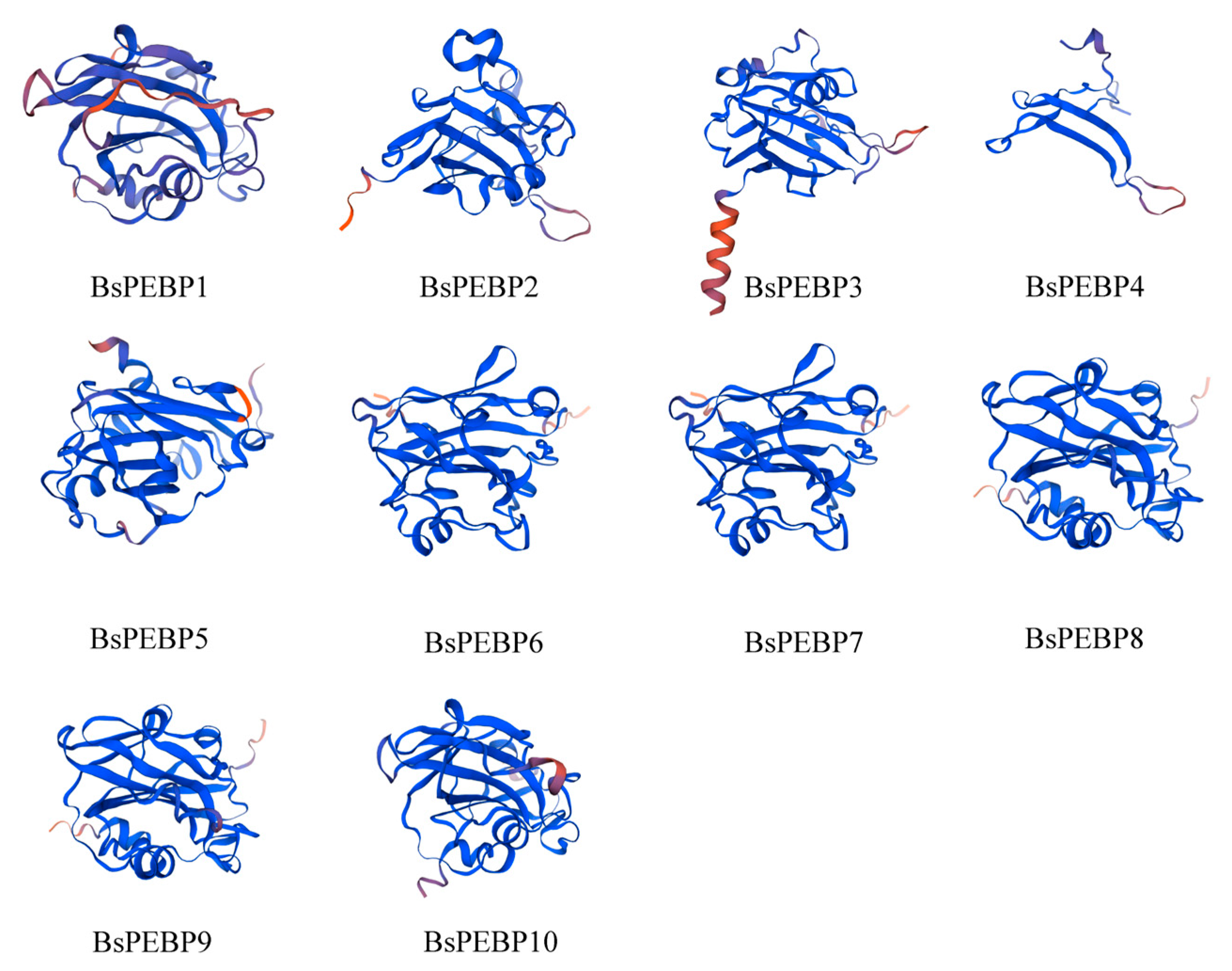
2.6. Prediction of Protein Secondary and Tertiary Structure
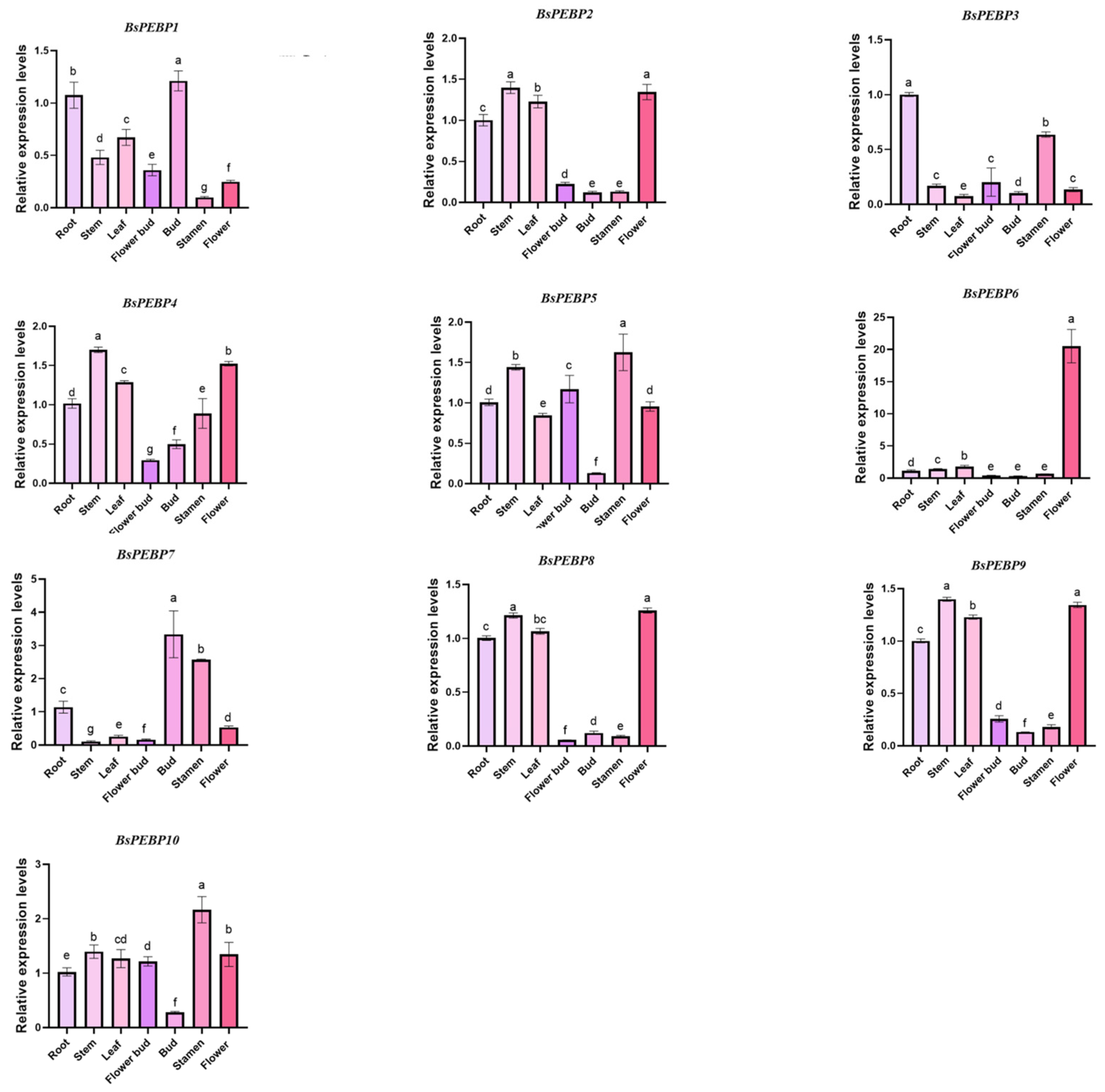
2.7. Tissue Expression Patterns of BsPEBP Genes
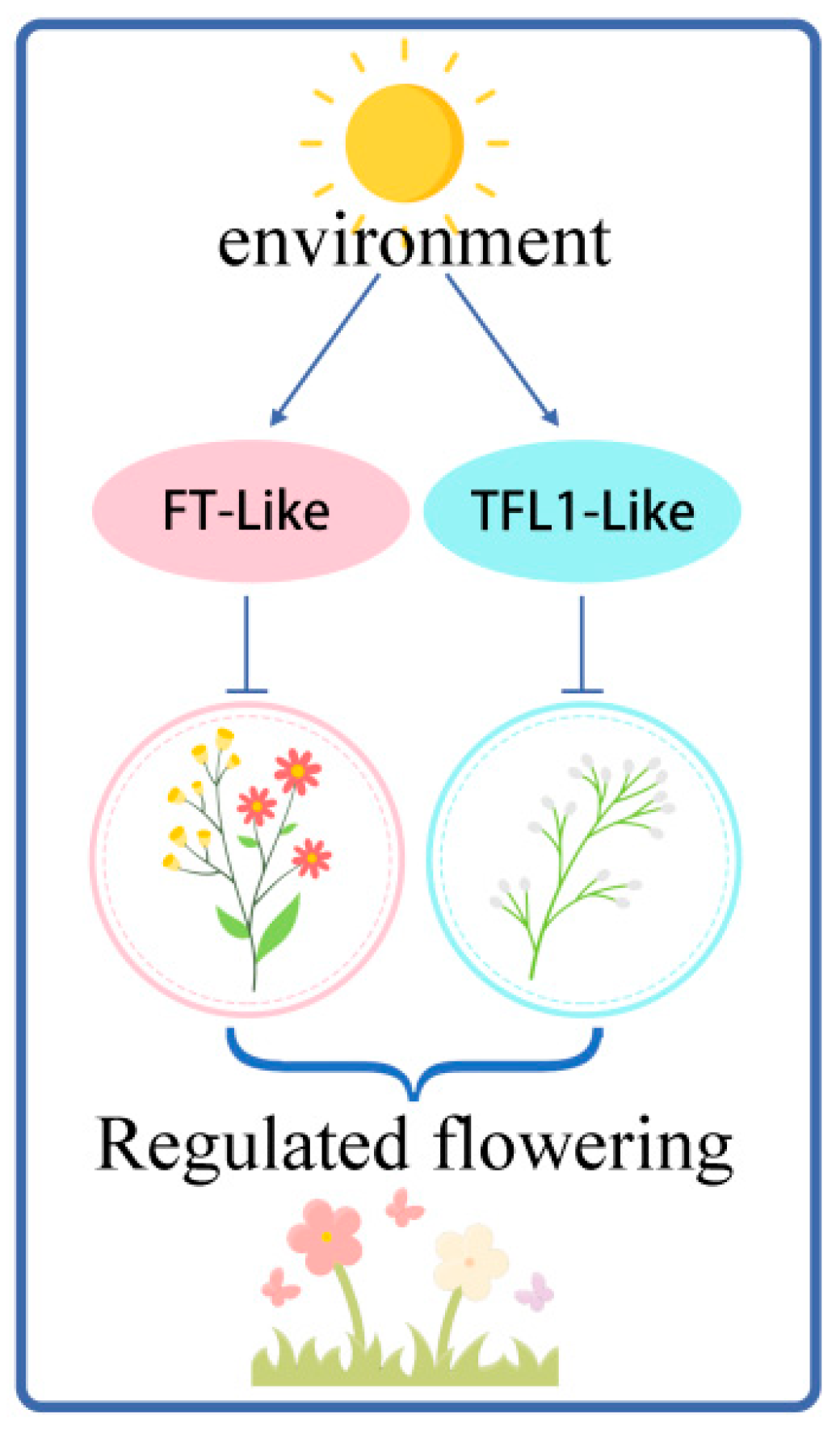
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of PEBP Proteins in B. semperflorens ‘Super Olympia’
4.3. Analysis of the Physicochemical Properties of PEBP Family Members in B. semperflorens ‘Super Olympia’
4.4. Sequence Alignment and Phylogenetic Analysis
4.5. Analysis of Promoter Cis-Acting Elements
4.6. BsPEBP Protein Interaction Network
4.7. Phylogenetic Evolution and Conserved Motif Analysis
4.8. Protein Secondary and Tertiary Structure Prediction
4.9. RNA Extraction, cDNA Synthesis, and qRT-PCR Experiments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chautard, H.; Jacquet, M.; Schoentgen, F.; Bureaud, N.; Bénédetti, H. Tfs1p, a member of the PEBP family, inhibits the Ira2p but not the Ira1p Ras GTPase-activating protein in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Hengst, U.; Albrecht, H.; Hess, D.; Monard, D. The phosphatidylethanolamine-binding protein is the prototype of a novel family of serine protease inhibitors. J. Biol. Chem. 2001, 276, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Banfield, M.J.; Barker, J.J.; Perry, A.C.; Brady, R.L. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 1998, 6, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Hedman, H.; Källman, T.; Lagercrantz, U. Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 2009, 70, 359–369. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Sundström, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP gene family in plants: Functional diversification in seed plant evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef]
- Chardon, F.; Damerval, C. Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 2005, 61, 579–590. [Google Scholar] [CrossRef]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef]
- Khan, M.R.; Ai, X.Y.; Zhang, J.Z. Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip. Rev. RNA 2014, 5, 347–359. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Roux, F.; Touzet, P.; Cuguen, J.; Le Corre, V. How to be early flowering: An evolutionary perspective. Trends Plant Sci. 2006, 11, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Kaneko-Suzuki, M.; Kurihara-Ishikawa, R.; Okushita-Terakawa, C.; Kojima, C.; Nagano-Fujiwara, M.; Ohki, I.; Tsuji, H.; Shimamoto, K.; Taoka, K.I. TFL1-Like Proteins in Rice Antagonize Rice FT-Like Protein in Inflorescence Development by Competition for Complex Formation with 14-3-3 and FD. Plant Cell Physiol. 2018, 59, 458–468. [Google Scholar] [CrossRef]
- Ospina-Zapata, D.A.; Madrigal, Y.; Alzate, J.F.; Pabón-Mora, N. Evolution and Expression of Reproductive Transition Regulatory Genes FT/TFL1 With Emphasis in Selected Neotropical Orchids. Front. Plant Sci. 2020, 11, 522623. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.; Ratcliffe, O.; Vincent, C.; Carpenter, R.; Coen, E. Inflorescence commitment and architecture in Arabidopsis. Science 1997, 275, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Patil, H.B.; Chaurasia, A.K.; Azeez, A.; Krishna, B.; Subramaniam, V.R.; Sane, A.P.; Sane, P.V. Characterization of two TERMINAL FLOWER1 homologs PgTFL1 and PgCENa from pomegranate (Punica granatum L.). Tree Physiol. 2018, 38, 772–784. [Google Scholar] [CrossRef]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef]
- Zheng, J.; Ma, Y.; Zhang, M.; Lyu, M.; Yuan, Y.; Wu, B. Expression Pattern of FT/TFL1 and miR156-Targeted SPL Genes Associated with Developmental Stages in Dendrobium catenatum. Int. J. Mol. Sci. 2019, 20, 2725. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, L.; Kohnen, M.V.; Xiong, B.; Zhen, X.; Liao, J.; Oka, Y.; Zhu, Q.; Gu, L.; Lin, C.; et al. Identification and Characterization of the PEBP Family Genes in Moso Bamboo (Phyllostachys heterocycla). Sci. Rep. 2019, 9, 14998. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Y.N.; Liu, H.L.; Huang, X.Z. Identifcation of PEBP Gene Family in Gossypium arboreum and Gossypium raimondii and Expression Analysis of the Gene Family in Gossypium hirsutum. Acta Agron. Sin. 2015, 41, 394–404. [Google Scholar] [CrossRef]
- Dong, L.; Lu, Y.; Liu, S. Genome-wide member identification, phylogeny and expression analysis of PEBP gene family in wheat and its progenitors. PeerJ 2020, 8, e10483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.F.; Xu, Y.W.; Dai, H.Y. Preliminary Analysis of the PEBP gene family in soybean (Glycine max). J. Plant Genet. Resour. 2015, 16, 151–157. [Google Scholar]
- Carmona, M.J.; Calonje, M.; Martínez-Zapater, J.M. The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 2007, 63, 637–650. [Google Scholar] [CrossRef]
- Zhang, M.; Li, P.; Yan, X.; Wang, J.; Cheng, T.; Zhang, Q. Genome-wide characterization of PEBP family genes in nine Rosaceae tree species and their expression analysis in P. mume. BMC Ecol. Evol. 2021, 21, 1–32. [Google Scholar] [CrossRef]
- Danilevskaya, O.N.; Meng, X.; Hou, Z.; Ananiev, E.V.; Simmons, C.R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008, 146, 250–264. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Yang, K.Z.; Wei, X.X.; Wang, X.Q. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUST gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 2016, 212, 730–744. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Hibrand Saint-Oyant, L.; Ruttink, T.; Hamama, L.; Kirov, I.; Lakhwani, D.; Zhou, N.N.; Bourke, P.M.; Daccord, N.; Leus, L.; Schulz, D.; et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 2018, 4, 473–484. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. In Current Protocols in Bioinformatics; John Wiley and Sons: Hoboken, NJ, USA, 2002; Chapter 2, Unit 2.3. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, C.; Zhao, M.; Xia, H.; Ren, P.; Liu, W.; Wang, Q.; Zhang, K. Genome-Wide Identification and Functional Analysis of the PEBP Gene Family in Begonia semperflorens ‘Super Olympia’ Reveal Its Potential Role in Regulating Flowering. Int. J. Mol. Sci. 2025, 26, 6291. https://doi.org/10.3390/ijms26136291
Fu C, Zhao M, Xia H, Ren P, Liu W, Wang Q, Zhang K. Genome-Wide Identification and Functional Analysis of the PEBP Gene Family in Begonia semperflorens ‘Super Olympia’ Reveal Its Potential Role in Regulating Flowering. International Journal of Molecular Sciences. 2025; 26(13):6291. https://doi.org/10.3390/ijms26136291
Chicago/Turabian StyleFu, Congcong, Mengru Zhao, Huiting Xia, Puyu Ren, Weichao Liu, Qirui Wang, and Kaiming Zhang. 2025. "Genome-Wide Identification and Functional Analysis of the PEBP Gene Family in Begonia semperflorens ‘Super Olympia’ Reveal Its Potential Role in Regulating Flowering" International Journal of Molecular Sciences 26, no. 13: 6291. https://doi.org/10.3390/ijms26136291
APA StyleFu, C., Zhao, M., Xia, H., Ren, P., Liu, W., Wang, Q., & Zhang, K. (2025). Genome-Wide Identification and Functional Analysis of the PEBP Gene Family in Begonia semperflorens ‘Super Olympia’ Reveal Its Potential Role in Regulating Flowering. International Journal of Molecular Sciences, 26(13), 6291. https://doi.org/10.3390/ijms26136291




